Abstract
MicroRNAs (miRNAs) are key regulatory RNAs that act in concert to coordinately repress messenger RNA (mRNA) translation through imperfect recognition of multiple specific binding sites (BS) located in their 3′ untranslated region (UTR). Here, we present a polymerase-chain reaction (PCR)-based cloning strategy that allows the rapid and efficient generation of regulatory elements harboring up to 10 miRNA BS. Amenable for the study of regulatory elements of any multiplicity, such as those recognized by miRNAs and transcription factors, this methodology will facilitate functional miRNA/miRNA BS studies and accelerate discoveries mainly in the field of gene regulation.
Keywords: Polymerase chain reaction, molecular cloning, microRNA, microRNA binding site, gene regulation
MicroRNAs (miRNAs) are key regulatory RNA species that act in concert to coordinately repress messenger RNA (mRNA) translation through imperfect recognition of multiple specific binding sites (BS) located mainly in their 3′ untranslated region (UTR) [for a recent review, see 1]. Made possible through the discovery and characterization of relevant mRNA targets, delineation of the cellular processes controlled by miRNAs and establishment of possible links between miRNA dysfunction and genetic diseases raises tremendous interest among the scientific community. Aided by the use of computational approaches, the task of validating the functionality and biological importance of candidate mRNAs remains challenging. Indeed, only a few miRNA:mRNA target combinations have been characterized in detail; the major limitations to these studies being (i) the multiplicity of the miRNAs involved and (ii) the plurality of the miRNA BS through which mRNA translation is regulated. The latter context is exemplified by the lin-14 mRNA 3′UTR, which harbors 7 binding sites complementary to miRNA lin-4 in C. elegans [2].
Although bioassays that closely mimic these aspects of miRNA regulation do exist [3; 4], they are not easily amenable for the multifaceted characterization of natural miRNA:miRNA BS interactions based on imperfect complementarity and initial translational repression, which is a prerequisite for functional validation and annotation of putative miRNA BS. This prompted investigators to favor the use of experimental models of low biological relevance that are based on miRNA-directed cleavage of mRNAs bearing unique, unnatural sequences of perfect complementarity. Currently implying the synthesis of several very long oligonucleotides to be used in a gene cloning scheme requiring multiple PCR and ligations reactions, and as many bacterial transformations, preparation of the molecular tools required for the study of a single miRNA:miRNA BS combination is lengthy, tedious and expensive.
In order to circumvent these issues, we have developed a simple, versatile, cost-effective and time-efficient PCR approach to generate regulatory elements bearing a various number of contiguous miRNA BS sequences. These inserts can be subcloned into any vector of interest for subsequent functional characterization (eg, gene regulation studies in a relevant cell culture system). In order to validate this strategy experimentally, we utilized mmu-miR-328, mmu-miR-298 and their respective putative BS located in the 3′ UTR of mouse β-amyloid precursor protein converting enzyme 1 (mBACE1) mRNA as model miRNA:miRNA BS combinations and studied the functionality of their BS in cultured N2a and NIH 3T3 cells. The sequence encoding the precursors of miR-328 (pre-miR-328) and mmu-miR-105 (pre-miR-105), which was used as a control, were cloned in the psiSTRIKE vector (Promega) according to the manufacturer’s protocol, whereas a synthetic miR-298 duplex (Integrated DNA Technologies) was used to study miR-298 BS function.
