Summary
The ESX-1 secretion system of M. tuberculosis delivers bacterial virulence factors to host cells during infection. The most abundant factor, the ESAT-6/CFP-10 dimer, is targeted for secretion via a C-terminal signal sequence on CFP-10 that is recognized by the cytosolic ATPase, Rv3871. However, the selection determinants for other ESX-1 substrates appear to be more complex. Some substrates, such as ESAT-6, are secreted despite lacking signal sequences. Furthermore, all substrates require targeting of the other ESX-1 secreted proteins, a distinguishing feature of this system. How ESX-1 substrates are selected and the basis for co-dependent secretion is unknown. Here we show that the EspC substrate interacts with Rv3868, a cytosolic AAA ATPase, through its C-terminus. Swapping the C-termini of EspC and CFP-10 revealed that these signals are functionally distinct, suggesting that the proteins are targeted via interactions with different ATPases. Surprisingly, biochemical purification experiments demonstrate that these substrates and ATPases form multi-protein complexes inside the cell and identified a new secreted substrate. By interfering with this protein interaction network, we have partially uncoupled co-dependent substrate secretion. Our results suggest that proper functioning of the ESX-1 pathway requires the interaction of multiple ESX-1 substrates and components prior to their secretion. Ultimately, understanding the details of ESX-1 targeting may allow for engineering of better vaccines to prevent tuberculosis.
Introduction
Gram-positive and mycobacterial pathogens use the ESX-1 (ESAT-6 system 1) secretion system (Type VII SS) to manipulate the host cell response during infection (Abdallah et al., 2007, Burts et al., 2005, Gao et al., 2004, Guinn et al., 2004, Hsu et al., 2003, Stanley et al., 2007, Stanley et al., 2003). While there are many proteins required for ESX-1 function in mycobacteria, how the majority of these proteins contribute to virulence factor secretion is unclear (Abdallah et al., 2007, Brodin et al., 2006, Gao et al., 2004, Guinn et al., 2004, Hsu et al., 2003, MacGurn et al., 2005, McLaughlin et al., 2007, Raghavan et al., 2008, Stanley et al., 2003). In M. tuberculosis, the core components of the ESX-1 system include Rv3877, and two AAA ATPases, including Rv3870 and Rv3871 (Guinn et al., 2004, Stanley et al., 2003). The two major substrates, ESAT-6 (early secreted antigen, 6kDa) and CFP-10 (culture filtrate protein, 10kDa), form a heterodimer, and are targeted for secretion by a C-terminal signal sequence on CFP-10 that is recognized by Rv3871 (Champion et al., 2006, Guinn et al., 2004, Hsu et al., 2003, Renshaw et al., 2005, Renshaw et al., 2002, Stanley et al., 2003). In addition to the ESAT-6/CFP-10 pair, there are at least four additional ESX-1 substrates, including EspR (Rv3849), EspA (Rv3616c), EspB (Rv3881c), and EspC (Rv3615c) (Fortune et al., 2005, MacGurn & Cox, 2009, McLaughlin et al., 2007, Raghavan et al., 2008, Xu et al., 2007). EspC was previously identified in genetic screens to identify M. tuberculosis genes required for growth during mouse infection and intracellular trafficking (Cox et al., 1999, MacGurn & Cox, 2007, MacGurn et al., 2005). EspC is required for the secretion of ESAT-6 and CFP-10 and is also an ESX-1 substrate that requires Rv3870, Rv3871, Rv3877, ESAT-6 and CFP-10 for secretion from M. tuberculosis (MacGurn & Cox, 2009, MacGurn et al., 2005).
One of the most distinctive aspects of ESX-1 secretion is that all of the substrates are mutually dependent upon each other for secretion (Fortune et al., 2005). For example, in strains lacking the ESAT-6/CFP-10 complex, none of the other known substrates are secreted by ESX-1 (Fortune et al., 2005, MacGurn & Cox, 2009, McLaughlin et al., 2007, Raghavan et al., 2008, Xu et al., 2007). Likewise, strains lacking EspA, EspB, EspC, or EspR fail to secrete ESAT-6/CFP-10 (Fortune et al., 2005, MacGurn & Cox, 2009, McLaughlin et al., 2007, Raghavan et al., 2008, Xu et al., 2007). One possible explanation is that substrates interact prior to, or during export by ESX-1, and that these interactions are critical for ESX-1 function (Fortune et al., 2005, Ize & Palmer, 2006). An alternative explanation of this phenomenon is that these proteins are not only substrates, but also components of the secretion machine itself (Fortune et al., 2005, Ize & Palmer, 2006).
M. marinum, a close relative of M. tuberculosis, causes a tuberculosis-like infection in ectotherms but rarely causes serious disease in humans (Stamm & Brown, 2004). The ESX-1 system is conserved and functional in M. marinum, and mutant M. marinum strains lacking ESX-1 components are attenuated in macrophage and animal infections (Gao et al., 2004, McLaughlin et al., 2007, Volkman et al., 2004). This attenuation can be complemented using the paralogous genes from M. tuberculosis suggesting that the functions of these proteins are the same in both species (Gao et al., 2004, McLaughlin et al., 2007). Importantly, the mechanism of ESX-1 substrate targeting is also likely conserved, as the CFP-10 signal sequence required for ESAT-6/CFP-10 secretion in M. tuberculosis is identical in M. marinum, making this organism an attractive model for studying ESX-1 protein secretion.
Here we report the results of our studies focused on the targeting of EspC to the ESX-1 secretion system in both M. marinum and M. tuberculosis. Like CFP-10, the C-terminus of EspC is required for proper targeting through ESX-1. However, these sequences are not functionally interchangeable with those of CFP-10 and bind to a different ATPase than the CFP-10 signal sequence. Furthermore, co-purification studies in M. marinum, together with yeast two-hybrid studies in M. tuberculosis, revealed interactions between EspC and other ESX-1 secreted proteins, supporting the model that the formation of substrate complexes inside the cell underlies the phenomenon of co-dependent secretion. Finally, identification of proteins in this complex revealed a new ESX-1 substrate, EspF.
