Abstract
The spatial organization of chromosomes inside the cell nucleus is still poorly understood. This organization is guided by intra- and interchromosomal contacts and by interactions of specific chromosomal loci with relatively fixed nuclear “landmarks” such as the nuclear envelope and the nucleolus. New molecular genome-wide mapping techniques have begun to uncover both types of molecular interactions, providing insights into the fundamental principles of interphase chromosome folding.
Introduction: 3D organization of chromosomes
The three-dimensional architecture of interphase chromosomes is one of the most fascinating topological problems in biology. Decades of microscopy studies have revealed several important general principles that govern chromosome architecture1–3. First, interphase chromosomes each occupy their own territory in the nucleus, with only a limited degree of intermingling. Second, genomic loci tend to be non-randomly positioned within the nuclear space and relative to each other, strongly suggesting that chromosomes adopt a configuration that is at least partially reproducible. Finally, the degree of compaction of the chromatin fiber varies locally, and is often, but not always, inversely linked to transcriptional activity and gene density.
These important insights have been mostly obtained by fluorescence in situ hybridization (FISH) and in vivo tagging of selected genomic loci1–3. The power of these methods lies in their ability to visualize individual loci inside single cell nuclei by light microscopy. However, the resolution limits of light microscopy and the practical restriction that only a few loci can be visualized simultaneously, have hampered the construction of detailed models of chromosome architecture. Fortunately, over the past few years several new molecular techniques have been developed towards this goal. These techniques directly probe molecular interactions and thereby offer exciting new views beyond the resolution limits of microscopy. Moreover, by taking advantage of genome-wide detection methods such as high-density microarrays and massively parallel sequencing, comprehensive measurements of structural parameters of chromatin are now feasible for entire genomes in a single experiment.
In essence, the new techniques focus on the detection of two distinct classes of molecular contacts of the chromatin fiber (Figure 1). One set of techniques identifies physical interactions of genomic loci with relatively fixed nuclear structures (landmarks) such as the nuclear envelope or the nucleolus. This can yield important information on the position of genomic loci in nuclear space. A second set of techniques monitors physical associations between linearly distant sequences that come together by folding or bending of the chromatin fiber. Such associations may also occur between loci on different chromosomes. Knowledge of intra- and inter-chromosomal contacts provides insight into the local or global folding of chromosomes, and into the positioning of chromosomes relative to one another. Various chromatin-landmark interactions as well as chromatin-chromatin contacts have now been mapped systematically. Here, we highlight these new technological developments and the biological understanding that they have yielded so far.
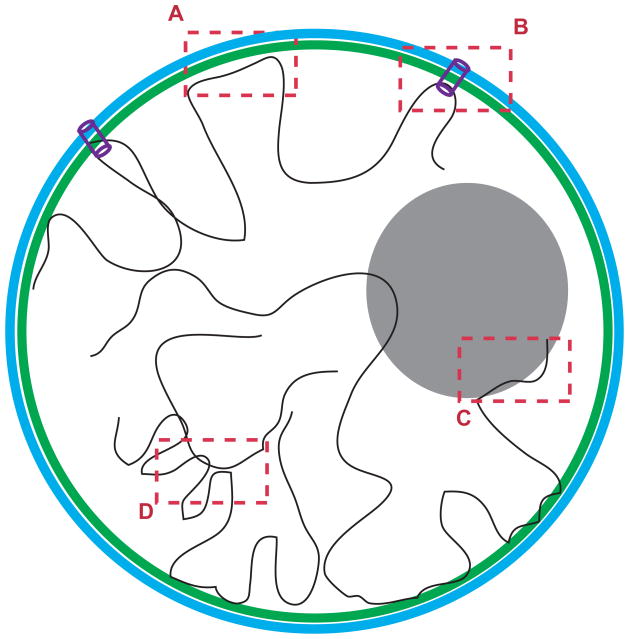
Figure 1.
Cartoon of nucleus depicting the spatial interactions that contribute to the overall architecture of interphase chromosomes. Table on the right summarizes the techniques that are currently used to map the respective interactions genome-wide.