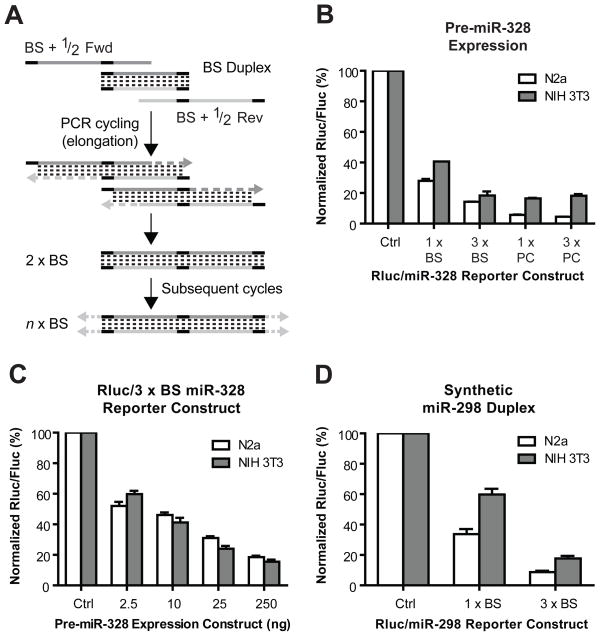
The DNA oligonucleotide sequences used to generate the PCR fragments containing various number of contiguous miR-328 BS are the following: mmu-miR-328 Fwd (5′-atctcgtccctgtggtaccctggcagagaaagggccaatctcaatctc-3′), mmu-miR-328 Rev (5′-gagattgagattggccctttctctgccagggtaccacagggacgagat-3′), BS + 1/2 Fwd (5′-atctcgtccctgtggtaccctggcagagaaagggccaatctcaatctcgtccctgtggtacc-3′) and BS + 1/2 Rev (5′-gagattgagattggccctttctctgccagggtaccacagggacgagattgagattggccc-3′). The nucleotides composing the miRNA binding sites (BS) are in bold, whereas the intervening sequences are underlined. Two oligonucleotides (mmu-miR-328 Fwd and mmu-miR-328 Rev) reconstituting a miRNA BS, flanked by a 5-nt intervening sequence, were annealed to obtain a BS duplex DNA template. When amplified by PCR using the same oligonucleotides, but extended at their 3′end by the 5′ half of the miRNA BS (BS + 1/2 Fwd and BS + 1/2 Rev), the PCR products were elongated further, yielding a mixture of blunt-ended DNA fragments harboring various numbers (n) of miRNA BS in a pre-determined orientation (Fig. 1A). The PCR reactions containing 30 ng/μl BS duplex, 0.5 μM of each BS+1/2 Fwd and BS+1/2 Rev oligonucleotides, 200 μM dNTPs and 2.5 U PFU polymerase (New England Biolabs) in reaction buffer were subjected to the following protocol: an initial 5-min denaturation period at 94°C was followed by 35 cycles of denaturation for 1 min at 94°C, annealing for 45 s at 55°C and elongation for 1 min at 72°C, followed by a final elongation period of 10 min at 72°C. The samples were preserved at 4°C. The PCR mixture is then used in a single ligation reaction and bacterial transformation. Alternatively, fragments of the desired lengths can be separated by agarose gel electrophoresis prior to ligation.
Figure 1. Experimental validation of the PCR-based strategy designed to generate regulatory elements bearing a various number of contiguous miRNA binding sites.
(A) Schematic representation of the PCR amplification strategy. (B–C) N2a and NIH 3T3 cells were cotransfected with a pre-miR-328 expression vector and either (B) the Rluc/miR-328 reporter construct harboring 1 or 3 copies of the miR-328 binding site (BS) or a sequence perfectly complementary (PC) to miR-328 (n = 2 experiments), or (C) the Rluc/miR-328 reporter construct harboring 3 copies of the miR-328 BS (n = 2 experiments). (D) Cells were cotransfected with the synthetic miR-298 duplex and an Rluc/miR-298 reporter construct harboring 1 or 3 copies of the miR-298 BS (n = 2 experiments). (B–D) The results of Rluc activity were normalized with Fluc reporter activity and expressed as a percentage of the results obtained with the empty reporter vector (set at 100%). Results are expressed as mean ± standard error of the mean.
After phenol/chloroform extraction and ethanol precipitation, the DNA products were phosphorylated using T4 polynucleotide kinase (New England Biolabs) and ligated blunt with the T4 DNA ligase (New England Biolabs) in the PmeI cloning site of psiCHECK vector (Promega), downstream of the Rluc coding region, as described previously [5] to generate Rluc/miRNA BS reporter constructs. The ligation products were transformed in DH5α competent bacteria and the constructs harboring the desired number of BS, as indicated by the length of their inserts, were selected by colony PCR screening of ampicillin resistant bacteria. The correct orientation and authenticity of the cloned PCR-amplified inserts were ascertained by DNA sequencing.
Analysis of the PCR reactions performed at various template concentrations by agarose gel electrophoresis and ethidium bromide staining revealed a ladder-like pattern of discrete bands of the size expected for DNA fragments differing by the length of a single miRNA BS (data not shown). Colony PCR screening of selected bacterial clones revealed the successful construction of Rluc/miRNA reporter constructs harboring up to 10 copies of miR-328 or miR-298 BS (data not shown), thereby validating our PCR-based cloning strategy. When considering both miR-328 and miR-298 BS reporter constructs combined, 52 (or 72%) of the 72 clones that were tested contained BS at the following frequencies: 11% (1 copy), 42% (2 copies), 23% (3 copies), 8% (4 copies), 6% (5 copies) and 10% (> 5 copies). The proportion of positive bacterial clones could be enriched further, if necessary, through specific digestion of the religated, blunt-ended empty vectors by the restriction enzyme PmeI prior to the bacterial transformation step.