Results
The C-terminus of EspC is necessary for secretion by ESX-1
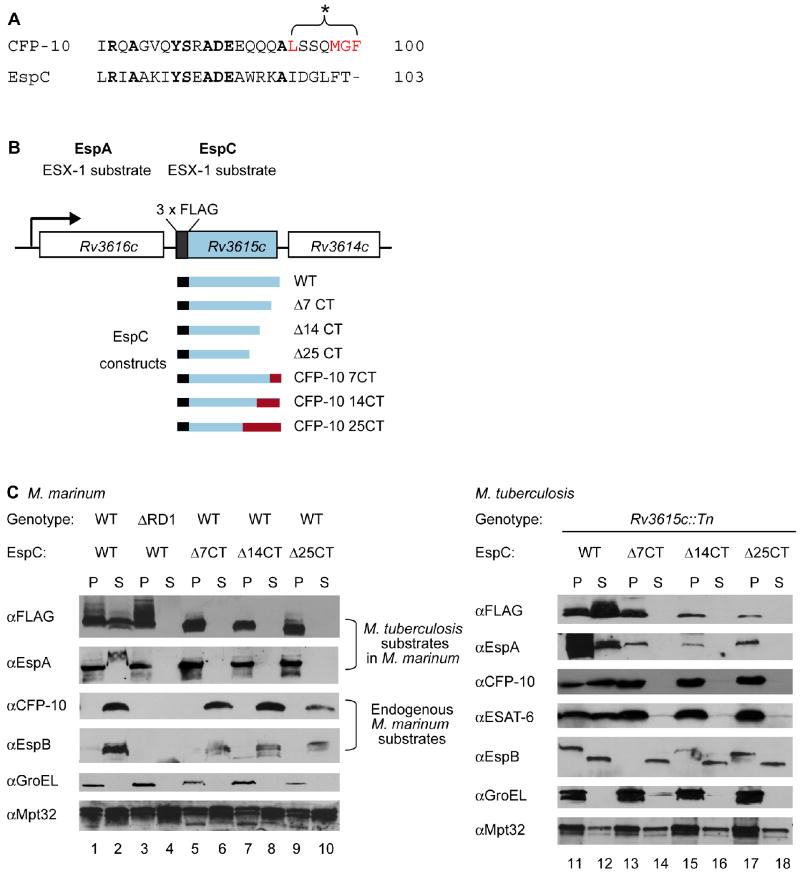
Previously we demonstrated that the C-terminus of CFP-10 functions as a signal sequence that targets the CFP-10/ESAT-6 substrate pair for secretion by the ESX-1 system in M. tuberculosis (Champion et al., 2006). Based on this discovery, we sought to identify additional proteins that bear an ESX-1 signal sequence. To this end, we used the FASTA algorithm and searched the last 25 amino acids of all predicted proteins in M. tuberculosis for similarity to the CFP-10 signal sequence. The protein with the highest similarity to the C-terminus of CFP-10 is EspC (Rv3615c) (Figure 1A). While there are some conserved residues within the last 25 amino acids of CFP-10 and EspC, the signal sequence region (L94-F100) is divergent between the two substrates (Figure 1A). Since these amino acids are critical for targeting CFP-10 and for its interaction with Rv3871 (Champion et al., 2006), this suggested that the targeting sequences for these two substrates are likely different.
Fig. 1. The C-terminus of EspC is required for secretion.
(A) Alignment of the C-terminal amino acids of CFP-10 and EspC. Residues required for CFP-10 signal recognition are in red. (B) Schematic of the M. tuberculosis Rv3616c operon. Rv3615c is FLAG-tagged at its N-terminus. Constructs for expression of EspC truncated or hybrid proteins are shown. (C) Immunoblot analysis of cell pellets (P) and supernatants (S) from (left) wild-type or ΔRD1 M. marinum or (right) Rv3615c∷Tn M. tuberculosis cells expressing FLAG-EspC or truncated forms of FLAG-EspC. GroEL is used as a control for autolysis, and Mpt32 is used as a loading control for all samples.
To test whether the C-terminus of EspC is required for targeting, we created constructs to express N-terminally FLAG-tagged M. tuberculosis EspC or truncated versions lacking the terminal seven, 14 or 25 amino acids. These genes were used to replace the wild-type espC gene in an integrating plasmid containing the Rv3616c-Rv3614c operon, such that the normal signals for transcription and translation were preserved (Figure 1B). In an effort to determine if M. marinum can be used as a model to study ESX-1 substrate targeting, these plasmids were introduced into M. marinum and FLAG-EspC secretion was monitored by western blot analysis (Figure 1B and 1C). The full-length M. tuberculosis FLAG-EspC protein (FLAG-EspCMt) was present in M. marinum cell pellets (P) and secreted into the culture supernatants (S) in an ESX-1 dependent manner (Figure 1C, lanes 1-4). In both M. tuberculosis and M. marinum, FLAG-EspCMt migrates aberrantly on SDS-PAGE gels, for unknown reasons (MacGurn & Cox, 2009). However, deletion of the terminal seven, 14, or 25 amino acids of FLAG-EspCMt resulted in stable production the mutant proteins, but blocked their secretion into the M. marinum culture supernatants (Figure 1C, lanes 5-10). The FLAG antibody specifically recognized the M. tuberculosis FLAG-EspCMt proteins, as there were no size appropriate cross reacting bands in wild-type M. marinum samples lacking the FLAG-EspCMt plasmid (Figure S1, S2).
We also monitored the secretion of the other known ESX-1 substrates from M. marinum in the presence of FLAG-EspCMt. M. marinum CFP-10 (CFP-10Mm), and EspB (Mh3881c, M. marinum homolog of Rv3881c, EspBMm) were secreted into the culture supernatant independently of FLAG-EspCMt secretion, although expression or stability of both was slightly diminished in some of these strains (Figure 1C, lanes 1-10). However, despite the presence of a functional ESX-1 secretion machine in M. marinum, the EspA protein from M. tuberculosis (EspAMt) was retained in the cell pellets of M. marinum strains bearing truncated FLAG-EspCMt proteins, while it was secreted from M. marinum strains expressing wild-type FLAG-EspCMt (Figure 1C, lanes 1 and 2 compared to lanes 3-10). Why the secreted form of the protein migrates slower through the gel is currently unclear, however, the EspA antibody (Fortune et al., 2005) specifically recognized the M. tuberculosis EspAMt protein, and did not cross react with any M. marinum protein (Figure S1). Together, these data indicate that expression of the truncated forms of FLAG-EspCMt had a specific, dominant effect on the secretion of M. tuberculosis EspAMt in M. marinum.
In an effort to both confirm the results from the M. marinum experiments, and to study EspC targeting in a single copy system, we expressed the same FLAG-EspCMt constructs in an M. tuberculosis strain bearing a transposon insertion in the espC gene (MacGurn et al., 2005). As in M. marinum, full length FLAG-EspCMt was secreted into the culture supernatant (Figure 1C, lanes 11 and 12), but the truncated forms of FLAG-EspCMt were retained in the cell pellet (Figure 1C, lanes 13-18). These results demonstrate that, as with CFP-10, the C-terminal region of EspCMt is required for secretion by the ESX-1 system in mycobacteria.
In the M. tuberculosis espC mutant strain, the ESX-1 secretion system is non-functional (MacGurn et al., 2005)(Figure S2), and this defect is complemented by expression of full-length FLAG-EspCMt (Figure 1C and S2). In contrast, the C-terminally truncated forms of FLAG-EspCMt failed to promote secretion of ESAT-6, CFP-10, and EspA (Figure 1C, lanes 11-18), though the levels of EspA were reduced in the FLAG-EspCMt deletion strains, indicating that EspA stability and/or expression requires EspC (Figure 1C, lanes 13-18). Surprisingly, despite variability in the levels of EspB in the cell pellets, EspB was secreted into the culture supernatants regardless of FLAG-EspCMt secretion (Figure 1C, lanes 11-18; Figure S3). Taken together, these data demonstrate that the C-terminal region of EspC is required for secretion by the ESX-1 system in both M. marinum and M. tuberculosis.