Molecular mapping of genome interactions with nuclear landmarks
The nuclear envelope is the main fixed structure of the nucleus, and has for a long time been thought to provide anchoring sites for interphase chromosomes, and thus help to organize the genome inside the nucleus. The nuclear envelope consists of a double lipid membrane punctured by nuclear pore complexes (NPCs), which act as channels for nuclear import and export4. In most metazoan cells, the nucleoplasmic surface of the inner nuclear membrane is coated by a sheet-like protein structure termed the nuclear lamina (NL). Its major constituents are nuclear lamins, which form a dense network of polymer fibers5–7. Both the NL and NPCs have been proposed decades ago to provide anchoring sites for interphase chromosomes8,9. Indeed, many FISH microscopy studies have supported this model: some genomic loci are preferentially located in close proximity to the nuclear envelope, while other loci are typically found in the nuclear interior3,10,11. However, due to resolution limits it was generally not possible to tell whether these loci are in fact in molecular contact with the NL or the NPCs. Recent genome-wide mapping techniques have begun to provide more global insights into the molecular interactions of chromosomes with components of the nuclear envelope.
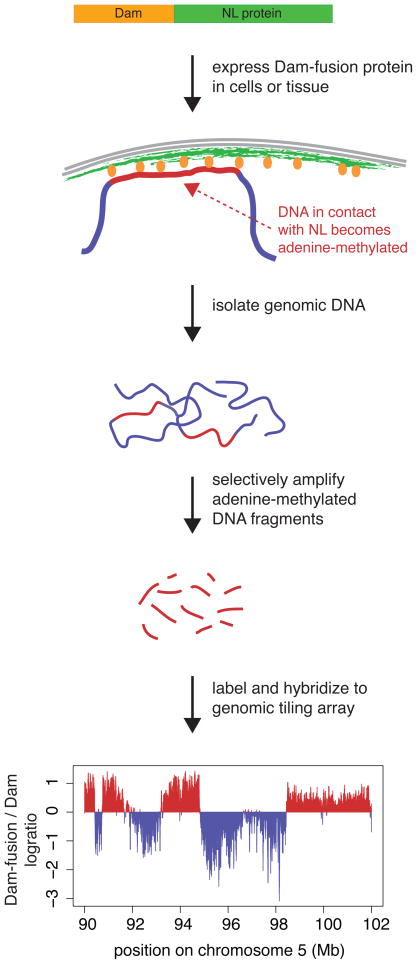
Interactions of the genome with the NL have been mapped by means of the DamID technology (Figure 2). Here, a protein of the NL (typically a lamin) is fused to DNA adenine methyltransferase (Dam) from E. coli. When expressed in cells, this chimaeric protein is incorporated into the NL. As a consequence, DNA that is in molecular contact with the NL in vivo becomes methylated by the tethered Dam. The resulting tags, which are unique because DNA adenine methylation does not occur endogenously in most eukaryotes, can be mapped using a microarray-based readout12,13. By this approach, NL interactions have been mapped in detail in Drosophila, mouse and human cells14–16. In all three species, interactions with the NL involve very large genomic domains, rather than focal sites. Mouse and human genomes have more than 1,000 lamina-associated domains (LADs) with a median size of ~0.5Mb. In human cells, several sequence elements demarcate the borders of many LADs, indicating that LAD organization is at least partially hard-coded in the genome15.
Figure 2.
Mapping of interactions of the genome with nuclear landmarks, here illustrated for the NL. See text for explanation. Adenine-methylated DNA is specifically amplified using a PCR-based protocol that employs restriction endonucleases that selectively digest DNA depending on the adenine-methylation state, as described elsewhere12,13.
Although LADs are relatively gene-poor, combined they nevertheless harbor thousands of genes. Interestingly, most of these genes are transcriptionally inactive15,16. This points to a repressive role of the NL in gene regulation. Consistent with this, deletion of the major Lamin in Drosophila causes upregulation of some NL-associated genes17. Moreover, artificial tethering to the NL can cause the downregulation of reporter and some endogenous genes, although this may depend on the reporter or its genomic integration site18–20. Furthermore, during differentiation hundreds of genes show altered interactions with NL. For many genes, detachment from the NL occurs concomitant with transcriptional activation; other detached genes initially remain silent but are more prone to activation in a second differentiation step, suggesting that interaction with the NL locks these genes in a stably repressed state16.
