Abstract
In bacteria, a subset of transcription termination events requires the participation of the transcription termination factor Rho. Rho is a homo-hexameric, ring-shaped, motor protein that uses the energy derived from its RNA-dependent ATPase activity to directionally unwind RNA and RNA-DNA helices and to dissociate transcription elongation complexes. Despite a wealth of structural, biochemical and genetic data, the molecular mechanisms used by Rho to carry out its biological functions remain poorly understood. Here, we briefly discuss the most recent findings on Rho mechanisms and function and highlight important questions that remain to be addressed.
Key words: transcription termination, helicase, RNA, molecular motor, hexamer
Many important cellular processes are carried out by molecular motor enzymes that use various energy sources to perform specific mechanical tasks such as to trigger dynamic rearrangements, or to move, assemble or dissociate macromolecular entities. Often, these motors cooperate or compete in complex regulatory loops. For instance, intricate interactions between several molecular machineries in charge of RNA metabolism are evident in prokaryotes where the absence of a nuclear membrane precludes the physical separation of the processes of DNA transcription (carried out by the RNA polymerase; RNAP), RNA translation (carried out by ribosomes) and RNA decay (carried out by the RNA-degradosome). How these highly-dynamic processes influence and regulate one another remains poorly understood at the molecular level. At the heart of this regulatory network is the occurrence of transcription termination events that can be favored by translational defects or kinetic uncoupling between RNAP and ribosomes (transcriptional polarity),1 or that may affect pathways used for mRNA decay.2 For a significant fraction of these termination events (20–50% in E. coli), supplemental dynamic complexity is brought by the involvement of transcription termination factor Rho, an ATP-dependent molecular motor.1 Rho is composed of six identical protein subunits arranged in a ring (Fig. 1A) and as such constitutes an unusual example of RNA-dependent ring-shaped helicase motor.1 Rho is an essential, general modulator of gene expression in E. coli3 (and, probably, in many other bacteria)1 where it also appears to control the expression of non-coding RNAs (sRNAs and antisense RNAs),4 suggesting a possible role in the bacterial response to environmental stresses or in bacterial virulence. Rho may also help to prevent formation of toxic RNA-DNA structures (R-loops) during transcription.5
Figure 1.
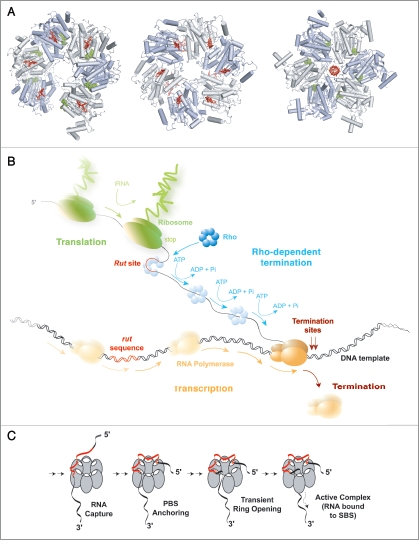
Rho-dependent termination of transcription. (A) The various crystal structures of the Rho hexamer6,10,12 with bound RNA segments in red and nucleotide cofactors in green, filled balls. Hexamers are shown from their top, N-terminal faces. In the first structure (PDB #1PVO; left),10 the hexamer forms a splint-open, “lock-washer” ring which may mimic the transient state required for RNA entry into the ring channel during formation of a catalytically-competent Rho-RNA complex (see C). The two other structures represent distinct, conflicting models of the translocating Rho enzyme. In one model (PDB #2HT1, middle),12 the Rho ring is a trimer of dimers and contains RNA bound to the composite, crown-like primary binding site (PBS). Electron density was insufficient to position RNA precisely in the central channel of the trimer of dimers.12 The other model (PDB #3ICE, right)6 shows an asymmetric Rho hexamer without RNA in the PBS but with an r(U)6 segment bound into the ring channel in a staircase-like arrangement reminiscent of that observed for single-stranded DNA in the crystal structure of the ring-shaped papillomavirus E1 helicase.29 (B) Schematic depiction of Rho-dependent termination of transcription in the context of