Abstract
C/EBPs are implied in an amazing number of cellular functions: C/EBPs regulate tissue and cell type specific gene expression, proliferation and differentiation control. C/EBPs assist in energy metabolism, female reproduction, innate immunity, inflammation, senescence and the development of neoplasms. How can C/EBPs fulfill so many functions? Here we discuss that C/EBPs are extensively modified by methylation of arginine and lysine side chains and that regulated methylation profoundly affects the activity of C/EBPs.
Key words: transcription, chromatin remodeling, differentiation, histone code, post-translational modification, signaling, methylation
Introduction
Many organ functions depend on C/EBP, such as the skin, liver, lung, bone or white and brown adipose tissue. In addition, many signaling pathways affect C/EBP activity. C/EBPs interact with a plethora of transcription factors, chromatin modifiers and remodeling complexes and components of the cell cycle machinery.1–4 It thus appears that extensive regulatory translational and post-transcriptional alterations downstream of signaling events serve to shape C/EBP functions in a way to seamlessly integrate into context specific cellular activities. In the following we provide a comprehensive survey of the available literature and some novel results on extensive C/EBP arginine and lysine methylation supporting this general assessment.
The CCAAT/enhancer binding proteins alpha and beta (C/EBPα,β) are founding members of the C/EBP-transcription factor family, initially characterized in adipogenesis and myelopoiesis. C/EBP modifications include the generation of protein isoforms of variable N-terminal length by regulated initiation of translation (C/EBPα,β) or alternative splicing and promoter usage (C/EBPδ,ε), respectively. Such translation isoforms with different N-termini have variable domain compositions and thus display distinct functions in gene regulation.5,6 In addition, a multitude of post-translational modifications (PTMs), including phosphorylation of serine, threonine and tyrosine residues,7,8 acetylation of lysine residues (Kac)9–11 and, as recently shown, methylation of lysine and arginine (Kme; Rme) occur on C/EBP N-termini.7,12,13
Structure and Function of C/EBPs
In order to conceptualize the multitude of C/EBP functions, it is important to consider some of the structural features of C/EBP transcription factors, as shown in Figure 1A and in Sup. Figure 1. Sequence alignments show that similarities in the N-termini are confined to short discretely conserved regions (CR1 to CR7, see also Sup. Fig. 1).14–16 The highly conserved and contiguous C-terminal ends encompass the basic DNA binding leucine zipper dimerization domains (bZip), that consist of a pre-bZip basic/acidic region (BA), the DNA binding part (DBD), the fork domain, the leucine zipper (LZ) dimerization domain and a post-bZip C-terminal peptide (CP). The alignment suggested that the family members C/EBPα, ε, on one hand and C/EBPβ, δ, on the other hand, are more closely related to each other and that all four family members may have arisen by consecutive gene duplications from an ancient vertebrate C/EBP. No significant homologies to CR1-7 were found in the inhibitory C/EBPγ and C/EBPζ, suggesting a more distant relationship between the latter C/EBP-related proteins and C/EBPα,β,δ,ε.
Figure 1.
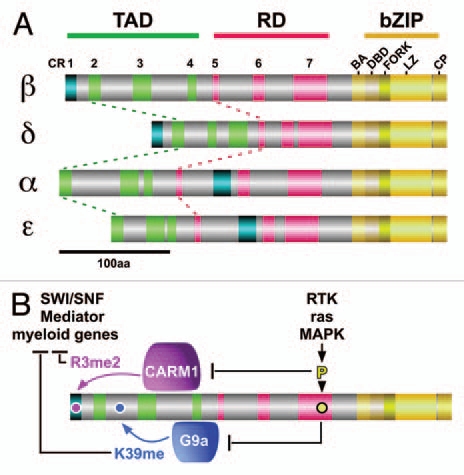
Scheme of the C/EBPα,β,δ,ε structure and crosstalk between phosphorylation and arginine/lysine methylation in C/EBPβ. (A) CR1-4 (green, turquoise), representing the transactivation domain (TAD) and CR5-7 the regulatory domain of C/EBPs. Towards the C-terminus a basic/acid motif (BA) precedes the DNA binding region (DB) followed by the fork and the leucine zipper (LZ) regions. A C-terminal peptide (CP) is separated from the LZ. (B) Model of signaling through receptor tyrosine kinase (RTK) / ras-MAPK pathways causes phosphorylation within CR7. Phosphorylation abrogates interaction with G9a and PRMT4 that methylate C/EBPβ K39 and R3, respectively. Methylation at K39 interferes with activation of myeloid genes and methylation of R3 interferes with recruitment of SWI/SNF that is required for the activation of a subset of myeloid genes.