The C-termini of EspC and CFP-10 are not functionally equivalent
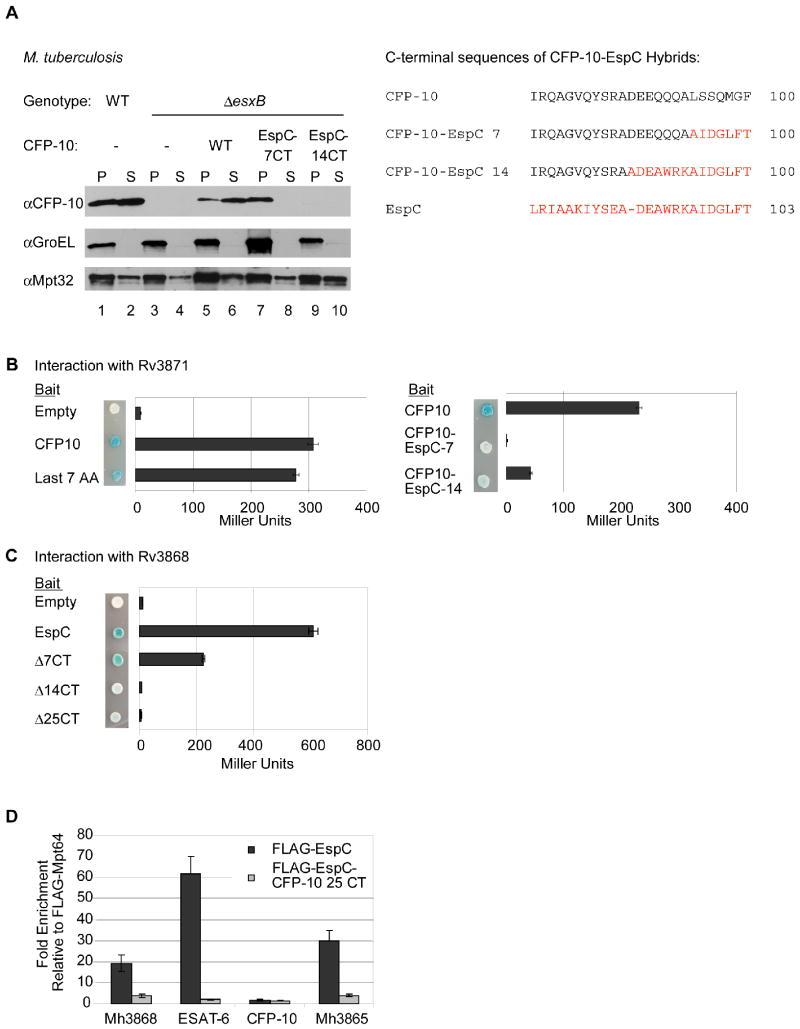
To determine if the C-terminus of EspC could substitute for the CFP-10 signal sequence, we replaced the last seven or 14 amino acids of CFP-10 with the corresponding amino acids from EspC, and expressed these hybrid proteins in esxB mutant M. tuberculosis cells (Figure 2A). The CFP-10-EspCMt hybrid protein bearing the last seven amino acids of EspC remained in the cell pellet and was not secreted into the culture supernatant (Figure 2A, lanes 7 and 8). Additionally, the CFP-10-EspCMt hybrid protein bearing the last 14 amino acids of EspC was undetectable (Figure 2A, lanes 9 and 10), likely due to instability as a result of disruption of the CFP-10/ESAT-6 complex (Guinn et al., 2004, Stanley et al., 2003). Indeed, we have shown previously that aspartate 87 of CFP-10, which is changed to alanine in this construct, is required for interaction with ESAT-6 and that disruption of the CFP-10/ESAT-6 dimer leads to instability of the proteins (Champion et al., 2006). Furthermore, whereas the terminal seven amino acids of CFP-10 are bound by Rv3871 (Champion et al., 2006), yeast two-hybrid analysis using proteins from M. tuberculosis demonstrated that the last seven amino acids of EspC failed to interact with this protein (Figure 2B). Taken together, these data demonstrate that although the C-termini of both CFP-10 and EspC are required for secretion, the EspC C-terminus does not mediate an interaction with Rv3871, and is not functionally equivalent to the CFP-10 signal sequence.
Fig. 2. Rv3868 interacts with the C-terminus of EspC in vitro and in vivo.
(A) Immunoblot analysis of cell pellets (P) and supernatants (S) from wild-type or ΔesxB M. tuberculosis cells harboring plasmids expressing ESAT-6 and either CFP-10 or CFP-10-EspC hybrid proteins. GroEL is used as a control for autolysis, and Mpt32 is used as a loading control for pellet and supernatant samples. The resulting C-terminal sequences of the CFP-10 EspC hybrid proteins are shown (right). (B) Yeast two-hybrid analysis of the interaction between CFP-10 or CFP-10-EspC hybrid proteins with Rv3871 (amino acids 248-591). (C) Yeast two-hybrid analysis of interaction between C-terminal truncations of EspC and Rv3868. For B and C, blue yeast colonies grown on solid media containing X-gal were positive for interaction. β-galactosidase activity is shown; error bars represent standard deviation. For all yeast two-hybrid experiments, M. tuberculosis constructs were used to study interaction. (D) Relative fold-enrichment of each protein selectively identified in the immunoprecipitation from M. marinum was assessed using quantitative mass spectrometry. The average enrichment over three injections is plotted for each protein. The fold enrichment was normalized to the average enrichment from immunoprecipitation with FLAG-Mpt64.
Rv3868 interacts with the C-terminus of EspC
Given the differences between the seven terminal amino acids of CFP-10 and EspC (Figure 1A), we reasoned that another cytosolic component of the secretion machine likely interacts with the C-terminus of EspC. A candidate for EspC recognition was Rv3868, a cytosolic AAA ATPase that is required for ESX-1 secretion but whose role in secretion is unknown (Brodin et al., 2006, Gao et al., 2004). Using directed yeast two-hybrid analysis with proteins from M. tuberculosis, we found that EspC interacted with Rv3868, and deletions from the C-terminus of EspC abrogated this interaction (Figure 2C). CFP-10 failed to interact with Rv3868 by this method, in agreement with previous biochemical interaction studies (Luthra et al., 2008), which further suggested that the Rv3868/EspC interaction is specific.
To test whether this interaction occurs in vivo in M. marinum, we expressed M. tuberculosis FLAG-EspCMt in M. marinum, performed immunoprecipitation on cell pellet extracts, and identified potential interacting proteins by mass spectrometry. As a control, we performed the same experiment using FLAG-Mpt64Mt, an unrelated protein secreted by the Sec secretion pathway. We surmised that interacting proteins would be enriched by immunoprecipitation with FLAG-EspCMt relative to the immunoprecipitation with FLAG-Mpt64Mt. Four ESX-1 associated proteins were specifically enriched by immunoprecipitation with FLAG-EspCMt, most notably Mh3868, the M. marinum homolog of Rv3868 (Figure 2D and Table S3). Using quantitative mass spectrometry, we found that Mh3868 was enriched at least 20-fold by immunoprecipitation with FLAG-EspCMt as compared to Flag-Mpt64Mt (Figure 2D). Mh3868 and the other ESX-1 associated proteins did not co-immunoprecipitate with the FLAG-EspC-CFP-10Mt hybrid protein bearing the last 25 amino acids of CFP-10Mt, indicating that the interaction between EspCMt and Mh3868 in vivo requires the C-terminus of EspCMt (Figure 2D).
Taken together, the genetic and biochemical data presented here suggest that the EspCMt C-terminus interacts with the Rv3868 AAA ATPase, and that this interaction also occurs in vivo in M. marinum. This indicates that the C-termini of two ESX-1 substrates from M. tuberculosis, CFP-10 and EspC, interact with different AAA ATPases, Rv3871 and Rv3868.