bacterial gene expression. Note that, contrary to what is shown here, Rho and RNA polymerase may form a binary complex throughout the transcription cycle21 and that RNA may also remain bound to the PBS during translocation.1 (C) The putative Rho activation pathway. Rho bears two separate RNA binding sites that have different but coordinated functions. On top (5′-end of RNA) of the hexameric ring, the PBS is made of six identical subunit clefts (white ovals), each capable of specifically holding the base residues of a 5′-YC dimer (PBS-bound dimers are shown in red on (A), left and center). PBS clefts are responsible for tight and preferential Rho binding to C-rich Rut sites (in red).1 Once bound to the PBS, the substrate presumably awaits transient opening of the Rho ring10,11 to enter its central channel and bind SBS components (SBS-bound RNA is shown in red on A, right). Only upon completion of this multi-step process, which is slow and rate-limiting in vitro does Rho become enzymatically active.1
The current model for Rho-dependent termination of transcription posits that Rho binds to ∼70 nucleotide (nt)-long unstructured and cytosine-rich Rut (Rho Utilization) regions of the nascent transcript and moves in an ATP-dependent fashion towards the RNA 3′-end where it eventually dissociates the transcription elongation complex (TEC; Fig. 1B).1 However, despite a wealth of genetic, biochemical and structural data to rely on,1,6 many central features of this process remain obscure. This is in part due to the great complexity of the architecture of the Rho hexamer, which bears multiple, functionally-coupled nucleic acid (NA) and ATP binding sites (Fig. 1A). Although this holds true for other multi-subunit ring-shaped motors, the situation is exacerbated in Rho because it also contains several specific structural modules for other functions, such as substrate recognition, loading and enzymatic activation (Fig. 1C), that are often ensured by accessory factors with other ring-shaped ATPases.1 Furthermore, there is a complex interplay between Rho's components responsible for substrate recognition (primary binding clefts; Fig. 1C, white ovals) and those in charge of 2′-OH-dependent mechanochemical work (secondary binding sites, SBS; Fig. 1C, right).7–9 Understanding how the multiple RNA and ATP binding sites communicate to dictate Rho action is thus a major challenge that has not yet been fully addressed.
Two key determinants of Rho function are the nucleotide sequence of its RNA track and the presence of 2′-OH groups within it. Sequence preference derives in large part from the presence of a composite primary binding site (PBS) which is made of 5′-YC-specific binding clefts (Y and C being a pyrimidine and cytosine, respectively) contained within the N-terminal domains of the Rho hexamer (Fig. 1A) and which favors Rho binding to single-stranded C-rich/G-poor DNA or RNA sequences (such as the few transcript Rut sites characterized in vitro).1 Rho binding to RNA sequences may be more promiscuous in vivo,3 although this conjecture is still debated.4 Once firmly bound to the PBS, the substrate presumably awaits transient opening of the Rho ring10,11 to enter its central channel and bind SBS components in a strict RNA-specific manner (Fig. 1C). Only upon completion of this multi-step process does Rho become enzymatically active.1 The enzyme then uses swinging loops or “levers” contained within its SBS to move directionally along RNA.6,12 Whether Rho maintains or requires RNA-PBS interactions for its RNA translocation activity is currently uncertain as both biochemical1 and structural6,12 studies provided contradictory evidence. It will thus be important to establish if RNA is released from the PBS following the initial activation steps (Fig. 1C) or if allosteric communication between the PBS and SBS7 is also an integral part of the translocation mechanism.
