Abstract
Background
No studies have evaluated roles of insulin-like growth factor binding protein 5 (IGFBP-5) polymorphisms in risk of squamous cell carcinoma of the head and neck (SCCHN).
Methods
A hospital-based study of 1082 SCCHN patients and 1120 cancer-free controls was performed to investigate associations between two functional polymorphisms -1195T>C and -709G>C in the IGFBP5 promoter region and SCCHN risk.
Results
We demonstrated that the transcription factor AP-1 differentially bound to T or C variants at -1195 in the promoter to regulate the IGFBP5 promoter activity and that the C variant genotypes were associated with deferential risk of late-stage SCCHN, compared with the TT genotype, particularly for HPV-unrelated sites (adjusted OR, 2.21; 95% CI, 1.19-4.11 for CC vs. TT).
Conclusion
The IGFBP5 -1195T>C polymorphism is functional and may potentially be a biomarker for susceptibility to late-stage SCCHN.
Keywords: IGFBP5, head neck cancer, TNM stage, polymorphism, association
Introduction
Squamous cell carcinoma of the head and neck (SCCHN) includes cancers of the oral cavity, oropharynx, hypopharynx and larynx (1). In 2009, it was estimated that 48,010 new SCCHN cases would be diagnosed and 11,260 deaths would occur (2). Although the cause of SCCHN is multifactorial, tobacco use and alcohol consumption have been well recognized as the major risk factors for SCCHN, and the population attributable risk of tobacco or alcohol use has been recently reported to be 72% for SCCHN (3). However, many SCCHN patients had never smoked or drunk, suggesting that other factors may play important roles in the etiology of SCCHN (4). Indeed, genetic factors include polymorphisms in genes involved in carcinogen metabolism, cell cycle regulation, DNA repair, and apoptosis have been reported to be associated with SCCHN risk (5, 6).
The insulin-like growth factor (IGF) signaling pathway plays a critical role in cell growth, differentiation and apoptosis (7, 8). The IGF axis includes two IGFs (IGF-I and IGF-II), two IGF receptors (IGF-IR and IGF-IIR), six well-characterized insulin-like growth factor binding proteins (IGFBPs), and several IGFBP proteases (9). The six conventional IGFBPs (IGFBP-1 to -6) are all cysteine-rich proteins and all bind to IGFs with a high affinity, either enhancing or inhibiting activation of the IGF signaling pathway, depending on different cell types (9, 10). Moreover, IGFBPs could also interact with other proteins, such as ECM (extracellular matrix) and GAGs (glycosaminoglycans), and functionally independent of the IGF signaling (11).
IGFBP-5 is one of the most conserved IGFBPs across the vertebrates, and human IGFBP-5 is a 29 kDa protein encoded by the IGFBP5 gene, which is located on chromosome 2q33-q36 and composed of four exons. Like other IGFBPs, IGFBP-5 binds to IGFs and either positively or negatively regulates the IGF signaling and by the IGF-independent pathway, through the interaction with ECM proteins (12, 13). Several studies have suggested that IGFBP-5 expression level may be associated with tumorigenesis and/or metastasis of several cancer types, including cancers of the breasts, prostate and ovaries (12). A recently published study showed that the food ingredient curcumin could up-regulate IGFBP-5 expression and suppress the xenograft tumorigenesis of oral cancer cells in mice(14). In addition, two other pathological studies suggested that down-regulation of IGFBP-5 is associated with the suppression of SCCHN tumorigenesis (15, 16). Taken together, these studies suggested that the IGFBP-5 may play a tumor suppressor role in the etiology of SCCHN.
Although the functions of IGFBP5 have been well studied, few studies have assessed the associations between genetic polymorphisms of IGFBP5 and cancer risk. To date, no published study has evaluated the associations between IGFBP5 polymorphisms and SCCHN risk. In this study, we hypothesized that polymorphisms located in the IGFBP5 promoter region are associated with risk of SCCHN. To test this hypothesis, we genotyped the only two known common, potentially functional polymorphisms in the IGFBP5 promoter regions [-1195T>C (rs1978346), and -709G>C (rs4480966)] and assessed their associations with SCCHN risk in 1082 SCCHN patients and 1120 cancer-free controls of a non-Hispanic white population. We further performed laboratory experiments to investigate functional relevance of any of the SNPs that may be associated with SCCHN risk.