ESX-1 substrates interact with at least two ATPase components of ESX-1
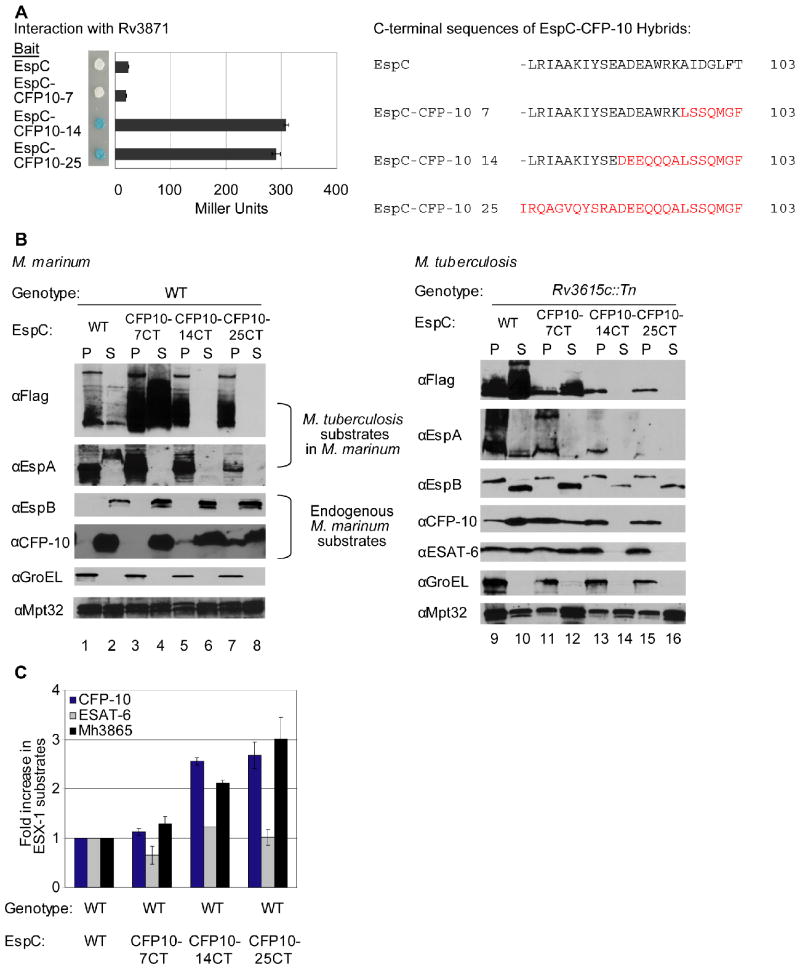
Given that the CFP-10 signal sequence is recognized by Rv3871, and the C-terminus of EspC interacts with Rv3868, we hypothesized that there are at least two ways in which substrates can interact with ESX-1 components. We tested if the C-terminus of CFP-10 could promote interaction of FLAG-EspCMt with Rv3871. We substituted the last seven, 14, or 25 amino acids of EspC with the corresponding amino acids of CFP-10 (Figure 1A), and monitored interaction with Rv3871 by yeast two-hybrid analysis using proteins from M. tuberculosis. Both EspC and EspC-CFP-10 bearing the last seven amino acids of CFP-10 failed to interact with Rv3871 by this method, while the EspC-CFP-10 hybrid proteins bearing the terminal 14 or 25 amino acids of CFP-10 interacted robustly with Rv3871 (Figure 3A). It was curious that the EspC-CFP-10 bearing the last seven amino acids of CFP-10 failed to interact with Rv3871, since the last seven amino acids of CFP-10 fused to LexA is sufficient for Rv3871 interaction (Champion et al., 2006). It is possible that the addition of EspC between LexA and the terminal seven amino acids of CFP-10 precludes interaction with Rv3871 by folding in such a way that makes the last seven amino acids unavailable for interaction.
Fig. 3. Interaction of substrates with at least two ESX-1 associated ATPases is required for secretion.
(A) Yeast two-hybrid analysis of the interaction between EspC or EspC-CFP-10 hybrid proteins with Rv3868. Blue yeast colonies grown on solid media containing X-gal were positive for interaction. β-galactosidase activity is shown; error bars represent standard deviation. M. tuberculosis constructs were used to study interaction. The resulting C-terminal sequences of the EspC-CFP-10 hybrid proteins are shown (right). (B) Immunoblot analysis of cell pellets (P) and supernatants (S) of wild-type M. marinum strains (left) or M. tuberculosis bearing a transposon insertion in Rv3615c (right) expressing FLAG-EspC or FLAG-EspC-CFP-10 hybrid proteins. GroEL is used as a control for autolysis, and Mpt32 is used as a loading control for pellet and supernatant samples (C) Accumulation of M. marinum ESX-1 substrates in M. marinum due to expression of FLAG-EspC-CFP-10 hybrid proteins. Shown is the fold-change of cytosolic ESX-1 proteins in strains expressing the FLAG-EspC with the CFP-10 targeting region relative to wild-type M. marinum. 3 peptides were averaged together for CFP-10 and ESAT-6, and 5 were used for Mh3865 (EspFMm). Error bars are the %CV for the average of three measurements per peptide. Peak areas are available in the supplementary data (Table S4) and confirmatory spectra can be found in Figures S6 and S7.
We expressed the EspC-CFP-10Mt hybrid proteins in wild-type M. marinum and found that FLAG-EspCMt bearing the last seven amino acids of CFP-10 was secreted into the culture supernatant (Figure 3B, lanes 1-4). This is consistent with our previous finding that the terminal seven amino acids of CFP-10 represent a portable signal sequence for ESX-1 (Champion et al., 2006). For unknown reasons, it also appeared that levels of FLAG-EspC-CFP-10-7CTMt protein present in both the cell pellet and supernatant samples were increased, despite loading equal amounts of protein sample (Figure 3B, lanes 1-4). The FLAG-EspC-CFP-10Mt hybrid proteins bearing the terminal 14 or 25 amino acids of CFP-10 were retained in the cell pellets and not secreted by ESX-1 (Figure 3B lanes 5-8).
We monitored the secretion of additional ESX-1 substrates from M. marinum to determine if EspCMt bearing the C-terminal sequence of CFP-10 affected ESX-1 secretion in the presence of a functional secretion machine. Although FLAG-EspC-CFP-10Mt bearing the last seven amino acids of CFP-10 was secreted into the culture supernatant, EspAMt from M. tuberculosis was retained in the M. marinum cell pellet (Figure 3B, lanes 3 and 4). EspAMt was also retained in the cell pellets of the strains expressing the “trapped” EspC-CFP-10Mt hybrid proteins (Figure 3B, lanes 5-8). So, as in Figure 1C, perturbing EspCMt secretion uniquely affected EspAMt secretion.
In the M. marinum strains expressing the FLAG-EspC-CFP-10Mt hybrid proteins, the endogenous M. marinum substrates, EspBMm and CFP-10Mm were secreted into the culture supernatants (Figure 3B). Interestingly, western blot analysis revealed that endogenous CFP-10Mm accumulated in the cell pellets of the strains expressing the “trapped” FLAG-EspC-CFP-10Mt hybrid proteins bearing the last 14 or 25 amino acids of CFP-10 (Figure 3B, lanes 5-8). One possible explanation for the accumulation of CFP-10Mm in the cell pellet is that the hybrid proteins are competing with CFP-10Mm for secretion, likely through the interaction with Mh3871 (M. marinum homolog of Rv3871). A second explanation is that the hybrid protein itself is slowing CFP-10Mm export.