Interactions of the genome with NPCs have been studied by both DamID and Chromatin Immunoprecipitation (ChIP). The latter technique employs cross-linking of protein-DNA interactions with formaldehyde (and sometimes other cross-linking chemicals), followed by mechanical fragmentation of the DNA and subsequent immunoprecipitation using antibodies, in this case against NPC proteins (Nups). Genome-wide tiling microarrays were used to identify the immunoprecipitated DNA sequences. In yeast, Drosophila and human cells, hundreds of genes are associated with various Nups21–25. Surprisingly, detailed analyses in Drosophila established that a substantial proportion of these binding events occurs in the nuclear interior, involving freely diffusing Nups23,24. Although this sheds interesting light on an NPC-independent regulatory role of certain Nups, it also implies that most genome-wide maps of Nup interactions cannot be easily interpreted in terms of spatial organization of the genome, unless one conducts ChIP or DamID experiments with Nups that are only present in the NPC and not in the nucleoplasm. Fornerod and colleagues compared DamID maps obtained with engineered Nups that are either exclusively NPC-associated or mostly nucleoplasmic23. True NPC-associated loci thus identified are rather short sequences of <2kb that do not overlap with the larger NL-associated domains, in agreement with the spatial separation of NPCs and the NL as seen by high-resolution microscopy26. The NPC-interacting sites tend to be located in genes that are transcribed at moderate levels23.
Both ChIP and DamID have some limitations. In its current implementation DamID has a low temporal resolution13 and is therefore unable to capture the dynamics of NL and NPC interactions, for example during cell cycle progression. Development of a rapidly switchable Dam enzyme should overcome this limitation. ChIP has a better temporal resolution because formaldehyde crosslinking occurs within minutes. However, it has so far proven to be difficult to generate ChIP maps of NL components, for reasons that are not understood.
Another nuclear landmark that acts as an anchoring site for DNA is the nucleolus. Originally it was thought that this nuclear compartment harbors only the rRNA-encoding genes, which are transcribed by RNA Polymerase I. In order to find other sequences that may interact with nucleoli, a recent study used simple sedimentation fractionation to isolate nucleoli from human cells. The associated DNA was then characterized by massively parallel sequencing and microarray hybridizations27. Besides rRNA genes, many large genomic regions named nucleolus-associated domains (NADs) were identified. NADs are large genomic segments (median size 750 kb) that are highly enriched in centromeric satellite repeats and specific inactive gene clusters, which is consistent with the preferential localization of centromeres around nucleoli27,28. Interestingly, the 5S and tRNA genes, which are transcribed by RNA Polymerase III, also preferentially associate with the nucleolus, in agreement with earlier microscopy observations29. Other NAD-embedded genes tend to take part in specific biological processes, such as odor perception, tissue development and the immune system, suggesting that nucleolus interactions may help to coordinate the expression of specific gene sets. Together, these results demonstrate that distinct sets of chromosomal regions interact specifically with the NL, NPCs and nucleoli.
Mapping of long-range chromatin interactions
Microscopic analysis of interphase chromosomes suggests that they form rather amorphously shaped territories, with seemingly little internal organization. Yet, chromosomes must be folded in intricate patterns, e.g. to accommodate association of silent loci to the nuclear periphery, while simultaneously allowing expressed loci to congregate at sites of active transcription (transcription factories). Further, gene expression is modulated by cis-regulatory elements, such as enhancers, that often are located hundreds of kb from their target genes. Many enhancers are thought to physically associate with the promoters they regulate, resulting in formation of chromatin loops. A human chromosome contains hundreds to thousands of genes and each interacts, when active, with a set of regulatory elements. This array of long-range interactions will constrain the chromatin fiber into a highly complex three-dimensional network. The precise topology of these chromatin interaction networks, and how these networks are embedded inside the nucleus, is still largely unknown, but new molecular and genome-wide approaches are now starting to bring the folding principles of chromosomes into view.
