Abstract
Transcription of eukaryotic genes by RNA polymerase II is typically accompanied by minimal exchange of histones H3/H4 carrying various covalent modifications. In vitro studies suggest that histone survival is accompanied by the formation of a small transient DNA loop on the surface of the histone octamer including a molecule of transcribing enzyme.
Key words: chromatin, nucleosome, RNA polymerase II, transcription, intermediates, structure, mechanism, elongation
Efficient recovery of histones associated with DNA during passage of RNA Polymerase II (Pol II) is essential for cell viability.1 Recovery of histones occurs by two different mechanisms. During intense transcription, partial loss2–6 and exchange7–12 of all core histones during passage of Pol II were observed. In contrast, on moderately transcribed genes, extensive transcription-dependent exchange of H2A/H2B, but not H3/H4, histones was detected.7–12
The Pol II-type mechanism of transcription through chromatin has been recapitulated in vitro and is conserved from yeast to human.13 It is characterized by the displacement of a single H2A/H2B dimer on 40–50% of templates14,15 and by the displacement of all histones from the remaining 50–60% of templates.14–16 H2A/H2B dimer displacement matches the apparent effect of Pol II passage in vivo.9 The subnucleosome (DNA-bound histone hexamer [hexasomes] formed upon release of H2A/H2B dimer from the octamer) survives Pol II passage and remains at the original position on DNA14 (also see ref. 16). A different mechanism (Pol III-type) involves obligatory transfer of a complete histone octamer from in front of the transcribing enzyme to behind it.17–19
More recently, we have shown that histone survival during Pol II transcription is accompanied by formation of a small transient DNA loop (zero-size or ∅-loop) on the surface of the histone octamer including a molecule of transcribing Pol II.20 During formation of the ∅-loop, the recovery of DNA-histone interactions behind Pol II is tightly coupled with their disruption ahead of the enzyme. This coupling is a distinct feature of the Pol II-type mechanism that allows recovery of H3/H4 histones bound at the original position on DNA during transcription.20 The accompanying displacement of H2A/H2B dimer is most likely induced by considerable partial uncoiling of DNA from the octamer. In the absence of H2A/H2BDNA interactions, an H2A/H2B dimer may spontaneously dissociate from the octamer.
The new model predicts that the octamer never leaves DNA during transcription through chromatin20 (Fig. 1A, partial DNA displacement pathway 1 → 2′ → 3). In this case, nucleosomes would remain at their original positions after transcription, even if these are not the preferential (equilibrated) positions on DNA before transcription (Fig. 1B, mechanism 2′). Alternatively, during transcription through a nucleosome, the histone octamer could be transiently and completely displaced from DNA (the transient displacement pathway 1 → 2 → 3).21 In this case, the octamer re-binds to DNA released behind the enzyme when Pol II proceeds further. In this case, nucleosome positions would be re-equilibrated during transcription because octamers would select thermodynamically preferred positions while re-binding to DNA (Fig. 1B, mechanism 2).17 If the original nucleosome positions are non-equilibrium, transcription would result in a change in nucleosome positions.
Figure 1.
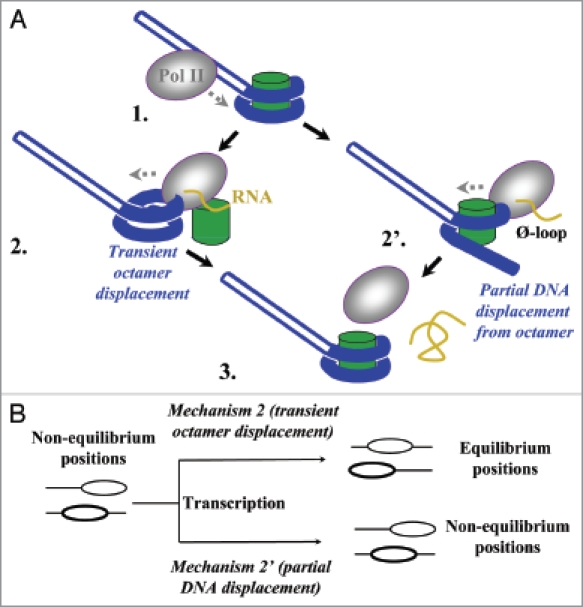
The experimental strategy for analysis of the mechanism of nucleosome survival during Pol II transcription. (A) Two possible mechanisms of nucleosome survival during transcription by Pol II.21 After Pol II approaches the nucleosome (1) and partially displaces proximal nucleosomal DNA,20 the octamer could either be completely (2) or partially (2′), transiently displaced from DNA. In the first case octamer re-binds to DNA released behind the enzyme when Pol II proceeds further (3). In the latter case transcription through the nucleosome proceeds without even transient complete dissociation of the octamer from nucleosomal DNA. Histone octamer, DNA and RNA are shown in green, blue and yellow, respectively. (B) The expected outcomes of transcription through a non-equilibrium population of mononucleosomes by Pol II. Transient histone octamer displacement (mechanism 2) would result in equilibration of nucleosome positions on DNA after transcription. In contrast, non-equilibrium position(s) of nucleosomes would be preserved if the octamer is never completely displaced from DNA (mechanism 2′).