Materials and Methods
Study Subjects
The study population consisted of 1111 patients with newly diagnosed cancers of SCCHN at The University of Texas M. D. Anderson Cancer Center between October 1999 and October 2007 and 1130 cancer-free controls recruited during the same period. Patients with second primary tumors, primary tumors of the skin, nasopharynx, sinonasal tract, and/or any histopathologic diagnoses other than squamous cell carcinoma were excluded. The cancer-free controls were recruited from among the cancer-free visitors who came along with patients to clinics of M. D. Anderson Cancer Center and were not genetically related to the patients. SCCHN patients and the cancer-free controls were non-Hispanic whites who were frequency matched by age (± 5 years) and sex, and their response rates were approximately 93% and 85%, respectively. The stages of SCCHN were classified according to the tumor-node-metastasis (TNM) staging system (17).
Having signed a written informed consent during the interview, all subjects provided information about age, sex, ethnicity, history of tobacco and alcohol use, family income, and the highest level of education. A 30-mL venous blood sample was collected from each subject and used for biomarker assays, including DNA extraction for genotyping. The research protocol was approved by the M.D. Anderson Institutional Review Board.
Genotyping of IGFBP5
The IGFBP5 gene reportedly has 268 variants (http://www.ncbi.nlm.nih.gov/projects/SNP/). In this study, we focused on genetic variants in the promoter region of IGFBP5 so that additional functional experiments could be performed, if necessary. In the IGFBP5 promoter region, only two common (MAF- minor allele frequency ≥ 0.05 in Caucasians), potentially functional SNPs [-1195T>C (rs1978346) and -709G>C (rs4480966)] were identified. We also searched the coding region of IGFBP5 but did not find any common SNPs. Therefore, the two promoter SNPs were genotyped by the restriction fragment length polymorphism (RFLP) method with the following primers: -1195T>C (forward) 5′-GACTTGCCTGTGAGGCTCATGTC -3′ and (reverse) 5′-GTCCTGACCAGGAACCGAATCA -3′; and -709G>C (forward) 5′-GGTTCTCTCTCTCTTTCTCTCTGTCT-3′ and (reverse) 5′-GTGTCCATCTCTGCAACTCACT-3′. The PCR reactions included 35 cycles with an annealing temperature of 62°C for both SNPs. When the PCR products were digested with BsmaI (New England Biolabs, Beverly, MA) at 55°C overnight, the 138-bp PCR products with the -1195T allele exhibited 118- and 20-bp bands, while the C allele remained uncut; and the 120-bp PCR products with the-709C allele exhibited 98- and 22-bp bands, while the G allele remained uncut. For both SNPs, approximately 10% of the samples were randomly selected and repeated with RFLP, and the error rate was less than 1%.
Construction of reporter plasmids
The 1337-bp IGFBP5 promoters (from -1280 to +56 relative to the translation start site) were cloned by PCR with the primers of (forward) 5′- AAGGTACCAAGGTGTAACAGCCCACACC -3′ and (reverse) 5′-AAGCTAGCCTGCTTTGCAGCTCTTTCCT-3′, in which the underlined sequences represented the KpnI and NheI restriction enzyme sites in forward and reverse primers, respectively. The difference between T and C alleles in the promoters was obtained by using the DNA template from subjects homozygous (TT and CC) for the -1195 variant. The PCR products were cloned into the basic-pGL3 firefly luciferase vector (Promega, Madison, WI) at the KpnI and NheI restriction sites. The -1195T and -1195C constructs were sequenced to confirm the orientation and integrity of each insert.
Transient transfection and luciferase reporter gene assay
The human colon cancer cell line HCT116 (a gift from Dr. Bert Vogelstein of John Hopkins University School of Medicine) and human head and neck carcinoma cell line MDA-1386Ln (obtained from Dr. Jeffrey N. Myers at M. D. Anderson Cancer Center) were cultured in 1× Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (Sigma-Aldrich, MO), and UMSCC-17B (obtained from Dr. Ruben Lotan at M. D. Anderson Cancer Center) was cultured in DMEM/F12 medium with 10% fetal bovine serum at 37 °C in 5% CO2. The cultured cells were transiently transfected with 1.0 μg of -1195T or -1195C reporter constructs by FuGENE 6 (Roche Applied Science, IN) in 24-well plates. The p-TK renilla luciferase (pRL-TK) (Promega) was co-transfected as an internal control. The Luciferase activities of both firefly and renilla luciferase were quantified by a Dual-Luciferase Reporter Assay System (Promega) 48 hr after transfection, and the relative luciferase activity was obtained as the ratio of firefly to renilla luciferase activities, according to the manufacturer's instructions (TD-20/20 Luminometer, Promega). Fold induction of the relative luciferase activity was measured by setting reporter gene activity of the construct with the T allele as 1. The experiments were performed in triplicate and repeated at least once, the average means and standard errors of fold induction were calculated and tested with Student t test in Stata 10.0 (StataCorp LP, College Station, TX).
