Abstract
Folding of the trigger loop of RNA polymerase promotes nucleotide addition through creating a closed, catalytically competent conformation of the active center. Here, we discuss the impact of adjacent RNA polymerase elements, including the F loop and the jaw domain, as well as external regulatory factors on the trigger loop folding and catalysis.
Key words: RNA polymerase, transcription elongation complex, trigger loop, nucleotide discrimination, temperature adaptation
Conformational Plasticity of the RNAP Active Site
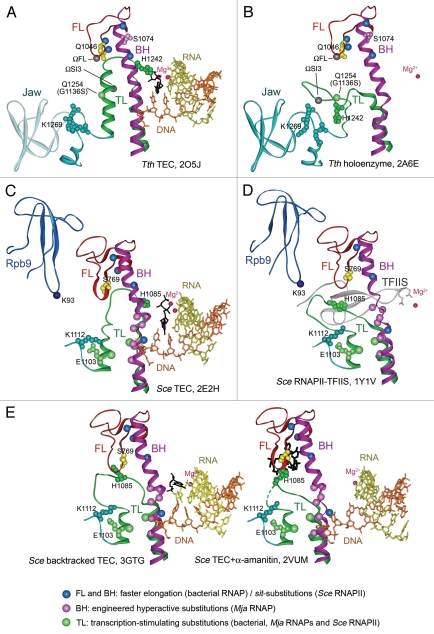
Multisubunit RNAPs (an α2ββ'ω penta-subunit core complex in bacteria) are responsible for transcription of all cellular genomes. They share a common catalytic mechanism in which initial binding of an NTP substrate is followed by crucial conformational changes in the active site that involve two structural elements, the trigger loop (TL) and the bridge helix (BH).1–4 The incoming NTP binds in the i+1 site of the active center while the TL is relaxed and induces its transition (Fig. 1B and D) into an α-helical hairpin, which packs against the BH in a three-helix bundle (Fig. 1A and C), resulting in closure of the active site. In the helical conformation, the TL directly contacts the substrate and facilitates nucleotidyl transfer.1,2,5,6 Refolding of the TL is required for efficient and accurate RNA synthesis and has been proposed to constitute a rate-limiting step in the nucleotide addition cycle.1,2,5–8 Following nucleotide addition, RNAP moves one nucleotide downstream; the TL relaxation and PPi release may precede3 or be coupled with8 translocation that is likely accompanied by changes in both the BH and the TL. For example, the BH has been observed in an intermediate, shifted conformation stabilized by a wedged TL;3 the shifted BH partially occupies the i+1 site and thus could favor translocation of the RNA 3′-end into the i site. Recent findings suggest that conformational changes during catalysis by RNAP may be a target for extensive transcription regulation.
Figure 1.
Structure of the RNAP active center and conformational transitions of the TL. (A) and (B) Structures of the Tth transcription elongation complex (TEC) with NTP (black) in the i+1 site2 and Tth holoenzyme RNAP (a core bound to the promoter-specific σ subunit),28 respectively. Catalytic Mg2+ ions are shown as red spheres, the template DNA is in orange, RNA is in yellow, the FL is in red, the BH is in magenta, the TL is in green. Q1046, S1074 and H1242 in the FL, BH and TL are shown as CPK models in yellow, light violet and dark green, respectively. Light green spheres indicates position of the G1136SEco substitution; blue spheres indicate positions of the M747IEco, P750LEco, F773VEco and R780HEco substitutions; gray spheres—insertions in the TL in Eco RNAP (ΩSI3) and the FL (ΩFL) in various bacterial lineages. A part of the jaw-domain (semitransparent in A) is not visible in the TEC structure; variable Taq/Dra residues at the base of this domain are in turquoise. (C) and (D) Structures of the Sce TEC with NTP in the i+1 site1 and RNAPII bound to TFIIS.25 The TL E1103Sce and an adjacent K1112Sce in the jaw-domain are in light green and turquoise, respectively; S769Sce (yellow) in the FL corresponds to Q1046Taq. Positions of sit-substitutions (blue) and hyperactive Mja variants in the BH (pink) and the TL (light green) are indicated. The Zn-ribbon domains of Rpb9 and TFIIS are dark blue and gray, respectively; Rpb9 K93 residue (a dark blue sphere) is adjacent to the open TL. (E) Structure of a backtracked Sce TEC11 (RNA 3′-nucleotide is in black, the TL is partially open) and of TEC in complex with α-amanitin (black)3 with the TL in a wedged conformation.