The most widely used molecular method to probe the spatial folding of chromatin is chromosome conformation capture30 (3C). 3C allows the determination of the relative frequency with which pairs of genomic loci are in direct physical contact. Chromatin is cross-linked with formaldehyde after which DNA is digested and then re-ligated under dilute conditions that favor intra-molecular ligation of cross-linked fragments (Figure 3 and Table 1). This results in a genome-wide library of 3C ligation products, each of which is composed of a pair of restriction fragments that were in sufficiently close spatial proximity to become cross-linked. Interactions detected by 3C can be mediated by proteins that bridge the two loci, but can also reflect co-association of loci with larger protein complexes, or perhaps even larger sub-nuclear structures such as nucleoli and transcription factories. Combined, the 3C library reflects the population-averaged folding of the entire genome, at a resolution of several kb.
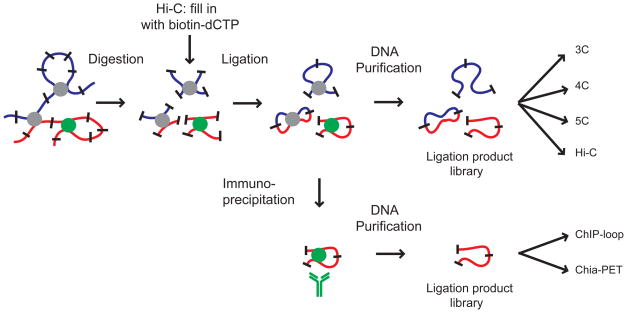
Figure 3.
Principles of the major 3C-based technologies. All protocols start with treatment of cells with formaldehyde (not shown), resulting into crosslinking of DNA segments that are in close proximity of one another. After digestion with one or more restriction enzymes linked restriction fragments are intramolecularly ligated. In the case of Hi-C the ends of the restriction fragments are first filled in with biotinylated dNTPs prior to ligation to facilitate purification of ligation junctions using streptavidin-coated beads. Either single or multiple ligation events are detected directly (3C, 4C, 5C and Hi-C), or first immunoprecipitation is used to enrich for DNA that is associated with a protein of interest (ChIP-loop, Chia-PET). See table 1 for an overview of the different detection strategies and their scope.
Table 1.
Scope and detection methods for techniques shown in Figure 3.
| Method | Scope | Detection | Example reference |
|---|---|---|---|
| 3C | interaction between two selected loci | quantitative PCR | 30 |
| 4C | genome-wide interactions of one selected locus | inverse PCR followed by detection with microarray or sequencing | 35 |
| 5C | all interactions among multiple selected loci | multiplex LMA followed by detection with microarray or sequencing | 37 |
| Hi-C | unbiased genome-wide interaction map | making of junctions with biotin, shearing, ligation junction purification, followed by sequencing. | 48 |
| ChIP-loop | interaction between two selected loci bound by a particular protein | quantitative PCR | 38 |
| Chia-PET | unbiased genome-wide interaction map of loci bound by a particular protein | insertion of linker into junction, followed by sequencing | 40 |
In conventional 3C the relative abundance of individual ligation products is determined using (semi-) quantitative PCR. Initial 3C analyses in yeast revealed long-range interactions between telomeres, and between centromeres located on different chromosomes, consistent with prior microscopic observations30. The first 3C studies that demonstrated long-range looping interactions between genes and their enhancers focused on the well-studied beta-globin locus31. Long-range interactions have now been identified in a large number of candidate loci, e.g. the Igf2 locus32, the TH2 cytokine locus33, the alpha-globin locus34, and in a variety of species, establishing that looping between genes and regulatory elements is a common mechanism for gene regulation. In many cases gene promoters interact with multiple elements, and these elements often also interact with each other leading to the formation of complex looped structures, sometimes referred to as chromatin hubs31.
To start to map chromatin interactions at a genome-wide scale several detection methods have been developed that allow more comprehensive interrogation of 3C libraries. 4C and 5C methods detect targeted subsets of 3C ligation products (Table 1)35–37. In 4C inverse PCR is used to amplify all fragments ligated to a single “anchor” fragment to obtain a genome-wide interaction profile for the anchor locus. 5C uses multiplexed ligation mediated amplification to amplify millions of pre-selected 3C ligation junctions in parallel, e.g. between a set of promoters and a set of enhancers. Chip-loop (also referred to as 6C) and CHIA-PET methods include a chromatin immunoprecipitation step to selectively identify 3C ligation products that are bound by a protein of interest, e.g. a transcription factor38–40. All these high-throughput methods use microarrays or deep sequencing to analyze the amplified ligation junctions. We note that careful experimental design of 3C-based methods is crucial to avoid artefacts and mis-interpretations, as discussed in detail elsewhere41,42.