Two recent studies addressed different aspects of Rho's mechanochemical cycle. First, Thomsen et al. unveiled a new RNA- and nucleotide-bound crystal structure of Rho (Fig. 1A, right).6 This structure shows a closed, asymmetric hexamer in which the RNA-binding motifs of the different subunits contact RNA in a unique manner by spiralling out-of-plane and around the RNA chain following a right-handed helix (as seen from the 5′-end; Fig. 2A).6 Interestingly, the subunit interfaces bind nucleotide mimic [ADP•BF3] using distinct allosteric networks, implying the presence of four different catalytic intermediates in the ATPase cycle. This study suggested that Rho hydrolyses ATP in a rotary, strictly sequential manner in which each turnover event produces the displacement of 1 nt (‘escort model’, Fig. 2A).6 A second study used a new helicase-based Nucleotide Analog Interference Mapping (NAIM) assay to show that Rho requires productive 2′-OH contacts every ∼7 nt during its translocation cycle.13 Several models were put forward to account for this observation. First, similarly to what was proposed by Moffitt and coworkers for the ring-shaped DNA packaging motor of bacteriophage Φ29,14 a small subset of subunits could contact RNA and function as a lever and latch during translocation. Translocation would occur in bursts of ∼7 nt that involve either a large out-of-plane deformation of the ring (‘open-ring model’, Fig. 2B) or the long-range movement of an internal lever device (‘large-lever model’, Fig. 2C). Former Rho structures showing a split-open, “lock-washer” ring conformation (Fig. 1A, left)10 or large movements of internal channel Q-loops (Fig. 1A, center),12 and the latest Rho structure displaying a notable asymmetry in nucleotide-binding states (Fig. 1A, right)6 are compatible with these models. Alternatively, RNA translocation could occur in an inchworm-like mechanism in which RNA is ‘scrunched’ in a succession of small translocation steps and released in ∼7 nt steps (‘RNA scrunching model’, Fig. 2D). This model would also be consistent with the latest structure of Rho, where the RNA is highly compressed at the SBS (Fig. 1A, right).6 The structural and NAIM data were later used by Patel15 to suggest a hybrid mechanism in which bursts of translocation of 7 nt lead to more than 1 nt translocated/ATP hydrolyzed. At present, these models cannot be discriminated based solely on available structural and biochemical data.15 They also cannot be tested easily with classical ensemble kinetics assays because the activation of the Rho-RNA complex (Fig. 1C) is slow and rate-limiting under all tested conditions.16–18 The quantitative analysis of Rho translocation and ATP hydrolysis mechanisms will therefore likely require experimentation at the single molecule level.
Figure 2.
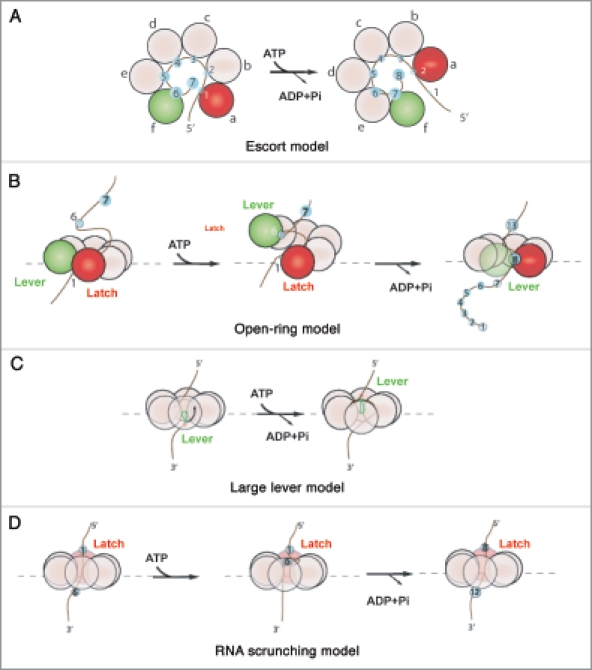
The various models proposed for the mechanochemical cycle of Rho. (A) The escort model6,29 posits that each subunit binds and translocates a single RNA contact sequentially through the ATP cycle. Lower-case letters indicate the conformational states that each subunit successively adopts to escort a nucleotide through the ring. Green and red circles represent, respectively, the two extreme “distal” and “proximal” subunit conformations. (B) The open-ring model14 proposes that ATP binding leads to uniform, out-of-plane conformational changes at the subunit interfaces that ultimately lead to large (∼40 A) distance movements between the latch and the lever subunits. Release of product leads to a reset in the hexamer that triggers a burst of RNA translocation. (C) The large-lever model13 suggests that an internal lever binds and translocates RNA. (D) the RNA scrunching model13 indicates that the RNA is internally compacted during the ATP binding phase (RNA is translocated while at the same time fixed to a latch subunit) and released in a rapid burst in the product release phase.