To further understand this, we monitored the accumulation of endogenous CFP-10Mm and ESAT-6Mm in the M. marinum pellets of strains expressing either FLAG-EspCMt, or the FLAG-EspC-CFP-10Mt hybrid proteins using quantitative mass spectrometry (Figure 3C). Relative to the strain expressing FLAG-EspCMt, M. marinum strains expressing the FLAG-EspC-CFP-10Mt hybrid proteins with the terminal seven, 14 or 25 amino acids of CFP-10 accumulated 1.2, 2.6, and nearly 3-fold amounts of endogenous CFP-10Mm in the cell pellets, but ESAT-6Mm secretion was unaffected (Figures 3C). Mh3865 (also named EspFMm), an ESX-1 substrate identified in this study (Figure 4) accumulated 1.4, 2.1, and 3.0 fold, respectively. It is also notable, that although CFP-10Mm accumulated in the cell pellets of the strains bearing the FLAG-EspC-CFP-10Mt hybrid proteins, it was still secreted into the culture supernatants to levels similar to strains bearing wild-type FLAG-EspCMt (Figure 3B, lanes 5-8). This suggests that expression of FLAG-EspCMt bearing the CFP-10 signal sequence leads to the accumulation of some ESX-1 substrates in the M. marinum cell pellet perhaps through interaction of the hybrid proteins with Mh3871. It is unclear why ESAT-6Mm does not accumulate in the M. marinum cell pellet (see Discussion).
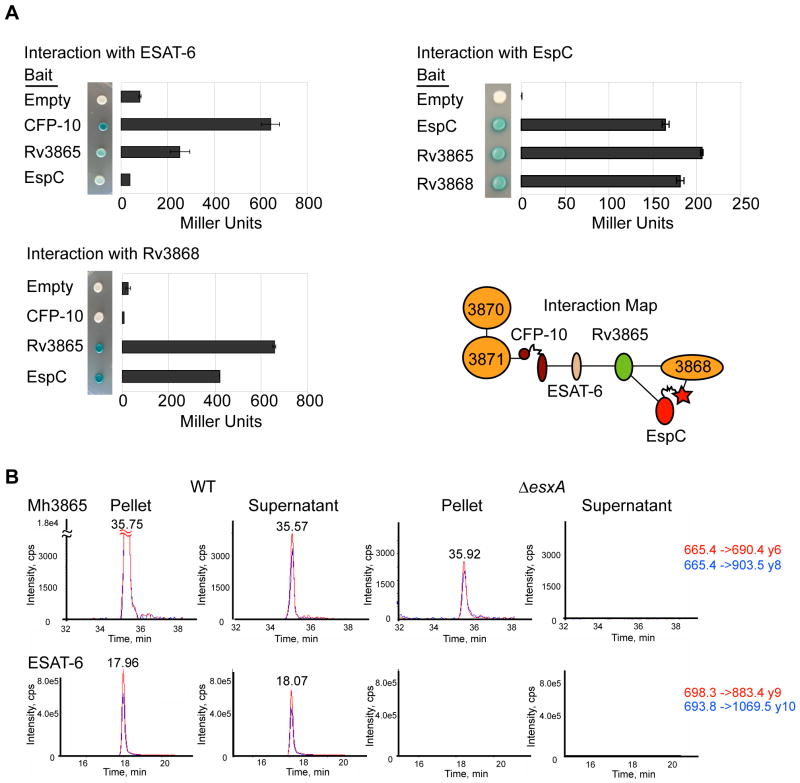
Fig. 4. At least three ESX-1 substrates interact for form a multi-protein complex in the cytosol.
(A) Yeast two-hybrid analysis measuring the interaction of M. tuberculosis ESX-1 substrates with Rv3868. Error bars represent standard deviation. Interaction map of ESX-1 associated AAA ATPases and substrates is shown. (B) Mh3865 (EspFMm) is an ESX-1 substrate. Shown are the MRM elution profiles for EspFMm and ESAT-6Mm peptides in digested M. marinum pellets (P) or supernatants (S). Two transitions (Red&Blue) for each peptide in WT or ΔesxA M. marinum cells are shown. For Mh3865 (EspFMm) and ESAT-6Mm, the transitions are shown as parent mass (m/z) -> fragment mass (m/z), and are shown to the right in red and blue. Additional peptides, including those for GroES and confirmatory spectra are in the supplementary data (Figures S6 and S7).
To test if the C-terminus of CFP-10 could promote the secretion of FLAG-EspCMt in the absence of endogenous EspC, we expressed the FLAG-EspC-CFP-10Mt hybrid proteins in espC mutant M. tuberculosis cells. Although FLAG-EspCMt bearing the last seven amino acids of CFP-10 was expressed at lower levels than FLAG-EspCMt, the hybrid protein was still secreted into the culture supernatant (Figure 3B, lanes 9-12). Again, this is consistent with the terminal seven amino acids of CFP-10 functioning as a portable ESX-1 signal sequence (Champion et al., 2006). As in M. marinum, the FLAG-EspC-CFP-10Mt hybrid proteins bearing the terminal 14 or 25 amino acids of CFP-10 were retained in the cell pellets and not secreted by ESX-1 (Figure 3B, lanes 13-16). Interestingly, CFP-10 and ESAT-6 secretion mirrored that of EspC; CFP-10 and ESAT-6 were secreted into the culture supernatant of strains expressing FLAG-EspCMt or the EspC-CFP-10 Mt hybrid protein bearing the terminal seven amino acids of CFP-10 (Figure 3B, lanes 11 and 12). The hybrid proteins that interacted with Rv3871 by yeast two-hybrid analysis (Figure 3A) blocked secretion of CFP-10 (Figure 3B, lanes 13-16).
EspA was not secreted from any of the M. tuberculosis strains expressing the FLAG-EspC-CFP-10 Mt hybrid proteins (Figure 3B lanes 9-16), and the levels of EspA in the cell pellets were greatly diminished. This is again consistent with the requirement of EspC for either stability or expression of EspA. In contrast, although EspB was expressed at lower levels, it was secreted from M. tuberculosis independently of the FLAG-EspC-CFP-10 Mt hybrid proteins (Figure 3B, lanes 11-16 and Figure S3).
These data argue that there are intrinsic differences between the targeting of ESX-1 substrates and that the interaction with at least two ESX-1 associated ATPases is required. Specifically, while EspA requires the proper targeting of EspC for secretion, likely though Rv3868, CFP-10 and EspB are targeted independently of EspC and EspA.
At least four ESX-1 substrates form a multi-protein complex in vivo
The co-immunoprecipitation studies of FLAG-EspCMt from M. marinum, which led to the identification of Mh3868, also identified three other ESX-1 associated proteins that were specifically enriched for interaction with FLAG-EspCMt, namely ESAT-6Mm, EspFMm and Mh3864. ESAT-6Mm was enriched approximately 60 fold, while a second protein EspFMm (Mh3865) was enriched almost 30-fold relative to co-immunoprecipitation using FLAG-Mpt64Mt (Figure 2D, Table S3). As with Mh3868, the co-immunoprecipitation of these proteins required the C-terminus of EspCMt, since the Flag-EspC-CFP-10Mt hybrid protein failed to enrich for these proteins. The interactions we observed in M. marinum were confirmed using yeast two-hybrid analysis with proteins from M. tuberculosis, which revealed direct interactions between ESAT-6 and EspF, EspF and EspC, and EspF and Rv3868 (Figure 4A).