Results obtained with these methods confirm that long-range interactions are widespread and also identified several new phenomena. First, long-range interactions can occur over very large genomic distances, up to tens of Mb, suggesting that chromosomes are extensively folded back upon themselves. Second, interactions not only occur between specific short functional elements, such as enhancers and promoters, but also occur over larger chromosomal domains. Groups of genes can be found to display elevated levels of interactions with each other all along their lengths, suggesting these genes are in general in close spatial proximity, perhaps as a result of association with the same sub-nuclear structure such as the nuclear envelope, or a transcription factory. Third, interactions occur not only along chromosomes, but also between them. For instance, the X-chromosome inactivation center (Xic) of one X-chromosome transiently interacts with the Xic of the other X-chromosome during the process of establishing X chromosome inactivation43–45. Another example is the trans-association of imprinted genes, which may contribute to their regulation46.
Recently, it has become possible to determine chromatin interactions in a truly unbiased and genome-wide manner, i.e. without the need to limit the analysis to one or group of selected anchors, or to sites bound by a specific protein47–49. The Hi-C technology is again based on 3C but includes a step prior to ligation in which the staggered ends of the restriction fragments are filled in with biotinylated nucleotides48. As a result, ligation junctions are marked with biotin, allowing their subsequent purification after DNA shearing using streptavidin-coated beads. Ligation junctions are then analyzed by paired-end deep sequencing to identify the interacting loci. Hi-C data can be used to study the overall folding of genomes. Presently, for large genomes such as those of human and mouse, Hi-C analysis will produce an interaction map with a resolution of around 0.1 to 1 Mb. This resolution is only limited by the number of sequence reads that current platforms can produce, and expected future increases in throughput and decreases in cost will allow the generation of interaction maps with significantly higher resolution.
The first Hi-C maps for the human genome confirm several features of nuclear organization that were also detected by microscopy and have also already uncovered several new interesting aspects of chromosome architecture and nuclear organization48. First, chromosomes extensively interact with each other, with some chromosome pairs showing preferred associations. Thus, chromosomes appear to occupy preferred locations with respect to each other. Second, chromosomes are spatially compartmentalized to form two types of nuclear neighborhood, referred to as A- and B-type compartments. The A-type compartments contain active loci (as indicated by gene expression level and the presence of chromatin features associated with active chromatin such as DNAseI hypersensitive sites) whereas B-type compartments are composed of inactive chromatin. Spatial separation of active and inactive domains is consistent with earlier observations obtained for individual loci by microscopy50 and by 4C35. Third, Hi-C data, as any 3C-based data, can be modeled using polymer models to uncover folding states of chromatin [e.g.30,51]. Computational modeling of Hi-C data revealed that at the length scale of up to several Mb, human chromatin may be folded in a polymer state that is referred to as a fractal globule48. This is a densely packed state that is characterized by the absence of knots and entanglements. This unique conformation allows easy folding and unfolding of sections of chromosomes, which may be relevant for activating and repressing genes.
A variant of Hi-C was also described that marks ligation junction with a biotinylated oligonucleotide to facilitate their purification49. This method was applied to analysis of the 3D structure of the yeast genome. The data confirmed all the known hallmarks of nuclear organization, including clustering of centromeres and telomeres52. Further, it was found that inter-chromosomal interactions occur between tRNA genes, and between early firing origins of replication.
Combined, 3C-based studies point to a bewildering complexity in long-range communication between a variety of genomic elements across chromosomes and the genome. There is still room for further technological improvements. For instance local, there may be some biases in the interaction maps caused by differences in cross-linkability between chromatin types, and differential access of sequences to the enzymes used in the protocol. Refinement of the technology may overcome some of these potential limitations. Clearly, we are only just starting to explore the spatial folding of chromosomes, and the new genome-wide 3C methods will likely provide a wealth of new insights.