Consistent with their key role in catalysis, many substitutions in the TL and the BH severely disrupt RNAP function.2,5,6,9–11 More interestingly, a number of variants that stimulate RNA synthesis in vitro have been described (see Fig. 1 and Suppl. Fig. 1); BH F773VEco,12 R780HEco,13 TL G1136SEco14 in Escherichia coli (Eco) β' RNAP subunit,14 TL E1103GSce in Saccharomyces cerevisiae (Sce) Rpb17,15 and several engineered substitutions in both the BH and the TL in Methanocaldococcus jannaschii (Mja) RNAP.10 In addition, an in vivo screen for gain-of-function variants of Sce RNAPII identified several substitutions in the BH and the TL (Fig. 1C and D, Suppl. Fig. 1).16 These sit-substitutions were selected for increased transcription of the HIS4 gene in an activator-deficient strain and their phenotypes could be due to altered promoter function, increased RNA chain elongation, etc. The properties of sit-variants have not been tested in vitro, but two of the affected residues were also substituted in hyperactive Mja RNAPs.
In support of the importance of the coordinated BH/TL transitions, the effects of substitutions therein on nucleotidyl transfer appear to correlate with the predicted changes in stability of the triplehelix bundle: substitutions that favor the bundle formation increase the rate of catalysis whereas those that destabilize it inhibit catalysis.1,2,5,10,14,15 Thus, the primary role of the TL in nucleotide addition was proposed to be in proper positioning and stabilization of the substrate in the active center. However, certain TL residues (in particular, R933Eco/R1239Taq and H936Eco /H1242Taq that interact with the triphosphate moiety of the incoming NTP, see Fig. 1) were also shown to directly participate in catalysis.5,6
Role of the FL in Catalysis by Thermophilic and Mesophilic RNAPs
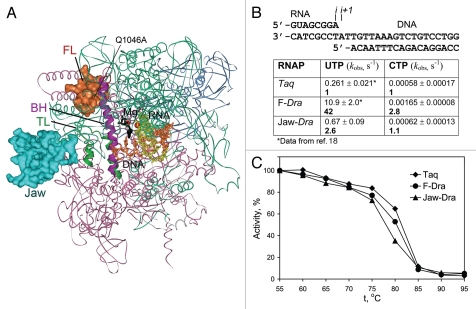
Recent studies of RNAP catalysis primarily focused on analysis of amino acid substitutions in the TL and the BH. However, additional RNAP regions may affect TL folding, in turn modulating catalysis. Comparison of RNAPs from thermophilic Thermus aquaticus (Taq) and its closest mesophilic relative Deinococcus radiodurans (Dra), which significantly differ in the rates of RNA synthesis,17 identified one such element, the F loop (FL, shown in red in Fig. 1). The FL is located near the N-terminus of the BH and directly contacts the closed TL in the NTP-bound transcription elongation complex (TEC) (Fig. 1A), suggesting that it may affect the TL folding and/or stabilize the closed TL conformation. Replacement of the Taq FL with a homologous sequence from Dra (eight amino acid substitutions, with one additional substitution in the BH, (Suppl. Fig. 1) stimulated transcription elongation (Suppl. Fig. 2) and increased the rate of nucleotide addition by a hybrid F-Dra enzyme (∼40-fold at 20°C, Fig. 2B).18 Since this rate was measured in the post-translocated TEC (Fig. 2B), the observed stimulation is likely due to the effects on catalysis, at least at temperatures which are suboptimal for the thermophilic Taq RNAP. Consistent with the hypothesis that the FL promotes the TL folding, deletion of the central part of the FL decreased the catalytic rate of Taq RNAP ∼70-fold at 20°C and conferred its resistance to streptolydigin (Stl),18 an RNAP inhibitor that blocks TL folding.2
Figure 2.
Transcription properties of the mosaic RNAP variants. (A) Positions of the β′ substitutions in mosaic RNAPs shown on the structure of Tth TEC in complex with Stl (2PPB2). RNAP orientation corresponds to Figure 1A. The β (pink), β′ (green), α (blue) and ω (gray) subunits are shown as ribbons; the FL and the jaw-domain are in red and turquoise, respectively. (B) Rates of single nucleotide addition by mosaic Taq TECs assembled on the minimal scaffold (top) were measured at 20°C and 1 mM NTP as described previously.18 Numbers in bold indicate fold differences in comparison with wild-type Taq RNAP. (C) Thermostability of mosaic Taq RNAPs. RNAPs were incubated for 3 min at indicated temperatures and their activity was measured on the T7A1 promoter in the presence of a CpA primer and UTP (10 µM each) for 5 min at 55°C.