The nucleosome positions on DNA are preserved after transcription by Pol II.22 To discriminate between the two mechanisms, it has to be established whether the original nucleosome positions were equilibrium or non-equilibrium. If the original positions were non-equilibrium, preservation of these positions during transcription would strongly support the partial DNA displacement mechanism (Fig. 1).
In previous experiments, differently positioned mononucleosomes were assembled on the 204 bp pVT1 5S-containing DNA fragment, ligated to the 50 bp promoter fragment and transcribed.22 To evaluate whether nucleosomes occupy equilibrium conformations after ligation, nucleosome reconstitution was conducted before and after ligation of histone-free 204 bp and 50 bp DNA fragments (Fig. 2A). The positions of nucleosomes ligated after nucleosome assembly were the same as observed previously22 and clearly distinct from the equilibrium positions of nucleosomes ligated before reconstitution (Fig. 2B). Thus, the original positions of nucleosomes used in previous experiments were non-equilibrium. Therefore, the original non-equilibrium positions of nucleosomes before transcription are preserved during transcription.22 These data suggest that the octamer never leaves DNA during Pol II transcription through a nucleosome (Fig. 1A, pathway 1 → 2′ → 3) and are consistent with the ∅-loop-mediated mechanism of transcription through chromatin20 and with the in vivo observations on the lack of H3/H4 exchange during moderate transcription.7–12
Figure 2.
Nucleosomes occupy non-equilibrium positions before transcription by Pol II. (A) The experimental approach for analysis of the mechanism of nucleosome survival during transcription by Pol II. In previous transcription experiments14 nucleosome assembly on the 204-bp DNA fragment was conducted before ligation to the promoter-containing 50-bp fragment and transcription (on the right). To evaluate whether after such assembly → ligation nucleosomes occupy equilibrium positions on DNA, nucleosomes were assembled after ligation (ligation → assembly, on the left). In this case nucleosome positions are equilibrated during the process of assembly. (B) Nucleosomes were assembled before or after ligation to the promoter-containing DNA fragment by dialysis from 2 M NaCl and analyzed by native PAGE.14 In the latter case nucleosome positions are equilibrated during reconstitution. Nucleosome mobility in the gel is dictated by nucleosome positioning on the 254 bp DNA fragment. Since nucleosomes occupy different positions after assembly before (right lane) and after ligation (left lane), nucleosomes occupied non-equilibrium positions before transcription by Pol II.
In all in vitro studies described above, short (250–300 bp) mononucleosomal templates were used. At the same time, it has recently been suggested that transcription of a long (∼3.9 kb) template containing a single mononucleosome by E. coli RNA polymerase (RNAP) could result in nucleosome translocation from in front to behind the transcribing enzyme.16 Our studies using a long (658 bp) mononucleosomal template also suggest that nucleosome translocation occurs on a considerable fraction of the templates transcribed by E. coli RNAP (Olga I. Kulaeva, data not shown). Since Pol II and E. coli RNAP share the same mechanism of transcription through chromatin,20,23 the data suggest that the fates of mononucleosomes formed on shorter (250–300 bp) and longer (658–3,900 bp) DNA templates during transcription by Pol II and E. coli RNAP are different. On the shorter templates nucleosomes remain at the original position on DNA; on the longer templates nucleosomes are translocated from in front to behind the transcribing enzyme.