Trans-activating and regulatory C/EBP modules.
Inspection of N-terminal CRs of C/EBPα,β,δ,ε revealed high conservation of primary structures in CR2, 3, 4 and 6 across species, whereas CR1 homologies were confined to C/EBPβ and C/EBPδ. More relaxed similarities between C/EBPs are found in CR5 and in CR7 (Sup. Fig. 1).
Data from many laboratories demonstrated that CR2, 3 and 4 make up a strong trans-activation domain (TAD). Removal or mutation of individual CR2, 3 or 4 in C/EBPβ strongly diminished its transactivation capacity. However, residual transactivation properties could be detected of individual TAD-CRs or any paired combination thereof, suggesting individual and co-operative functions of the CR2, 3 and 4 modules.14,17
CR1 of C/EBPβ contributes to the activation of a distinct set of chromatin embedded genes by recruiting the chromatin remodeling SWI/SNF complex and interacting with Mediator complexes.18,19 CR1 is restricted to C/EBPβ and sequence similarities to CR1 are only found in C/EBPδ, although an equivalent biological function in C/EBPδ has not yet been described. A functionally related but structurally distinct region was mapped in C/EBPα to reside between CR5 and CR6,20 (CR1 like, CR1L; Sup. Fig. 1). A structurally equivalent region can also be delineated in C/EBPε. C/EBPα CR1L affects cell differentiation and proliferation control and could be functionally replaced by CR1 of C/EBPβ.20,21
CR5 and CR7 of C/EBPβ display repressive and/or auto inhibitory functions.14–16 CR6 is a highly conserved short region present in all C/EBPs. CR6 contains a SUMOylation target that is involved in repressive functions.22–25 In summary, CR1-4 constitute transactivation domains (TAD) and CR5-7 may function as regulatory domains (RD).
Activation of C/EBPβ.
CR7 of C/EBPβ contains a conserved N-terminal motif followed by a poly-serine stretch and a proline embedded threonine residue, providing several phosphorylation target sites for various kinases.8 The proline embedded threonine residue serves as a target for mitogen activated kinase (MAPK) downstream of receptor tyrosine kinase (RTK) signaling. The CR7 MAPK site in C/EBPβ has been implied in de-repression and conditional trans-activation. Intra-molecular interactions between CR5 and CR7, as determined by yeast two-hybrid interactions were abrogated by a phospho-mimetic mutation of the MAP-kinase target site. Removal of either CR5 or CR7, or a phospho-mimicking mutation of the MAPK site, strongly activated C/EBPβ induced target gene expression.14,26 Moreover, CR5 was recently found to be phosphorylated and methylated on arginine and in conjunction with the TAD and bZip domain compact C/EBPβ in an inactive form.15 Several reports come to the conclusion that RTK signaling mediated C/EBPβ phosphorylation eliminates repressive functions of CR5-7 and that activation involves some structural conversion of the transcription factor. Along these lines, circumstantial evidence previously suggested that phosphorylation in CR7 of C/EBPβ entails a structural alteration that is allosterically transmitted to the Mediator complex which, together with CR1, converts Mediator into an active state.19,27 Accordingly, removal of CR5, 6 and 7 strongly activated gene expression, confirming that, at least in C/EBPβ, CR5-7 functions as a regulatory domain (RD) that conditionally inhibits transactivation or establishes repressor functions which are subject to signal induced negative regulation.7,14–16
C/EBP CRs: Beads on a string?