Even single lineage-specific substitutions in the FL and adjacent regions may confer adaptive differences in the catalytic properties of RNAPs. For example, FL Q1046ATaq and BH S1074TTaq substitutions (Fig. 1) increase the rate of RNA synthesis (Suppl. Fig. 2).18 Q1046Taq directly contacts the central part of the fully-folded TL (Fig. 1A) whereas the corresponding S769Sce residue in RNAPII interacts with a conserved TL H1085Sce (H1242Taq/H936Eco) residue, implicated in catalysis,1,5,6 in a backtracked TEC11 and in an α-amanitin-inhibited TEC3 (Fig. 1E), in which the TL adopts partially open and wedged conformations, respectively. Thus, changes in the FL may affect the pathway or the rate of the TL folding. In addition, several “stimulatory” substitutions in the FL have been isolated; the M747IEco and P750LEco substitutions increase the elongation rate in vitro,12,19 and some sit-mutations were also mapped to the FL (Fig. 1). Interestingly, RNAPs from several bacterial lineages contain 5–20 residue insertions at the FL tip (Fig. 1A and B, Suppl. Fig. 1); their significance for RNAP function remains to be tested.
Role of the TL and FL in Substrate Discrimination
The proper folding of the TL was shown to be important for the kinetic selection of incoming nucleotides by enhanced incorporation of the correct substrates.5–7,14,15 Similarly, the closure of dNTP substrates in the active site was shown to provide a basis for efficient nucleotide discrimination by DNA polymerases.20 Recent analysis of Taq RNAP demonstrated that the contribution of the TL to the overall nucleotide selectivity varies for different substrate-template base-pairs, playing a major role for some NTP-template combinations and for discrimination against 2′- and 3′-deoxyNTPs.6 Two kinds of substitutions in the TL were shown to decrease transcription fidelity. First, changes in Eco, Taq RNAPs and Sce RNAPII that likely impair TL folding or its contacts with the incoming NTP preferentially decrease the rate of incorporation of correct NTPs (as compared to the incorrect ones), resulting in an overall decrease in discrimination efficiency.5–7 Similarly, restricting the TL mobility by RNAP-specific inhibitors (Stl for bacterial RNAP and α-amanitin for Sce RNAPII) dramatically decreased substrate selectivity.6,7 Second, E1103GSce substitution at the base of the TL in RNAPII increases the misincorporation rate,15 perhaps by promoting closure of the TL, and thus sequestering NTP in the active center. Deletion of a non-essential Rpb9 subunit led to a similar decrease in Sce RNAPII fidelity;21 Rpb9 likely contacts the TL in an open conformation (Fig. 1D) and its deletion was shown to stimulate TL folding.21 Interestingly, a part of the Rpb9-binding site is formed by a 20-residue insertion in the FL which is specific to eukaryotic and archaeal lineages (Fig. 1C and D; Suppl. Fig. 1).
The FL also appears to play a role in transcription fidelity. Similarly to substitutions that impaired TL folding,6,7 deletion of the FL in Taq RNAP strongly impaired incorporation of correct nucleotides but had 10 to 100-fold smaller defects in incorporation of incorrect substrates.18 Conversely, the F-Dra replacement favored incorporation of correct over incorrect substrates (∼15-fold increase in selectivity for correct UTP vs. incorrect CTP, Fig. 2B) (see Suppl. text for discussion). To our knowledge, this is the first example of a RNAP substitution characterized by both faster elongation and increased substrate selectivity. We propose that, in contrast to fast RNAP variants with TL substitutions that stabilize the folded TL and decrease transcription fidelity,7,14,15 the F-Dra substitution may stimulate overall conformational mobility of the TL, thus increasing substrate selectivity.
Control of Catalysis by the Jaw Domain of RNAP
The changes in the FL alone cannot fully account for the decreased catalytic rate of Taq RNAP in comparison with Dra, and additional elements must contribute to the catalytic differences between these RNAPs.18 The β' jaw domain, which forms a part of the downstream DNA binding channel and likely contacts the TL in an open conformation (Fig. 1B and 2A), may be one of such elements. To test this hypothesis, we constructed a mosaic Taq RNAP variant with substitution of the jaw domain (Jaw-Dra, β' residues 1,267–1,325Taq) with a Dra sequence (Fig. 2A and Suppl. Fig. 3).