The Rluc/miRNA BS reporter constructs were functionally tested by performing dual luciferase assays in N2a and NIH 3T3 cultured murine cells transiently cotransfected with the pre-miR-328 expression (250 ng DNA) and Rluc/miRNA BS reporter (400 ng DNA) constructs using Lipofectamine 2000 (Invitrogen), as described in [5]. The pre-miR-105 expression vector was used as a control (Ctrl). In these assays, expression of the Rluc/miR-328 reporter gene was downregulated by ~60% to 80% upon coexpression of pre-miR-328, the extent of which depended on the number of miR-328 BS (Fig. 1B). These data support the notion that the extent of mRNA translational repression is proportional to the degree of miRNA BS occupancy [6]. Similar findings were obtained in HeLa cells (data not shown), suggesting that this bioassay may be applicable to cell lines of different origin, including human. The dosage response of the repression mediated by miRNAs of imperfect complementarity is revealed further by the dose-dependent, gene downregulatory effects of pre-miR-328 (Fig. 1C).
These observations are in contrast with the pronounced downregulation of Rluc/miR-328 reporter gene expression conferred by a single sequence of perfect complementarity to miR-328 (Fig. 1B) and reflect the nature of the silencing mechanism involved, i.e. initial mRNA translational repression versus mRNA cleavage. As illustrated in Fig. 1D for miR-298, our methodology is applicable to other miRNA BS sequences and can be used in combination with synthetic miRNA duplexes. This latter strategy becomes necessary when pre-miRNA sequences, such as that of pre-miR-298, cannot be adapted to meet the requirements for U6 promoter-based vector expression without significant loss of function: (i) the transcription template should start with a guanosine (G), the preferred transcription initiation nucleotide for the U6 promoter; and (2) the U6 termination sequence is TTTTT, so that after transcription, each transcript will have two 3′ Us.
Together, these findings attest of the suitability and applicability of our PCR-based cloning strategy to the study of mRNA translational repression mediated by natural miRNA:mRNA interactions in mammals. This methodology will facilitate experimental miRNA target validation and ease characterization of miRNA:mRNA interactions, including the role and importance of the intervening sequences, all of which may be utilized to further improve the accuracy of computational miRNA target predictions. Amenable for microRNA studies, the principle of our cloning strategy is also applicable to functional studies of coding (eg, epitopes) or non-coding (eg, regulatory elements) DNA sequences of any multiplicity, such as those recognized by transcription factors. As for any PCR-based approaches, oligonucleotides need to be carefully designed and selected so to avoid the formation of primer dimers or base pairing to unspecific sites, and the PCR conditions to be adjusted and optimized for any specific pairs of oligonucleotides.
Applicable to any organisms for which the relevant molecular tools have been developed and used in conjunction with other tools (eg, transfection of synthetic or expression of mature miRNAs, miRNA duplexes or pre-miRNAs), techniques (eg, in vitro analytical methods, library construction, site-directed or random mutagenesis), and biological systems (eg, gel shift assays, in vitro translation experiments or reporter gene activity assays in mammalian cell culture) currently available, this methodology may facilitate and accelerate the elucidation of the mechanisms underlying gene expression in eukaryotes.
Acknowledgments
P.P. is a New Investigator of the Canadian Institutes of Health Research (CIHR) and Junior 2 Scholar from the Fonds de la Recherche en Santé du Québec. This study was supported by the CIHR (grant HOP-83069 to P.P.) and NARSAD: The Mental Health Research Association (Young Investigator Award to P.P.).
Abbreviations
- miRNA
microRNA
- PCR
polymerase-chain reaction
- mRNA
messenger RNA
- UTR
untranslated region
- BS
binding site
References
- 1.Ouellet DL, Perron MP, Gobeil LA, Plante P, Provost P. MicroRNAs in Gene Regulation: When the Smallest Governs It All. J Biomed Biotechnol. 2006;2006:69616. doi: 10.1155/JBB/2006/69616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–65. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plante I, Davidovic L, Ouellet DL, Gobeil LA, Tremblay S, Khandjian EW, Provost P. Dicer-Derived MicroRNAs Are Utilized by the Fragile X Mental Retardation Protein for Assembly on Target RNAs. J Biomed Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ouellet DL, Plante I, Landry P, Barat C, Janelle MÈ, Flamand L, Tremblay MJ, Provost P. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008:1–13. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA::target interaction. Chem Biol. 2004;11:1619–23. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]