The highly conserved C/EBP CR1-7 are connected by “low complexity regions” (LCRs) that may be intrinsically unfolded and that are characterized by an accumulation of prolines and glycines.28–31 This may raise the possibility that proline isomerases are involved in altering the C/EBP structure. The C/EBP LCRs display relaxed interspecies conservation. Amino acid polymorphisms within the same C/EBP sub-type preferentially occur within LCRs (Sup. Fig. 1), although some primary structures are retained between all corresponding LCRs. As the activating motifs CR2, 3 and 4 may interact with components of the basic and inducible gene regulatory and epigenetic machinery,18,19,32,33 the LCRs might merely serve as flexible hinges to connect linear CR2-4 “docking” sites for co-factors or provide the scaffolds for complex assembly. Alternatively, the CRs may form a structural core and the semi-conserved, intervening LCRs may accommodate accessory docking sites for co-regulators in a C/EBP-family member specific fashion. Both models for LCR functionality are not mutually exclusive. Another possibility is that the CRs might exist as an intrinsically destabilized “molten globule” that may adopt an ordered structure only transiently and/or only after inter-molecular interactions with other proteins, depending on the signaling state and on PTMs.34,35 The fact that residual modular transactivating functions were observed with individual CR1-4 or combinations thereof currently supports a model of modular, linear functionality of CRs and tethering functions of LCRs, without ruling out other possibilities. Modular functionality is also supported by several other experiments: replacement of the internal C/EBPα SWI/SNF recruiting region with the N-terminal C/EBPβ derived CR1,20 abrogation of C/EBP-dependence of Myb regulated genes by a chimeric C/EBP-Myb protein,36 or even functional gene replacement of C/EBPα by C/EBPβ.37,38 It remains difficult to distinguish between several structural possibilities, since the intrinsic disorder prevents 3D structural elucidation of the C/EBP N-termini, indicating that C/EBP functions are based on an unusual degree of structural plasticity.
Multi-Site Methylation of C/EBPα,β
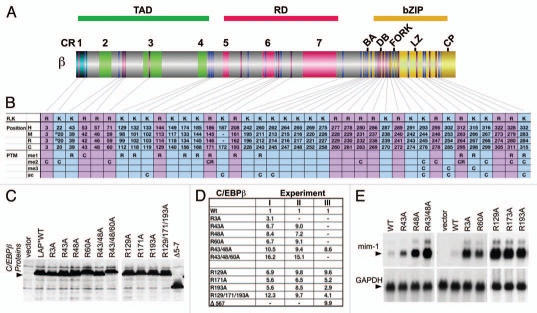
Considering the structural/functional C/EBP features further and seeking for an explanation of how C/EBPβ toggles between repressor/activator functions, it came to our attention that several basic residues (R, K), located at the borders close to CRs or within LCRs, are fairly conserved between C/EBPα,ε and C/EBPβ,δ or even between all C/EBPs (Sup. Fig. 1). Previous results obtained by metabolic labeling of full-length and truncated C/EBPα,β with 3H-SAM and immunoreactivity towards methyl-arginine/lysine specific antibodies12,13 suggested that C/EBPα,β are extensively post-translationally modified by K,R-methylation in their N-termini. In an effort to determine whether C/EBP modifications are more complex than previously anticipated, we examined C/EBP PTMs in a systematic approach. Comprehensive mass spectrometry of C/EBPβ and C/EBPα revealed more than 50 novel alterations, including phosphorylation (data not shown), acetylation (on K-residues) and methylation occurring on K,R-residues in C/EBPα,β from chicken, rat and human (Fig. 2 and Sup. Fig. 1). More than 30 methylation patterns, including mono- and di-methylation of arginine residues (Rme1, Rme2) and mono-, di- and trimethylation of lysine residues (Kme1, Kme2, Kme3) were uncovered, suggesting that extensive methylation on K,R-moieties occurs on both, C/EBPα,β (and probably on all C/EBPs).