Towards an integrated view of chromosome architecture
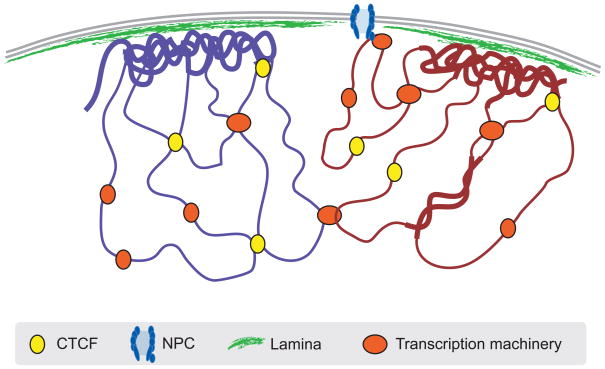
With several new genome-wide detection methods in place, an integrated picture of chromosome architecture seems within reach. Unfortunately, the maps produced so far are derived from diverse cell lines or from different species, so direct comparisons are not yet possible. Nevertheless, some conclusions and reasonable speculations can be derived. At least in Drosophila, NPCs and the NL clearly interact with different chromosomal regions, and thus provide two distinct sets of anchoring points. In human cells, LADs and NADs both tend to include centromeric regions15,27, suggesting that centromeres in each nucleus are distributed between the NL and nucleoli. LADs and B-type domains show some striking similarities (size range and an overall lack of gene activity), suggesting that they must overlap at least in part. If true, this has the interesting implication that LADs may interact or intermingle with other LADs and form aggregates of compacted chromatin near the NL (Figure 4). This model would explain the substantial amounts of heterochromatin in close contact with the NL, as observed by microscopy.
Figure 4.
Speculative cartoon model of chromatin organization. LADs may consist of relatively condensed chromatin (thick lines) and aggregate at the NL. Other repressed regions may interact with each other in the nuclear interior, as do active regions. Complexes formed by components of the transcription machinery (“transcription factories”) and CTCF may tether active regions together. Parts of only two chromosomes are depicted, each drawn in a different color for clarity. Most interactions occur within chromosomes, and relatively few between chromosomes.
Evidence is accumulating that some epigenetic marks are linked to nuclear organization. The timing of DNA replication along the genome shows a block-like structure of alternating large early and late-replicating segments53,54. A genome-wide comparison indicates that late-replicating domains roughly correspond to LADs16, consistent with the enrichment of late-replicating sequences at the nuclear periphery53,55. However, LADs and late-replicating domains do not overlap perfectly16, indicating that they are related but not identical. Late-replicating domains also show striking similarities to the B-type domains as identified by Hi-C56. Furthermore, the histone modification H3K9me2 exhibits a domain pattern with strong similarities to LADs15,57 and to segments of late-replicating DNA56,58. Taken together, LADs, late-replicating DNA, H3K9me2 domains, and B-type domains all appear to be closely related, but more systematic comparisons are needed in order to understand their precise relationships.
The active compartments of the genome, e.g. the A-domains identified by Hi-C, may also have cytological correlates. Expressed genes have been observed to cluster at sub-nuclear foci enriched in transcription machineries, which are sometimes referred to as transcription factories (Figure 4). In addition, these domains appear to correlate with open chromatin that is replicated early in S-phase56,59.
Another theme that is emerging is the critical role of the CTCF protein, which is a multi-functional DNA-binding protein60. Extensive 3C-based evidence indicates that CTCF can mediate long-range interactions, both in cis32,60–62 and in trans45 (Figure 4). In addition, borders of human LADs are frequently demarcated by CTCF binding sites15, suggesting that CTCF helps to control LAD organization. How these observations are linked remains to be elucidated, but it is clear that CTCF is an important factor in the regulation of chromosome topology.
Stochastic nature of interactions
So far, all genome-wide datasets that describe chromosome architecture are derived from large pools of cells. Yet microscopy studies have shown that the location of individual genomic loci is highly variable from cell to cell, even in clonal cell lines. This variability has two biological sources. First, within each nucleus, chromatin is mobile to a certain degree63,64. Second, in a newly formed nucleus after mitosis, the relative positioning of chromosomes may be substantially driven by stochastic processes65.