The Jaw-Dra substitution increased the average elongation rate (Suppl. Fig. 2) and the rate of nucleotide addition (Fig. 2B). The interactions of the jaw domain with an open TL may therefore delay its folding, while the Jaw-Dra substitution may stimulate catalysis by destabilizing an open TL conformation. Indeed, a deletion of the jaw in Eco RNAP led to an increased elongation rate and decreased pausing.22 The evolutionary conserved N-terminal part of the jaw domain contains several substitutions in Taq/Dra RNAPs, and these residues assume different conformations in different Tth RNAP structures (Fig. 1A and B, Suppl. Fig. 1). One of the corresponding residues (K1112Sce) was previously hypothesized to mediate E1103Sce-dependent stabilization of the open TL in Sce RNAPII (Fig. 1D).15 However, in contrast to the E1103GSce substitution, the Jaw-Dra substitution does not increase misincorporation, suggesting that it may have a distinct effect on catalysis (Fig. 2B and Suppl. text). Future analysis of species-specific substitutions will help to understand the role of the jaw-TL interactions in RNAP catalysis and substrate discrimination.
Catalytic Activity and Thermostability
In comparison with their mesophilic counterparts, thermophilic enzymes display reduced catalytic efficiencies at low temperatures. These differences are usually attributed to the increased structural rigidity, which confers thermostability yet at the same time decreases conformational flexibility of the active center. To test whether the effects of substitutions in Taq/Dra RNAPs on the catalytic rate are indeed connected with structural stabilization of RNAP, we analyzed the thermostability of the F-Dra and Jaw-Dra mosaic RNAPs. Both enzymes were fully active at 55°C whereas Dra RNAP was thermoin-activated at temperatures above 45°C (data not shown). We then measured the stability of the mosaic RNAPs at higher temperatures. The thermo-inactivation profiles of the F-Dra and Jaw-Dra RNAPs were very similar to that of the wild-type Taq RNAP (with only a slightly lower stability of the Jaw-Dra RNAP at 80°C, Fig. 2C), suggesting that species-specific substitutions in the FL and the jaw-domain are not dictated by the thermostability of RNAP per se but rather serve to fine-tune the catalytic properties of RNAP at different ambient temperatures.
Regulation of Catalysis through TL Folding
Due to its key role in catalysis and its strategic positioning at the junction of the main and the secondary RNAP channels, the TL is an attractive target for transcription regulation. Our data demonstrate that substitutions in RNAP regions that contact the TL during the catalytic cycle may serve as an evolutionary mechanism that modulates the RNAP catalysis in various organisms, through affecting the conformational changes of the conserved active center elements. Many auxiliary ligands may also regulate catalysis through the TL folding.
The nascent RNA may stabilize a partially open TL conformation during RNAP pausing,23 TEC backtracking,11 and perhaps RNA cleavage. The Rpb9 subunit of Sce RNAPII that is thought to delay the TL folding (see above) may be positioned similarly to the lineage-specific SI3 insertion in the Eco RNAP TL (Fig. 1A and B) which modulates nucleotide addition and RNAP pausing.2,5,23 Regulators that bind within the secondary channel may also control transcription through the TL. In particular, transcript cleavage factors (GreA/B in bacteria, TFIIS in eukaryotes) closely approach the TL in the TEC.24,25 The stimulation of the RNA cleavage by Eco RNAP by GreB was shown to be dependent on the presence of the SI3 insertion but not on the TL folding, although the requirement for the TL itself could not be tested.5 The effects of another secondary channel factor, DksA, on transcription initiation by Eco RNAP require the TL but not SI3; furthermore, folding-affecting substitutions at the tip of the TL were shown to facilitate DksA action.26 TFIIS may stabilize an open TL conformation as observed in the RNAPII-TFIIS structure (Fig. 1D).11,25 Interestingly, mutations in a loop of Rpb9 that approaches the open TL weaken the TFIIS effects on transcription,27 suggesting that Rpb9 and TFIIS may together stabilize a functionally important conformation of the TL. Other yet unknown regulators may bind at the same location as Rpb9, SI3 and TFIIS/Gre to modulate transcription by bacterial, archaeal and eukaryotic RNAPs.
Restricting the TL mobility would lead to an efficient inhibition of RNAP catalysis. Two well-known RNAP inhibitors, Stl and α-amanitin, have been shown to trap the TL in inactive states (Fig. 1E).2,3 Since accessible parts of the TL and the FL display sequence variability (Suppl. Fig. 1), a search for small molecules that selectively target these elements in bacterial RNAP may lead to development of novel potent antibiotics.
Supplementary Material
Acknowledgements
We thank D. Pupov for help. This work was supported in part by the Russian Academy of Sciences Presidium Program in Molecular and Cellular Biology (to A.K.), the National Institutes of Health grant GM67153 (to I.A.) and the Russian Foundation for Basic Research grant 10-04-00925.