Figure 2.
Distribution, methylation, and functional evaluation of conserved arginine residues in C/EBPβ. (A) Schematic representation of the C/EBPβ structure showing the distribution of evolutionary conserved arginine (R, magenta) or lysine (K, blue) in relation to the domain and CR structure. (B) List of R and K residues in human (H) mouse (M), rat (R), and chicken (C) C/EBPβ. Numbers refer to equivalent amino acid positions. Underneath: identification by mass spectrometry and summary of data of mono-(me1), di-(me2), tri-(me3) methylation, and acetylation (ac) in rat (R) and chicken (C) C/EBPβ. (C) Protein expression controls in QT6 fibroblasts of chicken C/EBPβ LAP* arginine to alanine (R to A) mutants or wild type (WT), as indicated. (D) Reporter activation by C/EBPβ mutants. Expression constructs encoding wild type or R to A C/EBPβ mutants were transfected together with a cMGF promoter luciferase construct and reporter expression was determined after 36 hours. Numbers show reporter activation as means of duplicates from 3 independent experiments (I, II, III). (E) Endogenous gene activation by C/EBPβ mutants. RNA blots show expression of the resident myeloid C/EBP target gene mim1 in QT6 fibroblasts. C/EBPβ R to A mutants or wild type C/EBPβ LAP* were transfected and expression levels of endogenous genes were monitored by Northern blotting. GAPDH expression serves as control.
Although modifications were found clustered in the bZip domain (that also harbor the most K,R-residues), many of the R- and K-residues in the N-terminus were modified by methylation. To unravel the functional consequences of methylation, we concentrated on the N-terminal “working end” of C/EBPβ. As shown in Figure 2B, methylation occurred on K39 that had previously been described as a target for acetylation9 and was then identified as a target of G9a K-methyltransferase.13 Moreover, the highly conserved K-residue within the SUMOylation site in CR6 (rat K134) may also become methylated (Fig. 2B), which would interfere with SUMO conjugation. The N-terminal R3 residue in CR1 that is involved in SWI/SNF recruitment, was found to be di-methylated and interaction analysis showed that methylation of R3 abrogates interaction with both SWI/SNF and Mediator. Activation of ras-MAPK signaling inhibited methylation of both R3 and K39 and mutation of either residue into alanine (R3A, K39A) enhanced C/EBPβ induced gene expression. Interestingly, mutation of R3 into leucine (to mimic R3-methylation) also strongly diminished interaction with SWI/SNF and Mediator and interfered with SWI/SNF dependent target gene activation.12 These data suggested an intramolecular crosstalk between signal dependent phosphorylation and K,R-methylation (Fig. 1B). In addition, it also indicated that methylation in the N-terminus might be involved in repression of C/EBP-mediated transactivation. Thus, the data also implied the existence of a signal dependent negative intra-molecular crosstalk between RTK induced phosphorylation and K,R-methylation. This notion was confirmed by inactivation of the methyltransferases PRMT4/CARM1 (methylating R3) or G9a (methylating K39) by dominant-negative enzyme mutants that resulted in activation of myeloid, C/EBPβ-mediated gene expression. Moreover, activation of the MAPK pathway or a phosphomimetic T to D mutation of the C/EBPβ MAPK site abrogated the interaction between C/EBPβ and PRMT4/CARM1 or G9a, as summarized in Figure 1B. It is plausible to assume that an allosteric alteration previously found to affect the N-terminus of C/EBPβ might also regulate the dynamic interaction with protein methyl-transferases.12,13,19,39
Mutational Analysis of C/EBPβ R-Methylation Sites