It is difficult to calibrate the genome-wide interaction datasets in terms of absolute contact frequencies. Currently this can only be approximated by FISH, which is hampered by insufficient resolution and the possible disruption of chromosome folding by the harsh denaturation conditions required for FISH. It is likely however, that most long-range interactions between chromosomal loci, as detected by 3C based methods, occur in less than 10–20% of cells at a given time point35,66–68. Contacts of individual LADs and NADs with their respective landmarks may occur in 10–50% of cells14,27. We emphasize that these are only rough estimates, subject to arbitrary definitions of contacts as used in the respective studies.
The stochastic nature of chromosome architecture raises important questions related to gene regulation. For example, if LADs contact the NL only transiently, or only in a subpopulation of cells, then how can such interactions contribute to robust gene repression? One possibility is that a transient contact with the NL causes a long-lasting change in the chromatin, for example by a histone-modifying enzyme that is embedded in the NL. Except for enhancer-promoter interactions, the functional relevance of stochastic, relatively low-frequency contacts between linearly distant genes (“gene kissing”) is largely unclear. In some cases these contacts have been observed to correlate with gene expression66, but in order to establish causal relationships it will be necessary to experimentally modulate these contacts, e.g. specifically disrupting them, and assessing the impact of expression and regulation of the genes.
Future outlook
A remarkable recurrent theme emerging from the studies so far is that metazoan genomes are linearly segmented into large multi-gene domains, which have specific interactions with nuclear landmarks and each other. This raises the interesting possibility that chromosomal aberrations such as translocations and inversions, which are found in a variety of human genetic disorders69 and in many types of cancer70, can disrupt the spatial organization of the affected chromosomes and perhaps thereby alter gene expression71. Interestingly, it was recently shown that this logic can also be turned around: 3C-derived techniques can identify chromosomal aberrations based on altered spatial relationships between loci72. Inversely, the spatial organization of the genome may also impact the spectrum of any translocations that could occur in that cell. Loci that are spatially proximal may more frequently engage in translocation than more distant ones73–75.
Another class of human disorders that may be of interest in the context of chromosome architecture are so-called laminopathies. These disorders are caused by congenital defects in proteins of the NL. For example, mutations in Lamin A/C cause a remarkably diverse spectrum of disorders including progeria, muscular dystrophy, and cardiomyopathy76. It is possible that some of these disorders involve changes in chromosome architecture due to altered interactions with the NL. Indeed, in cells from patients suffering from Hutchinson Gilford Progeria Syndrome (HPGS), which show abnormal accumulation of Lamin A at the NL, changes have been observed in the morphology and localization of heterochromatin77,78, although this may be an indirect effect of misregulation of certain chromatin proteins79. Mapping of genome - NL interactions and chromosome conformation in cells from laminopathy patients may provide important insights into the etiology of this class of disorders.
The initial results of various new genome-wide approaches have already uncovered some important principles of chromosome architecture. Higher resolution views, particularly for Hi-C, will become available when sequencing throughput continues to ramp up. Yet the probabilistic and dynamic nature of chromatin organization poses practical and conceptual challenges. It would be extremely helpful if techniques for the molecular mapping of chromatin architecture could be scaled down to single cells, as this would directly capture cell-to-cell variation. While this will be technically demanding, the rapid advances in high-throughput single-molecule DNA sequencing technologies combines with further development of interaction detection methods may offer new opportunities towards this goal.
Supplementary Material
Acknowledgments
We thank members of the van Steensel and Dekker labs and Marian Walhout for helpful suggestions. Supported by the Netherlands Genomics Initiative and an NWO-ALW VICI grant to B.v.S., and a grant from the National Institutes of Health (HG003143) and a W.M. Keck Foundation Distinguished Young Scholar Award to J.D.
Contributor Information
Bas van Steensel, Email: b.v.steensel@nki.nl.
Job Dekker, Email: job.dekker@umassmed.edu.