Nuclear Extract Preparation and Electrophoretic Mobility Shift Assay (EMSA)
The nuclear extracts from human head and neck carcinoma cell line UMSCC-17B were prepared according to the method of Andrews and Faller (18). Complementary single-stranded oligonucleotides (5′-TGTGAGGCTCATGGCTCACTCAGGTCAAT-3′ for the T allele and 5′-TGTGAGGCTCATGGCCCACTCAGGTCAAT-3′ for the C allele) were biotin-labeled using the 3′-end biotin labeling kit (Thermo Scientific, Rockford, IL) and were re-annealed to the double strand. DNA binding assays for AP-1 proteins were performed by incubation of 10 μg of nuclear extracts with 3′ biotin–labeled double strand oligonucleotides at room temperature for 30 min using the LightShift Chemiluminescent EMSA kit (Thermo Scientific, Rockford, IL). The DNA-protein complex was separated on 6% polyacrylamide gel, and the products were detected by Stabilized Streptavidin-Horseradish Peroxidase Conjugate (Thermo Scientific). The competition was performed with a 50-fold excess of unlabeled oligonucleotides. Supershift experiments were performed with anti-cfos antibody or nonspecific rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA). The gel density assessment was performed with ImageJ software from NIH (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/). This experiment was repeated once to ensure the consistent results.
Chromatin immunoprecipitation assay (ChIP)
The ChIP assay was performed with the ChampionChIP™ one day kit (SABiosciences, Frederick, MD), according to the manufacturer protocol. Briefly, UMSCC-17B cells were cross-linked with 1% formaldehyde at 37°C for 10 min. Then the cells were lysed and sonicated to generate DNA fragments. After a pre-clearing by protein A beads, the lysates were incubated with antibodies against c-fos or rabbit IgG (Santa Cruz Biotechnology) for immunoprecipitation at 4°C for overnight with rotation. Immune complexes were precipitated with 60 μl of protein A beads, washed and eluted with elution buffer. The eluted DNA was used for PCR verification using the primers for the genotyping of -1195T>C.
Statistical Analysis
The differences in the distributions of categorical variables, including demographic characteristics, tobacco smoking and alcohol use, and genotypes of the IGFBP5 polymorphisms between cases and controls were evaluated by Chi-square tests. Crude or adjusted (for age, sex, smoking and drinking status) odds ratios (ORs) and 95% confidence intervals (CIs) were obtained from unconditional univariate and multivariable logistic regression analyses to evaluate associations between IGFBP5 polymorphisms and SCCHN risk in the case-control analysis and between IGFBP5 polymorphisms and TNM stages in the case-only analysis. Associations between IGFBP5 genotypes and SCCHN risk were also stratified by age, sex, smoking and drinking status, education level, income level, tumor site, and TNM stage. Homogeneity of ORs between different strata was evaluated using a Cochrane-Mantel-Haenszel test (19), followed by analyses of gene-environment interactions, which were evaluated by the P value for the product of two variables of interest in multivariable logistic regression model with adjustment for age, sex, smoking and drinking status. All tests were two-sided, and all P values were further evaluated with Bonferroni correction for multiple testing, i.e., a more stringent P value cut-off point by considering the tests for two SNPs simultaneously (P≤0.05/2 or 0.025) was considered as significant after correction. All data were analyzed with SAS statistical software program (SAS/STAT version 9.1.3; SAS Institute Inc., Cary, NC).
Results
Characteristics of the study population
In this study, 1111 SCCHN cases and 1130 controls of non-Hispanic whites were recruited. Among these subjects, 28 cases and nine controls failed in the genotyping assays, and one case and one control showed inconsistent results from repeated genotyping. These samples were all excluded from the final analysis that included 1082 SCCHN patients and 1120 controls.