The possibility that arginine methylation is critical for the function of C/EBPβ was addressed by a mutagenesis approach. Figure 2A shows the conserved R,K-residues among vertebrate C/EBPβ (see also Sup. Fig. 1). The matrix in Figure 2B lists the respective amino acid sequence positions in human (H), rat (R), mouse (M) and chick (C) C/EBPβ. Underneath, mass spectrometric results indicate methylation or acetylation as found in chicken (C) or in rat (R). Arginines in the N-terminus of chicken and rat C/EBPβ were mutated to alanine (to abrogate modification of the side chain while retaining secondary structure) and mutants were tested in reporter and endogenous gene activation assays. All individual R to A mutations in chicken (Fig. 2C) and rat (data not shown) displayed enhanced reporter activation. Combinations of mutations in the TAD or the RD cooperated to further amplify reporter and endogenous gene expression (Fig. 2D and E). This indicates that both trans-activation and auto-inhibition rely on the methylation state of conserved arginines and that the extensive PTM methylation pattern seen by mass spectrometry appears to govern C/EBP functionality. It is noteworthy that ras activation of C/EBPβ was recently found to correlate with phosphorylation and methylation in CR5 that displays auto-inhibitory functions.7 These data suggest that the conserved R-residue in CR5 is involved in intra-molecular interactions that may depend on the methylation status. Pilot experiments with C/EBPα showed that R to A mutations also enhance its transactivation (data not shown) and thus suggest that other C/EBPs are regulated by arginine methylation.
Differential activity of individual C/EBPβ R to A point mutants was, however, observed on different chromatin embedded target genes (data not shown). This raises the possibility that, in addition to intra-molecular regulation, context-specific interactions within the local chromatin at cis-regulatory C/EBP sites may selectively distinguish between C/EBP modification patterns. Whether and how methylation/de-methylation of R and K C/EBPβ residues exactly modify intra-molecular interactions and context-specific functions will require systematic exploration of both a presumably signal-dependent PTM “code” on C/EBPs and PTM dependent protein interactions.
Outlook
C/EBPβ emerges as a nodal transcription factor that integrates and coordinates signals from several onco-developmental pathways, including Stat3, TGFβ and RTK-signaling.26,40,41 It seems reasonable to assume that signal integration occurs through a multitude of modification sites and the structural plasticity of C/EBPs. Multi-site PTM of C/EBPβ at the receiving end of signaling cascades may navigate and organize co-factor interactions, thus consolidating epigenetic functions in a loci-specific fashion.12,42–44 PTM specific antibodies and proteomic screening approaches will be required to identify such networks involved in the conversion of cell peripheral signals into gene regulation and epigenetics. We anticipate that unraveling the PTM-dependent C/EBPβ interaction network might help to elucidate how C/EBPβ regulates genes involved in such diverse settings as myogenic vs. adipogenic differentiation,45 cooperation between C/EBPβ, Stat3 and TGFβ in epithelial-mesenchymal/tumorigenic transition,40,41 and in complex C/EBPβ functions in the skin and lung, innate immunity, fertility, mammary gland formation, myelopoiesis and development of leukemia. In any case, the extensive PTM pattern seen with C/EBPs, FoxO proteins42 or p53,46 are reminiscent of the “histone code” and suggest that multi-site modifications of transcription factors might be central for integration of signals and gene expression.42,43,47,48
Acknowledgements
We thank Manfred Gossen for critical reading of the manuscript and suggestions, Maria Knoblich for protein expression and purification and Karolin Friedrich for technical assistance. Ole Pless is supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) to Achim Leutz (LE 770/4-1). Elisabeth Kowenz-Leutz, Maria Knoblich and Gunnar Dittmar are supported by institutional funds of the Helmholtz Association and the DFG (TRR54, TP6).