Abbreviations
- RNAP
RNA polymerase
- TL
trigger loop
- BH
bridge helix
- FL
F loop
- Stl
streptolydigin
- Eco
Escherichia coli
- Taq
Thermus aquaticus
- Tth
Thermus thermophilus
- Dra
Deinococcus radiodurans
- Sce
Saccharomyces cerevisiae
- Mja
Methanocaldococcus jannaschii
Footnotes
Previously published online: http://www.landesbioscience.com/journals/transcription/article/12544
References
- 1.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 3.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 4.Nudler E. RNA polymerase active center: the molecular engine of transcription. Annu Rev Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuzenkova Y, Bochkareva A, Tadigotla VR, Roghanian M, Zorov S, Severinov K, et al. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8:54. doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kireeva M, Kashlev M, Burton ZF. Translocation by multi-subunit RNA polymerases. Biochim Biophys Acta. 2010;1799:389–401. doi: 10.1016/j.bbagrm.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Epshtein V, Mustaev A, Markovtsov V, Bereshchenko O, Nikiforov V, Goldfarb A. Swing-gate model of nucleotide entry into the RNA polymerase active center. Mol Cell. 2002;10:623–634. doi: 10.1016/s1097-2765(02)00640-8. [DOI] [PubMed] [Google Scholar]
- 10.Tan L, Wiesler S, Trzaska D, Carney HC, Weinzierl RO. Bridge helix and trigger loop perturbations generate superactive RNA polymerases. J Biol. 2008;7:40. doi: 10.1186/jbiol98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artsimovitch I, Chu C, Lynch AS, Landick R. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science. 2003;302:650–654. doi: 10.1126/science.1087526. [DOI] [PubMed] [Google Scholar]
- 13.Severinov K, Markov D, Severinova E, Nikiforov V, Landick R, Darst SA, et al. Streptolydigin-resistant mutants in an evolutionarily conserved region of the beta' subunit of Escherichia coli RNA polymerase. J Biol Chem. 1995;270:23926–23929. doi: 10.1074/jbc.270.41.23926. [DOI] [PubMed] [Google Scholar]
- 14.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 15.Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, et al. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Archambault J, Jansma DB, Kawasoe JH, Arndt KT, Greenblatt J, Friesen JD. Stimulation of transcription by mutations affecting conserved regions of RNA polymerase II. J Bacteriol. 1998;180:2590–2598. doi: 10.1128/jb.180.10.2590-2598.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulbachinskiy A, Bass I, Bogdanova E, Goldfarb A, Nikiforov V. Cold sensitivity of thermophilic and mesophilic RNA polymerases. J Bacteriol. 2004;186:7818–7820. doi: 10.1128/JB.186.22.7818-7820.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miropolskaya N, Artsimovitch I, Klimasauskas S, Nikiforov V, Kulbachinskiy A. Allosteric control of catalysis by the F loop of RNA polymerase. Proc Natl Acad Sci USA. 2009;106:18942–18947. doi: 10.1073/pnas.0905402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weilbaecher R, Hebron C, Feng G, Landick R. Termination-altering amino acid substitutions in the beta' subunit of Escherichia coli RNA polymerase identify regions involved in RNA chain elongation. Genes Dev. 1994;8:2913–2927. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- 20.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 21.Walmacq C, Kireeva ML, Irvin J, Nedialkov Y, Lubkowska L, Malagon F, et al. Rpb9 subunit controls transcription fidelity by delaying NTP sequestration in RNA polymerase II. J Biol Chem. 2009;284:19601–19612. doi: 10.1074/jbc.M109.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ederth J, Artsimovitch I, Isaksson LA, Landick R. The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J Biol Chem. 2002;277:37456–37463. doi: 10.1074/jbc.M207038200. [DOI] [PubMed] [Google Scholar]
- 23.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Vassylyeva MN, Svetlov V, Dearborn AD, Klyuyev S, Artsimovitch I, Vassylyev DG. The carboxy-terminal coiled-coil of the RNA polymerase beta'-subunit is the main binding site for Gre factors. EMBO Rep. 2007;8:1038–1043. doi: 10.1038/sj.embor.7401079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 26.Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemming SA, Edwards AM. Yeast RNA polymerase II subunit RPB9. Mapping of domains required for transcription elongation. J Biol Chem. 2000;275:2288–2294. doi: 10.1074/jbc.275.4.2288. [DOI] [PubMed] [Google Scholar]
- 28.Artsimovitch I, Vassylyeva MN, Svetlov D, Svetlov V, Perederina A, Igarashi N, et al. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell. 2005;122:351–363. doi: 10.1016/j.cell.2005.07.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.