In addition to these proteins, Mh3864, a recently identified ESX-1 substrate in M. marinum (Carlsson et al., 2009) was also specifically enriched by immunoprecipitation with FLAG-EspCMt (Table S3). We were unable to directly link this protein to known components of the ESX-1 secretion system using yeast two-hybrid analysis with proteins from M. tuberculosis (data not shown). It is likely that Mh3864 interacts indirectly with the known ESX-1 components and substrates identified here, and the direct binding partners of this substrate await discovery.
Surprisingly, while ESAT-6Mm was enriched by immunoprecipitation with FLAG-EspCMt from M. marinum cell lysates, CFP-10Mm was not, despite being stable and detectable by western blot analysis in this strain (Figure 2D, Figure 1C, lane 1). This was not expected since in M. tuberculosis, ESAT-6 and CFP-10 are known to form a heterodimer, and are dependent upon each other for stability in vivo (Hsu et al., 2003, Renshaw et al., 2005, Renshaw et al., 2002, Stanley et al., 2003). Given that we detected a strong interaction between M. tuberculosis ESAT-6 and EspF, it is possible that ESAT-6Mm forms heterodimers with both EspFMm and CFP-10Mm in vivo, which may explain why ESAT-6Mm but not CFP-10Mm co-immunoprecipitated with FLAG-EspCMt from M. marinum in these experiments.
EspF was previously identified in proteomic studies as a secreted mycobacterial protein (Bahk et al., 2004). Additionally, EspF is paralogous to EspC, and the two M. tuberculosis proteins share 52% similarity and 37% identity at the amino acid level. We therefore hypothesized that EspFMm is an ESX-1 substrate in M. marinum. We monitored the production and secretion of EspFMm using mass spectrometry and found that EspFMm was present in the cell pellet and the culture supernatant of wild-type M. marinum (Figures 4B and S4). In contrast, EspFMm was retained in the cell pellet, and no longer secreted into the culture supernatant of an M. marinum strain bearing a deletion of the esxA gene, the M. marinum homolog of ESAT-6 (Figures 4B and S4). Additionally, we found that EspFMm was not secreted into the culture supernatant of strains lacking Mh3871, the M. marinum homolog of Rv3871 (Figure S5). This demonstrates that EspFMm is an ESX-1 substrate in M. marinum.
We attempted similar experiments to determine if EspF is an ESX-1 substrate in M. tuberculosis. While it is a highly abundant in M. marinum, EspF was barely detectable in cell pellet and culture supernatants of M. tuberculosis, making it difficult to distinguish between active secretion of EspF and release due to autolysis (data not shown). These data demonstrate a distinct difference between ESX-1 secretion in M. marinum and M. tuberculosis.
Discussion
Our data support a model in which different C-terminal sequences of ESX-1 substrates function to target proteins to cognate ATPases (Figure 5). Specifically, the CFP-10 signal sequence targets substrates to Rv3871, while the C-terminal amino acids of EspC targets substrates to Rv3868. We previously showed that the terminal seven amino acids of CFP-10 constitute a portable signal sequence (Champion et al., 2006), and here we find that this signal can promote the secretion of EspC. These data, considered alone, would suggest a simple model in which targeting substrates to ATPases is sufficient for secretion. However, our data from this study strongly suggests that the cis-acting sequences required for substrate targeting and for translocation across the membrane and cell well is more complicated. Specifically there are four results which prompt us to elaborate on our model: 1) the C-termini of CFP-10 and EspC are not functionally equivalent, 2) adding more than the terminal seven amino acids of CFP-10 to EspC blocks secretion, 3) EspA secretion is uniquely blocked in the presence of the EspC-CFP-10-7CT hybrid protein, and 4) multiple ESX-1 substrates interact with each other inside the mycobacterial cell prior to secretion. We propose a model for ESX-1 targeting that incorporates these new complexities (Figure 5). Specifically, we posit that the substrates have multiple signals to facilitate substrate targeting and translocation across the cell wall, and that the interactions between substrates and components are critical for ESX-1 function.
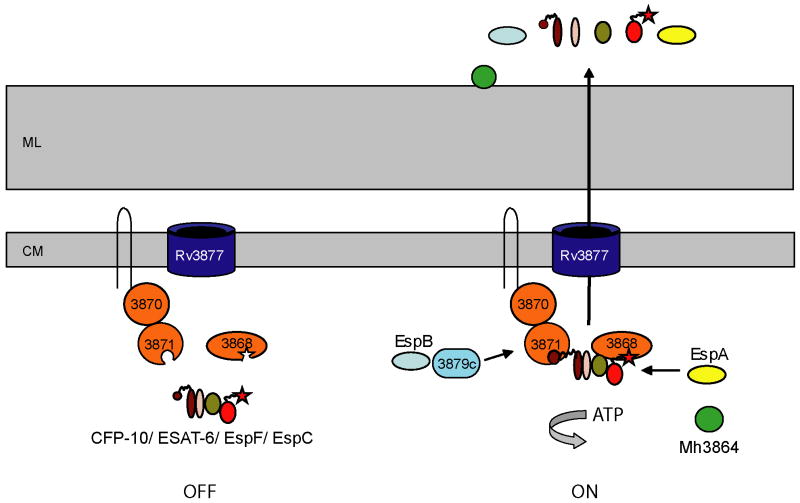
Fig. 5. A model for ESX-1 substrate targeting in mycobacteria.
Our work supports a model in which the C-terminal regions of ESX-1 substrates function to target them to cognate ATPases, either directly or through protein interaction with other substrates. The CFP-10 signal sequence targets substrates to Rv3871, while the C-terminal amino acids of EspC targets substrates to Rv3868. One possibility is that prior to engagement of these sequences by Rv3871 and Rv3868, the secretion machine is non-functional (left). Either prior to or after the formation of a multi-substrate complex (likely including CFP-10, ESAT-6, EspF and EspC), engagement of the C-termini by the ESX-1 associated ATPases activates the machine for secretion (right). EspB likely is indirectly recognized through Rv3879c by Rv3871 (McLaughlin et al., 2007), while EspA is likely targeted through Rv3868, though the mechanism by which this occurs is unknown thus far. Mh3864, a recently identified substrate that localizes to the cell wall, is also secreted by ESX-1, but the way that it is targeted remains unknown (Carlsson et al., 2009). ML is mycolate layer, CM is cytoplasmic membrane.
First, although the EspC C-terminus is required for secretion, it is not sufficient, suggesting that other sequences within the protein are important for secretion. Furthermore, unlike the CFP-10 signal sequence, the C-terminus of EspC is necessary but not sufficient for interaction with Rv3868 (Figure S8) and for translocation. We hypothesize that once directed to the ATPases by the C-terminal amino acids, other sequences present on the substrates are required for further steps in translocation. This would also explain why simply targeting proteins to ATPases is not sufficient for secretion. For example, EspC bearing the last 25 amino acids of CFP-10 still interacts with Rv3871, but is not secreted.
Second, the interactions between the substrates and components of the ESX-1 system prior to secretion are likely important for function. We envision a scenario where Rv3871 and Rv3868 are brought together via interactions between the various substrates (including CFP-10, ESAT-6, EspF and EspC), activating the ATPases for secretion. Here we found that the EspC-CFP-10-7CT protein was able to complement the secretion of known ESX-1 substrates in an espC mutant strain, but specifically blocked the secretion of EspA. We refer to this as substrate uncoupling. One possibility is that the EspC-CFP-10-7CT hybrid protein is able to interact with both Rv3868 and Rv3871, bringing these ATPases together and activating them, possibly rendering EspA unnecessary for secretion. Of course other models are possible, including one in which EspA is uniquely dependent on proper targeting of EspC. The dependence of ESX-1 on the formation of these protein complexes inside the cell either simultaneously or sequentially, likely explains the mechanism of co-dependent substrate secretion.