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/13510
Supplementary Material
References
- 1.Johnson PF. Molecular stop signs: regulation of cell cycle arrest by C/EBP transcription factors. J Cell Sci. 2005;118:2545–2555. doi: 10.1242/jcs.02459. [DOI] [PubMed] [Google Scholar]
- 2.Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318–324. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Sebastian T, Johnson PF. Stop and go: anti-proliferative and mitogenic functions of the transcription factor C/EBPbeta. Cell Cycle. 2006;5:953–957. doi: 10.4161/cc.5.9.2733. [DOI] [PubMed] [Google Scholar]
- 4.Zahnow CA. CCAAT/enhancer-binding protein beta: its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med. 2009;11:12. doi: 10.1017/S1462399409001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calkhoven CF, Muller C, Leutz A. Translational control of gene expression and disease. Trends Mol Med. 2002;8:577–583. doi: 10.1016/s1471-4914(02)02424-3. [DOI] [PubMed] [Google Scholar]
- 6.Wethmar K, Smink JJ, Leutz A. Upstream open reading frames: molecular switches in (patho)physiology. 2010. In press. [DOI] [PMC free article] [PubMed]
- 7.Lee S, Shuman JD, Guszczynski T, Sakchaisri K, Sebastian T, Copeland TD, et al. RSK-mediated phosphorylation in the C/EBP{beta} leucine zipper regulates DNA binding, dimerization and growth arrest activity. Mol Cell Biol. 2010;30:2621–2635. doi: 10.1128/MCB.00782-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nerlov C. C/EBPs: recipients of extracellular signals through proteome modulation. Curr Opin Cell Biol. 2008;20:180–185. doi: 10.1016/j.ceb.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Cesena TI, Cardinaux JR, Kwok R, Schwartz J. CCAAT/enhancer-binding protein (C/EBP)beta is acetylated at multiple lysines: acetylation of C/EBPbeta at lysine 39 modulates its ability to activate transcription. J Biol Chem. 2007;282:956–967. doi: 10.1074/jbc.M511451200. [DOI] [PubMed] [Google Scholar]
- 10.Joo M, Park GY, Wright JG, Blackwell TS, Atchison ML, Christman JW. Transcriptional regulation of the cyclooxygenase-2 gene in macrophages by PU.1. J Biol Chem. 2004;279:6658–6665. doi: 10.1074/jbc.M306267200. [DOI] [PubMed] [Google Scholar]
- 11.Xu M, Nie L, Kim SH, Sun XH. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPbeta. EMBO J. 2003;22:893–904. doi: 10.1093/emboj/cdg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowenz-Leutz E, Pless O, Dittmar G, Knoblich M, Leutz A. Crosstalk between C/EBPbeta phosphorylation, arginine methylation and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J. 2010;29:1105–1115. doi: 10.1038/emboj.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pless O, Kowenz-Leutz E, Knoblich M, Lausen J, Beyermann M, Walsh MJ, et al. G9a-mediated lysine methylation alters the function of CCAAT/enhancer-binding protein-{beta} J Biol Chem. 2008;283:26357–26363. doi: 10.1074/jbc.M802132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowenz-Leutz E, Twamley G, Ansieau S, Leutz A. Novel mechanism of C/EBPbeta (NF-M) transcriptional control: activation through derepression. Genes Dev. 1994;8:2781–2791. doi: 10.1101/gad.8.22.2781. [DOI] [PubMed] [Google Scholar]
- 15.Lee S, Miller M, Shuman JD, Johnson PF. CCAAT/enhancer binding protein {beta} DNA binding is auto-inhibited by multiple elements that also mediate association with p300/CBP. J Biol Chem. 2010 doi: 10.1074/jbc.M110.128413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams SC, Baer M, Dillner AJ, Johnson PF. CRP2 (C/EBPbeta) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J. 1995;14:3170–3183. doi: 10.1002/j.1460-2075.1995.tb07319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nerlov C, McNagny KM, Doderlein G, Kowenz-Leutz E, Graf T. Distinct C/EBP functions are required for eosinophil lineage commitment and maturation. Genes Dev. 1998;12:2413–2423. doi: 10.1101/gad.12.15.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowenz-Leutz E, Leutz A. A C/EBPbeta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- 19.Mo X, Kowenz-Leutz E, Xu H, Leutz A. Ras induces mediator complex exchange on C/EBPbeta. Mol Cell. 2004;13:241–250. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C. Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 2001;15:3208–3216. doi: 10.1101/gad.209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller C, Calkhoven CF, Sha X, Leutz A. The CCAAT enhancer-binding protein alpha (C/EBPalpha) requires a SWI/SNF complex for proliferation arrest. J Biol Chem. 2004;279:7353–7358. doi: 10.1074/jbc.M312709200. [DOI] [PubMed] [Google Scholar]