References
- 1.Pombo A, Branco MR. Functional organisation of the genome during interphase. Curr Opin Genet Dev. 2007;17:451–5. doi: 10.1016/j.gde.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–9. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hetzer MW, Wente SR. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell. 2009;17:606–16. doi: 10.1016/j.devcel.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- 6.Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu Rev Biochem. 2004;73:749–89. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 7.Prokocimer M, et al. Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franke WW. Structure, biochemistry, and functions of the nuclear envelope. Int Rev Cytol Suppl. 1974;4:71–236. [PubMed] [Google Scholar]
- 9.Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci U S A. 1985;82:8527–9. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fedorova E, Zink D. Nuclear genome organization: common themes and individual patterns. Curr Opin Genet Dev. 2009;19:166–71. doi: 10.1016/j.gde.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Greil F, Moorman C, van Steensel B. DamID: mapping of in vivo protein-genome interactions using tethered DNA adenine methyltransferase. Methods Enzymol. 2006;410:342–59. doi: 10.1016/S0076-6879(06)10016-6. [DOI] [PubMed] [Google Scholar]
- 13.Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2:1467–78. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 14.Pickersgill H, et al. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–14. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 15.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 16.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome - nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–13. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shevelyov YY, et al. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc Natl Acad Sci U S A. 2009;106:3282–7. doi: 10.1073/pnas.0811933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–7. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 19.Finlan LE, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casolari JM, et al. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–39. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 22.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–39. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–71. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Capelson M, et al. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–83. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaquerizas JM, et al. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schermelleh L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–6. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemeth A, et al. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stahl A, Hartung M, Vagner-Capodano AM, Fouet C. Chromosomal constitution of nucleolus-associated chromatin in man. Hum Genet. 1976;35:27–34. doi: 10.1007/BF00295616. [DOI] [PubMed] [Google Scholar]
- 29.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 31.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–65. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 32.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–93. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 33.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–27. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 34.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. Embo J. 2007;26:2041–51. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 37.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 39.Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 2008;18:1171–9. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 42.Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 43.Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–52. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 44.Bacher CP, et al. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–9. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 45.Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–6. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- 46.Sandhu KS, et al. Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development. Genes Dev. 2009;23:2598–603. doi: 10.1101/gad.552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodley CD, Bertels F, Jones B, O’Sullivan JM. Global identification of yeast chromosome interactions using Genome conformation capture. Fungal Genet Biol. 2009;46:879–86. doi: 10.1016/j.fgb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 48.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–7. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shopland LS, et al. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol. 2006;174:27–38. doi: 10.1083/jcb.200603083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dekker J. Mapping in vivo chromatin interactions in yeast suggests an extended chromatin fiber with regional variation in compaction. J Biol Chem. 2008;283:34532–40. doi: 10.1074/jbc.M806479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taddei A, Schober H, Gasser SM. The Budding Yeast Nucleus. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hiratani I, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwaiger M, et al. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev. 2009;23:589–601. doi: 10.1101/gad.511809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J Cell Biol. 1992;116:1095–110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryba T, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010 doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–50. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yokochi T, et al. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci U S A. 2009;106:19363–8. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilbert N, et al. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–66. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 60.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–54. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–98. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soutoglou E, Misteli T. Mobility and immobility of chromatin in transcription and genome stability. Curr Opin Genet Dev. 2007;17:435–42. doi: 10.1016/j.gde.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chuang CH, Belmont AS. Moving chromatin within the interphase nucleus-controlled transitions? Semin Cell Dev Biol. 2007;18:698–706. doi: 10.1016/j.semcdb.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bolzer A, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osborne CS, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–71. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 67.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 68.Miele A, Bystricky K, Dekker J. Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet. 2009;5:e1000478. doi: 10.1371/journal.pgen.1000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet. 2004;13(Spec No 1):R57–64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- 70.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 71.Harewood L, et al. The effect of translocation-induced nuclear reorganization on gene expression. Genome Res. 2010;20:554–64. doi: 10.1101/gr.103622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simonis M, et al. High-resolution identification of balanced and complex chromosomal rearrangements by 4C technology. Nat Methods. 2009;6:837–42. doi: 10.1038/nmeth.1391. [DOI] [PubMed] [Google Scholar]
- 73.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–91. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- 74.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–83. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mani RS, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Worman HJ, Fong LG, Muchir A, Young SG. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Invest. 2009;119:1825–36. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goldman RD, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2004;101:8963–8. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taimen P, et al. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0911895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pegoraro G, et al. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–7. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.