Our studies also provide insight into how EspB and EspA are differentially targeted for secretion by ESX-1. EspB secretion is not dependent upon EspC secretion, but is dependent upon the secretion of ESAT-6/CFP-10. The dependence of EspB on the secretion of ESAT-6/CFP-10 is consistent with the findings by Xu et al., but in contrast to those observed by McLaughlin et al. (McLaughlin et al., 2007, Xu et al., 2007). This is the first report of an ESX-1 substrate (EspB) that is dependent on the secretion of a subset of ESX-1 substrates (ESAT-6 and CFP-10, but not EspC). Although the precise molecular mechanisms of EspA secretion remain unclear, our data suggest that EspA requires the proper secretion of EspC. Clearly further work is required to dissect how EspA is targeted for secretion.
This study raises new questions about the ESAT-6/CFP-10 complex in vivo. It was previously shown that ESAT-6 and CFP-10 from M. tuberculosis interact to form a heterodimer, and require each other for stability in vivo (Guinn et al., 2004, Renshaw et al., 2005, Stanley et al., 2003). However, we found two instances in which ESAT-6 and CFP-10 were independent of each other in M. marinum. First, immunoprecipitation of FLAG-EspCMt from the M. marinum cytosol resulted in enrichment of ESAT-6Mm, but not CFP-10 Mm. Second, targeting EspC using the CFP-10 signal sequence led to the accumulation of CFP-10 Mm in the M. marinum cytosol but not ESAT-6 Mm. Consistent with this, certain mutations in the M. marinum ESX-1 system (Mh3876, Mh3878 and Mh3879) lead to the secretion of CFP-10 Mm but block ESAT-6 Mm secretion (Gao et al., 2004, McLaughlin et al., 2007). Together with the yeast two-hybrid analysis performed with proteins from M. tuberculosis, which demonstrated an interaction between ESAT-6 Mt and EspF Mt, these data argue that ESAT-6 is promiscuous, and interacts with multiple substrates in vivo. Indeed, ESAT-6 Mm also interacts directly with EspB Mm (Xu et al., 2007), further suggesting that the promiscuous nature of ESAT-6 allows for stability in the absence of interaction with CFP-10. Clearly this phenomenon suggests that the ESX-1 secretion system, and the interaction between substrates, is more complex than previously thought.
The question remains as to why the ESX-1 substrates are dependent upon each other for secretion. In most other bacterial protein secretion systems, each substrate is secreted independently; in the absence of one substrate, the remaining substrates are secreted. What benefit is gained by coupling the secretion of ESX-1 substrates? One possibility is that co-dependent secretion ensures the correct stoichiometry or timing of substrate secretion during infection. Since at least two of the substrates (EspA and EspC) are regulated at the level of transcription by EspR in M. tuberculosis (Raghavan et al., 2008), it could be that all of the substrates must be present in the cytosol for secretion to occur, allowing coordinated secretion of all substrates by controlling the expression of a few. The fact that two substrates interact with distinct AAA ATPases further indicates that coordination is important for ESX-1 protein secretion.
Finally, this work directly compared the molecular mechanisms of ESX-1 secretion in M. marinum and M. tuberculosis. While the majority of our findings were identical in both mycobacterial species, a curious difference is that EspF is secreted at much higher levels from M. marinum than its homolog in M. tuberculosis. This may have functional consequences that could reflect differences in the function of ESX-1 in these pathogenic mycobacteria. For example, M. marinum is known to escape the phagosome more robustly that M. tuberculosis during macrophage infection, and ESX-1 is thought to play a role in phagosomal escape (de Jonge et al., 2007, Stamm et al., 2003, van der Wel et al., 2007). It is possible that greater levels of EspFMm, or other ESX-1 substrates, could lead to greater amounts of phagosomal membrane damage.
Experimental Procedures
Strains and Plasmids
All strains, plasmids and primers are listed in Tables S1 and S2. M. marinum (M) was grown in Middlebrook 7H9 liquid broth plus glycerol and .05% Tween-80. M. tuberculosis (Erdman) was grown as previously described. Plasmid containing strains were grown in liquid media with 20ug/ml Kanamycin (Sigma). Constructs for M. tuberculosis FLAG-EspC expression were created using pJAM69 as a template for site-directed mutagenesis as in with primers in Table S2. Constructs were verified by DNA sequence analysis. Constructs for EspC-CFP-10 protein expression were constructed using fusion PCR with primers in Table S2.
Protein Preparation and Analysis
M. tuberculosis proteins were prepared for secretion analysis as in (Stanley et al., 2003). M. marinum proteins were prepared as M. tuberculosis proteins, with the following differences. 50ml cultures of M. marinum were grown to saturation in 7H9 liquid media with .05% Tween-80. The bacteria were diluted to .800 OD600 in 50 ml Sauton's liquid media with .05% Tween-80 and Kanamycin when appropriate. After growth for 48 hours at 30°C shaking at ∼200 rpm, the strains were harvested by centrifugation. For both M. tuberculosis and M. marinum, 10ug of cell lysates and supernatants, as determined by a MicroBCA assay (Promega) were separated using 4-20% Criterion polyacrylamide gels (BioRad). Proteins were visualized by immunoblot analysis as in (Champion et al., 2006, Stanley et al., 2003), Rv3881c (Arizona State University CIM Antibody Core), or Rv3616c (kind gift of S. Fortune).
Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was performed as in (Champion et al., 2006). Bait constructs were created by PCR amplification from either plasmids in Table S1 or M. tuberculosis genomic DNA using primers in Table S2, and introduced into pEG202 by yeast homologous recombination. Prey constructs were created by PCR amplification from M. tuberculosis genomic DNA using primers in Table S2, and introduced into pJSC401 by yeast homologous recombination. All constructs were confirmed by DNA sequence analysis and expression was confirmed by immunoblot analysis as in (Champion et al., 2006). Bait and prey constructs were introduced into EGY48 bearing the pSH18-34 lacZ+ reporter. Yeast strains were grown on agar containing X-gal (5-bromo-4-chloro-3-indolyl β-D-galactosidase). Liquid β-galactosidase assays were performed in triplicate as in (Champion et al., 2006). All error bars represent the standard deviation.
Co-immunoprecipitation Experiments
M. marinum strains were grown as described for secretion analysis above. Strains expressing FLAG-Mpt64, FLAG-EspC, or FLAG-EspC-CFP-10 25CT from M. tuberculosis were harvested by centrifugation, resuspended in lysis buffer (Stanley et al., 2003), and lysed. Lysates were clarified by centrifugation. Ezview FLAG affinity beads (Sigma) were blocked for 1 hour in 5% Milk in PBST (PBS + .1% tween-20), and washed 3 times in PBST. Lysates were incubated with the FLAG-beads at 4°C overnight. Samples were washed five times in PBST, and the co-immunoprecipitating proteins were removed from the beads by three elutions with FLAG peptide.
Mass Spectrometry
M. marinum were quantified using a Micro BCA assay (Promega) and frozen. Samples were prepared for digestion with trypsin as described elsewhere (Champion et al., 2006).