- 22.Berberich-Siebelt F, Berberich I, Andrulis M, Santner-Nanan B, Jha MK, Klein-Hessling S, et al. SUMOylation interferes with CCAAT/enhancerbinding protein beta-mediated c-myc repression, but not IL-4 activation in T cells. J Immunol. 2006;176:4843–4851. doi: 10.4049/jimmunol.176.8.4843. [DOI] [PubMed] [Google Scholar]
- 23.Eaton EM, Sealy L. Modification of CCAAT/enhancer-binding protein-beta by the small ubiquitin-like modifier (SUMO) family members, SUMO-2 and SUMO-3. J Biol Chem. 2003;278:33416–33421. doi: 10.1074/jbc.M305680200. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Cantwell CA, Johnson PF, Pfarr CM, Williams SC. Transcriptional activity of CCAAT/enhancerbinding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J Biol Chem. 2002;277:38037–38044. doi: 10.1074/jbc.M207235200. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian L, Benson MD, Iniguez-Lluhi JA. A synergy control motif within the attenuator domain of CCAAT/enhancer-binding protein alpha inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J Biol Chem. 2003;278:9134–9141. doi: 10.1074/jbc.M210440200. [DOI] [PubMed] [Google Scholar]
- 26.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, et al. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci USA. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eastburn D, Han M. When Ras signaling reaches the mediator. Dev Cell. 2004;6:158–159. doi: 10.1016/s1534-5807(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 28.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 29.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tompa P. Intrinsically unstructured proteins evolve by repeat expansion. Bioessays. 2003;25:847–855. doi: 10.1002/bies.10324. [DOI] [PubMed] [Google Scholar]
- 32.Mink S, Haenig B, Klempnauer KH. Interaction and functional collaboration of p300 and C/EBPbeta. Mol Cell Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nerlov C, Ziff EB. CCAAT/enhancer binding protein-alpha amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 35.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mo X, Kowenz-Leutz E, Laumonnier Y, Xu H, Leutz A. Histone H3 tail positioning and acetylation by the c-Myb but not the v-Myb DNA-binding SANT domain. Genes Dev. 2005;19:2447–2457. doi: 10.1101/gad.355405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen SS, Chen JF, Johnson PF, Muppala V, Lee YH. C/EBPbeta, when expressed from the C/ebpalpha gene locus, can functionally replace C/EBPalpha in liver but not in adipose tissue. Mol Cell Biol. 2000;20:7292–7299. doi: 10.1128/mcb.20.19.7292-7299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiu CH, Lin WD, Huang SY, Lee YH. Effect of a C/EBP gene replacement on mitochondrial biogenesis in fat cells. Genes Dev. 2004;18:1970–1975. doi: 10.1101/gad.1213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higashimoto K, Kuhn P, Desai D, Cheng X, Xu W. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc Natl Acad Sci USA. 2007;104:12318–12323. doi: 10.1073/pnas.0610792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 42.Benayoun BA, Veitia RA. A post-translational modification code for transcription factors: sorting through a sea of signals. Trends Cell Biol. 2009;19:189–197. doi: 10.1016/j.tcb.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Sims RJ, 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- 44.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 47.Chi P, Allis CD, Wang GG. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang XJ. Multisite protein modification and intramolecular signaling. Oncogene. 2005;24:1653–1662. doi: 10.1038/sj.onc.1208173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.