The difference in the fates of nucleosomes on transcription of shorter and longer templates is most likely explained by differential interaction of the upstream DNA with the histone octamer surface that is transiently exposed after partial uncoiling of nucleosomal DNA during transcription.20 The exposed octamer surface is available to competitor DNA present at sufficiently high concentration.17 On shorter templates, the DNA behind the transcribing enzyme may be too rigid to reach the transiently exposed octamer surface with high probability. On longer templates, the local DNA concentration in the vicinity of the open octamer surface is likely to be higher. This would result in capturing of the octamer by DNA, DNA loop formation and, eventually, in octamer transfer on longer templates.17,18
These observations raise the following question: Which of the in vitro experimental mononucleosomal models better recapitulate the in vivo scenario? On moderately transcribed genes (representing the vast majority of eukaryotic genes), nucleosomes are present immediately in front and behind of transcribing molecules of Pol II.24 Therefore, the existence of extended histone-free DNA regions on moderately transcribed genes in vivo is unlikely, and shorter mononucleosomal templates are appropriate models for analysis of the mechanism of transcription through chromatin in vitro. At the same time, during intense transcription histones are largely removed from DNA in vivo.2–6 Under these circumstances, histone-free DNA could be available for binding to the transiently exposed octamer surface, and translocation of the nucleosomes remaining on the genes would become much more likely. This may explain the observed irregular positioning of nucleosomes remaining on actively transcribed genes.25
In summary, during in vitro transcription of short (250–300 bp) mononucleosomal templates by Pol II one H2A/H2B dimer is displaced, but the subnucleosome (DNA-bound histone hexamer) never leaves the DNA during transcription and remains at the original position on the DNA.14 These observations are consistent with the extensive transcription-dependent exchange of H2A/H2B, but not H3/H4, histones on moderately transcribed genes.7–12 In contrast, transcription of longer (658–3,900 bp) mononucleosomal templates by Pol II in vitro results in nucleosome translocation from in front of the transcribing enzyme to behind it. This mechanism cannot explain the lack of exchange of histones H3/H4 detected on moderately transcribed genes because, after several rounds of transcription, the histone octamer would be transferred on a promoter and all core histones would be displaced/exchanged by ATP-dependent chromatin remodelers that are present there.20 Furthermore, this mechanism could operate only on actively transcribed genes, where some nucleosomes are displaced2–6 and histone-free DNA is available for the octamer transfer.
Acknowledgements
This work was supported by NIH GM58650 grant to V.M.S.
Footnotes
Previously published online: http://www.landesbioscience.com/journals/transcription/article/12519
References
- 1.Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 4.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao J, Herrera-Diaz J, Gross DS. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol Cell Biol. 2005;25:8985–8999. doi: 10.1128/MCB.25.20.8985-8999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wirbelauer C, Bell O, Schubeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 2005;19:1761–1766. doi: 10.1101/gad.347705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 11.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 13.Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, et al. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 15.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 16.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studitsky VM, Clark DJ, Felsenfeld G. A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 1994;76:371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 18.Studitsky VM, Clark DJ, Felsenfeld G. Overcoming a nucleosomal barrier to transcription. Cell. 1995;83:19–27. doi: 10.1016/0092-8674(95)90230-9. [DOI] [PubMed] [Google Scholar]
- 19.Studitsky VM, Kassavetis GA, Geiduschek EP, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 20.Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, et al. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol. 2009;16:1272–1278. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Studitsky VM. Chromatin remodeling by RNA polymerase II. Mol Biol. 2005;39:639–654. [PubMed] [Google Scholar]
- 22.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II. Loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 23.Walter W, Kireeva ML, Studitsky VM, Kashlev M. Bacterial polymerase and yeast polymerase II use similar mechanisms for transcription through nucleosomes. J Biol Chem. 2003;278:36148–36156. doi: 10.1074/jbc.M305647200. [DOI] [PubMed] [Google Scholar]
- 24.De Bernardin W, Koller T, Sogo JM. Structure of in-vivo transcribing chromatin as studied in simian virus 40 minichromosomes. J Mol Biol. 1986;191:469–482. doi: 10.1016/0022-2836(86)90142-7. [DOI] [PubMed] [Google Scholar]
- 25.Cavalli G, Thoma F. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 1993;12:4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