IP IDA Analysis
Peptides were analyzed by ESI (electrospray ionization) LC/MS/MS (liquid chromatography tandem mass spectrometer) on a QqQ-LIT (Triple quadrupole hybrid linear ion trap, 4000QTrap Applied Biosystems) in IDA (information dependent acquisition) mode essentially as described elsewhere (Champion et al., 2006, Jorge et al., 2005). Protein Pilot 2.0.1 was used for ID, The decoy strategy used for false-positive assessment (local FDR =0% for proteins used from IP) is described here (Shilov et al., 2007, Tang et al., 2005).
Quantitative MRM and Relative MRM Analysis
Following identification, peptides were selected for MRM (multiple reaction monitoring) as in (Anderson & Hunter, 2006)(Specific transitions are shown in the respective chromatogram and in the supplementary confirmatory spectra. 250ng of each sample digest was analyzed by ESI LC/MS/MS on a QqQ-LIT (4000QTrap Applied Biosystems). Operation was in MRM-EPI mode for confirmation and determination of peptide transitions. About 50-100 transitions were monitored for development, which included a Q0-trapped EPI (enhanced product ion MS/MS) spectrum from mass 225-1500 for the top two transitions per cycle, excluded for 120sec after acquisition. Verified peptides and transitions were then monitored in MRM only mode for improved quantitative accuracy in substrate detection and accumulation experiments (Fig 3). Multiquant 1.1 (Applied Biosystems) was used for peak integration and quantification, the results of the peak areas are in Figures S6 and S7. Data were corrected by normalizing to the peak areas of common peptides unrelated to ESX-1 similar to the method described in (Champion et al., 2006). For the fold-enrichment in the IP analysis, peak areas were near-noise in the mock-bait experiments, thus the reported enrichment likely underestimates the degree of specificity.
Acknowledgments
We thank members of the Cox laboratory for critical reading of this manuscript, and members of the Cox, A. Sil and E. Brown laboratories for helpful discussion. We thank S. Fortune for the EspA antibody, and E. Brown for strains. This investigation was supported by the NIH under Ruth L. Kirschstein Research Service Award A105155 to P.A.C. This work was supported by National Institutes of Health grants (AI63302 and AI51667). J.S.C. acknowledges the support of the Sandler Family Supporting Foundation and the W.M. Keck Foundation.
References
- Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. Type VII secretion--mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- Bahk YY, Kim SA, Kim JS, Euh HJ, Bai GH, Cho SN, Kim YS. Antigens secreted from Mycobacterium tuberculosis: identification by proteomics approach and test for diagnostic marker. Proteomics. 2004;4:3299–3307. doi: 10.1002/pmic.200400980. [DOI] [PubMed] [Google Scholar]
- Brodin P, Majlessi L, Marsollier L, de Jonge MI, Bottai D, Demangel C, Hinds J, Neyrolles O, Butcher PD, Leclerc C, Cole ST, Brosch R. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun. 2006;74:88–98. doi: 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burts ML, Williams WA, DeBord K, Missiakas DM. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci U S A. 2005;102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson F, Joshi SA, Rangell L, Brown EJ. Polar localization of virulence-related Esx-1 secretion in mycobacteria. PLoS Pathog. 2009;5:e1000285. doi: 10.1371/journal.ppat.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion PA, Stanley SA, Champion MM, Brown EJ, Cox JS. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science. 2006;313:1632–1636. doi: 10.1126/science.1131167. [DOI] [PubMed] [Google Scholar]
- Cox JS, Chen B, McNeil M, Jacobs WR., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- de Jonge MI, Pehau-Arnaudet G, Fretz MM, Romain F, Bottai D, Brodin P, Honore N, Marchal G, Jiskoot W, England P, Cole ST, Brosch R. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J Bacteriol. 2007;189:6028–6034. doi: 10.1128/JB.00469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune SM, Jaeger A, Sarracino DA, Chase MR, Sassetti CM, Sherman DR, Bloom BR, Rubin EJ. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci U S A. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao LY, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol. 2004;53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, Eisenberg D, Russell RG, Derrick SC, Collins FM, Morris SL, King CH, Jacobs WR., Jr The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ize B, Palmer T. Microbiology. Mycobacteria's export strategy. Science. 2006;313:1583–1584. doi: 10.1126/science.1132537. [DOI] [PubMed] [Google Scholar]
- Jorge I, Navarro RM, Lenz C, Ariza D, Porras C, Jorrin J. The holm oak leaf proteome: analytical and biological variability in the protein expression level assessed by 2-DE and protein identification tandem mass spectrometry de novo sequencing and sequence similarity searching. Proteomics. 2005;5:222–234. doi: 10.1002/pmic.200400893. [DOI] [PubMed] [Google Scholar]
- Luthra A, Mahmood A, Arora A, Ramachandran R. Characterization of Rv3868: An essential hypothetical protein of the ESX-1 secretion system in M. tuberculosis. J Biol Chem. 2008 doi: 10.1074/jbc.M807144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn JA, Cox JS. A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect Immun. 2007;75:2668–2678. doi: 10.1128/IAI.01872-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn JA, Cox JS. 2009 Submitted. [Google Scholar]
- MacGurn JA, Raghavan S, Stanley SA, Cox JS. A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Mol Microbiol. 2005;57:1653–1663. doi: 10.1111/j.1365-2958.2005.04800.x. [DOI] [PubMed] [Google Scholar]
- McLaughlin B, Chon JS, MacGurn JA, Carlsson F, Cheng TL, Cox JS, Brown EJ. A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog. 2007;3:e105. doi: 10.1371/journal.ppat.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan S, Manzanillo P, Chan K, Dovey C, Cox JS. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature. 2008;454:717–721. doi: 10.1038/nature07219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw PS, Lightbody KL, Veverka V, Muskett FW, Kelly G, Frenkiel TA, Gordon SV, Hewinson RG, Burke B, Norman J, Williamson RA, Carr MD. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. Embo J. 2005;24:2491–2498. doi: 10.1038/sj.emboj.7600732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw PS, Panagiotidou P, Whelan A, Gordon SV, Hewinson RG, Williamson RA, Carr MD. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J Biol Chem. 2002;277:21598–21603. doi: 10.1074/jbc.M201625200. [DOI] [PubMed] [Google Scholar]
- Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- Stamm LM, Brown EJ. Mycobacterium marinum: the generalization and specialization of a pathogenic mycobacterium. Microbes Infect. 2004;6:1418–1428. doi: 10.1016/j.micinf.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Stamm LM, Morisaki JH, Gao LY, Jeng RL, McDonald KL, Roth R, Takeshita S, Heuser J, Welch MD, Brown EJ. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J Exp Med. 2003;198:1361–1368. doi: 10.1084/jem.20031072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci U S A. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WH, Halpern BR, Shilov IV, Seymour SL, Keating SP, Loboda A, Patel AA, Schaeffer DA, Nuwaysir LM. Discovering known and unanticipated protein modifications using MS/MS database searching. Anal Chem. 2005;77:3931–3946. doi: 10.1021/ac0481046. [DOI] [PubMed] [Google Scholar]
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- Volkman HE, Clay H, Beery D, Chang JC, Sherman DR, Ramakrishnan L. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol. 2004;2:e367. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Laine O, Masciocchi M, Manoranjan J, Smith J, Du SJ, Edwards N, Zhu X, Fenselau C, Gao LY. A unique Mycobacterium ESX-1 protein co-secretes with CFP-10/ESAT-6 and is necessary for inhibiting phagosome maturation. Mol Microbiol. 2007;66:787–800. doi: 10.1111/j.1365-2958.2007.05959.x. [DOI] [PubMed] [Google Scholar]