A similar, equivocal situation exists for the framing of the molecular events leading to RNAP dissociation. Currently, there is insufficient experimental data to favor a specific model for TEC destabilization by Rho but recent biochemical studies have started to address this issue by investigating the release of artificially-stalled TECs.19,20 Results suggest that the catalytic proficiency of the RNAP as well as the rewinding of the DNA template at the rear of the transcription bubble assist forward movement of the RNAP and/or bubble collapse during Rho-induced destabilization of the TEC.19,20 Transcript elongation by a few nucleotides may also take place during forward RNAP translocation.20 In another seminal study, it is proposed that Rho quickly inactivates RNAP and then slowly induces dissociation through the propagation of extensive destabilizing conformational changes across the TEC (allosteric destabilization).21 Interestingly, these observations and the various models proposed for Rho-induced termination are reminiscent of the conflicting forward-translocation22 and allosteric23 models proposed for intrinsic (i.e., RNA hairpin-dependent) termination. Since aspects of the mechanisms supporting intrinsic termination may vary with the identities of the terminators,24 it will be important to test whether such variations exist for Rho-dependent terminators. Furthermore, salient mechanistic features may be overlooked with a given type of in vitro assays (such as with those using artificially-stalled TECs) which, by essence (and necessity), do not fully replicate the conditions and dynamics of Rho-dependent termination.1 It will thus be useful to develop distinct experimental strategies to complement the initial studies of the mechanisms of Rho-induced termination.
Recent chromatin immunoprecipitation (ChIP-chip) data suggest that Rho associates early with TECs, probably during the synthesis of 5′-leader RNA regions and thus even before ribosomes bind mRNAs.25 Moreover, recent estimations of the in vivo frequency of translation initiation events seem inconsistent with a tight control of Rho-dependent termination by steric occlusion of Rut sites by ribosomes trailing behind RNAP.26 These observations may be explained if Rho and RNAP were in fact forming a stable binary complex throughout the transcription cycle as was recently shown in vitro.21 Acting as a hidden predator, Rho would sit and wait on RNAP and end the “game” opportunistically when favorable conditions appear. Such conditions may include the synthesis of a Rut site in the nascent transcript (which may be scanned continuously by the RNAP-bound Rho) and conditional loss of transcription factors (or ribosomes) acting as structural shields or as allosteric effectors of RNAP processivity. In this model, many gene transcription attempts would be unproductive (i.e., terminated by Rho) since only the minor fraction of RNAPs accompanied by a translating ribosome26 and by adequate processivity factors would produce full-length, translated mRNAs. It is also possible that another class of abundant Rho-dependent termination events occurring even earlier (i.e., during the synthesis of 5′-leader regions of mRNAs) has escaped notice until now because the short resulting transcripts are too unstable to be detected by conventional approaches. Similar “transcriptional noise” mechanisms have been evidenced recently in eukaryotes.27 Thus, it will be important to assess precisely when Rho comes into play during transcription (and how this influences the bacterial transcriptome), which molecular events exactly activate (or prevent) Rho function, or which step(s) of the motor cycle are truly important for transcription termination. Intriguingly, the Rho factor from gram-positive Mycobacterium tuberculosis (mtRho) can promote dissociation of an artificially-stalled TEC in the absence of ATP hydrolysis.28 Although the exact mechanisms used by mtRho remain undetermined, this observation suggests that different molecular solutions can evolve in distinct regulatory networks. One should thus use caution when drawing general conclusions on Rho mechanisms from the sole use of the abundant information obtained with the E. coli's enzyme.
Given the complexity of Rho-dependent termination and its interplay with the other steps of gene expression, it remains critical to develop a coherent framework for future investigations of Rho mechanisms in higher-order regulatory environments. Addressing all the important questions outlined in this point of view will likely require many additional investigations with sophisticated experimental tools such as NAIM-based chemogenetics,13 single-molecule observations and manipulations, genome- and transcriptome-wide analyses,3,21,27 as well as real-time, high-resolution imaging of molecular assemblies in live cells.
Acknowledgements
We gratefully acknowledge A.R. Rahmouni for helpful discussions as well as the Agence Nationale de la Recherche (PCV06), the Conseil Régional du Centre (AO2007), and the Institut National de la Santé et de la Recherche Médicale (INSERM, Avenir) for financial support.
Footnotes
Previously published online: http://www.landesbioscience.com/journals/transcription/article/12232
References
- 1.Rabhi M, Rahmouni AR, Boudvillain M. Transcription termination factor Rho: a ring-shaped RNA helicase from bacteria. In: Jankowsky E, editor. RNA helicases. Cambridge (UK): RSC Publishing; 2010. pp. 243–271. [Google Scholar]
- 2.Mohanty BK, Kushner SR. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–5704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci USA. 2009;106:15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gowrishankar J, Harinarayanan R. Why is transcription coupled to translation in bacteria? Mol Microbiol. 2004;54:598–603. doi: 10.1111/j.1365-2958.2004.04289.x. [DOI] [PubMed] [Google Scholar]
- 6.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalissery J, Banerjee S, Bandey I, Sen R. Transcription termination defective mutants of Rho: Role of different functions of Rho in releasing RNA from the elongation complex. J Mol Biol. 2007;371:855–872. doi: 10.1016/j.jmb.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinmetz EJ, Platt T. Evidence supporting a tethered tracking model for helicase activity of Escherichia coli Rho factor. Proc Natl Acad Sci USA. 1994;91:1401–1405. doi: 10.1073/pnas.91.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geiselmann J, Wang Y, Seifried SE, von Hippel PH. A physical model for the translocation and helicase activities of Escherichia coli transcription termination protein Rho. Proc Natl Acad Sci USA. 1993;90:7754–7758. doi: 10.1073/pnas.90.16.7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skordalakes E, Berger JM. Structure of the rho transcription terminator. Mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 11.Kim DE, Patel SS. The kinetic pathway of RNA binding to the Escherichia coli transcription termination factor Rho. J Biol Chem. 2001;276:13902–13910. doi: 10.1074/jbc.M011043200. [DOI] [PubMed] [Google Scholar]
- 12.Skordalakes E, Berger JM. Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell. 2006;127:553–564. doi: 10.1016/j.cell.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz A, Rabhi M, Jacquinot F, Margeat E, Rahmouni AR, Boudvillain M. A stepwise 2′-hydroxyl activation mechanism for the ring-shaped transcription termination Rho helicase. Nat Struct Mol Biol. 2009;16:1309–1316. doi: 10.1038/nsmb.1711. [DOI] [PubMed] [Google Scholar]
- 14.Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, et al. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel SS. Structural biology: Steps in the right direction. Nature. 2009;462:581–583. doi: 10.1038/462581a. [DOI] [PubMed] [Google Scholar]
- 16.Walmacq C, Rahmouni AR, Boudvillain M. Influence of substrate composition on the helicase activity of transcription termination factor Rho: reduced processivity of Rho hexamers during unwinding of RNA-DNA hybrid regions. J Mol Biol. 2004;342:403–420. doi: 10.1016/j.jmb.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 17.Walmacq C, Rahmouni AR, Boudvillain M. Testing the steric exclusion model for hexameric helicases: substrate features that alter RNA-DNA unwinding by the transcription termination factor Rho. Biochemistry. 2006;45:5885–5895. doi: 10.1021/bi0600648. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz A, Margeat E, Rahmouni AR, Boudvillain M. Transcription termination factor Rho can displace streptavidin from biotinylated RNA. J Biol Chem. 2007;282:31469–31476. doi: 10.1074/jbc.M706935200. [DOI] [PubMed] [Google Scholar]
- 19.Dutta D, Chalissery J, Sen R. Transcription termination factor Rho prefers catalytically active Elongation complexes for releasing RNA. J Biol Chem. 2008;283:20243–20251. doi: 10.1074/jbc.M801926200. [DOI] [PubMed] [Google Scholar]
- 20.Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–249. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santangelo TJ, Roberts JW. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol Cell. 2004;14:117–126. doi: 10.1016/s1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- 23.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Smit MH, Verlaan PW, van Duin J, Pleij CW. In vivo dynamics of intracistronic transcriptional polarity. J Mol Biol. 2009;385:733–747. doi: 10.1016/j.jmb.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 27.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 28.Kalarickal NC, Ranjan A, Kalyani BS, Wal M, Sen R. A bacterial transcription terminator with inefficient molecular motor action but with a robust transcription termination function. J Mol Biol. 2010;395:966–982. doi: 10.1016/j.jmb.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 29.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]