As shown in Table 1, the cases and controls were adequately matched by age and sex (P = 0.466 and 0.481, respectively) with a slightly higher mean age for patients (57.1±11.1 years with a range of 18-90) than for controls (56.7±11.0 years with a range of 20-87). There were about two thirds of men in both cases (75.3%) and controls (76.6%). Compared with the controls, the cases were more likely to be smokers and drinkers (P < 0.001 for both). Smoking and drinking were further adjusted for in subsequent multivariate logistic regression analyses. As expected, a vast majority (91%) of oropharyngeal cancer patients presented with late stage diseases (stage III or IV), while only 57% of SCCHN occurring at HPV-unrelated sites (i.e., the oral cavity and larynx/hypopharynx) presented with stage III or IV diseases (data not shown). Control subjects attained a higher level of education and had higher income than the cases (P < 0.001 for both variables). In addition to segregating by site (oropharynx vs. HPV-unrelated sites), education and income levels along with age, sex, and smoking/drinking were further adjusted for in the subsequent multivariable logistic regression analyses.
Table 1.
Demographic characteristics of SCCHN cases and cancer-free controls
| Variables | Cases No. (%) |
Controls No. (%) |
P value* |
|---|---|---|---|
| All subjects | 1082 (100.0) | 1120 (100.0) | |
| Age† group (y) | |||
| Range | 18-90 | 20-87 | 0.466 |
| Mean | 57.1 ±11.1 | 56.7±11.0 | |
| ≤ 57 | 580 (53.6) | 583 (52.1) | |
| > 57 | 502 (46.4) | 537 (47.9) | |
| Sex | 0.481 | ||
| Female | 267 (24.7) | 262 (23.4) | |
| Males | 815 (75.3) | 858 (76.6) | |
| Smoking Status | <0.001 | ||
| Never | 305 (28.2) | 545 (48.7) | |
| Former | 369 (34.1) | 412 (36.8) | |
| Current | 408 (37.7) | 163 (14.6) | |
| Alcohol Status | <0.001 | ||
| Never | 299 (27.6) | 489 (43.7) | |
| Former | 234 (21.6) | 180 (16.1) | |
| Current | 549 (50.7) | 451 (40.3) | |
| Education Level§ | <0.001 | ||
| ≤ High School | 379 (35.3) | 299 (27.9) | |
| Certificate/Associate degree/some college | 321 (29.9) | 285 (26.6) | |
| Bachelor/Advanced degree | 373 (34.8) | 486 (45.4) | |
| Income Level§ | <0.001 | ||
| <$50,000 | 399 (38.4) | 328 (31.5) | |
| $50,000-$74,999 | 230 (22.1) | 212 (20.4) | |
| $75,000-$99,999 | 148 (14.2) | 174 (16.7) | |
| >$100,000 | 262 (25.2) | 327 (31.4) | |
| Tumor Sites | |||
| Oropharynx | 551 (50.9) | ||
| Oral Cavity | 319 (29.5) | ||
| Larynx | 170 (15.7) | ||
| Hypopharynx | 42 (3.9) | ||
| TNM Stages‡ | |||
| I | 117 (10.8) | ||
| II | 156 (14.4) | ||
| III | 182 (16.8) | ||
| IV | 626 (57.9) |
Two-sided χ2 test.
The median age of the controls.
There was one patient had missing data for TNM stages.
The numbers for education and income level may not add up to the total number due to the missing information from some subjects.
Association between IGFBP5 polymorphisms and SCCHN risk
The locations of two IGFBP5 promoter SNPs (-1195T>C and -709G>C) and the RFLP genotyping results are presented in Figure 1A. The distribution of three genotypes of -1195T>C in control subjects agreed with the Hardy-Weinberg equilibrium (HWE) (P = 0.604), but the genotype distribution of -709G>C in controls appeared to be departed from HWE (P = 0.013), which is most likely due to the extremely low frequency of the CC genotype. Compared with the reference homozygous genotypes (-1195TT and -709GG), SCCHN risk was not associated with variant genotypes, nor haplotypes/diplotypes, of these two SNPs, after adjustment for age, sex, smoking and drinking status (Table 2). No association in any of the subgroups was observed in further stratified analysis by age (young versus ≥ 45 years), sex, smoking and drinking status (never vs. ever), education level (low vs. > high school), income (low vs. ≥ $50,000), tumor site (oropharynx vs. HPV-unrelated sites), and TNM stage (Table 3); however, we did detect some borderline significant interactions between levels of education and income and the -1195T>C polymorphism in SCCHN risk, even after Bonferroni correction (interaction P value: 0.024 and 0.039, respectively).
Figure 1.
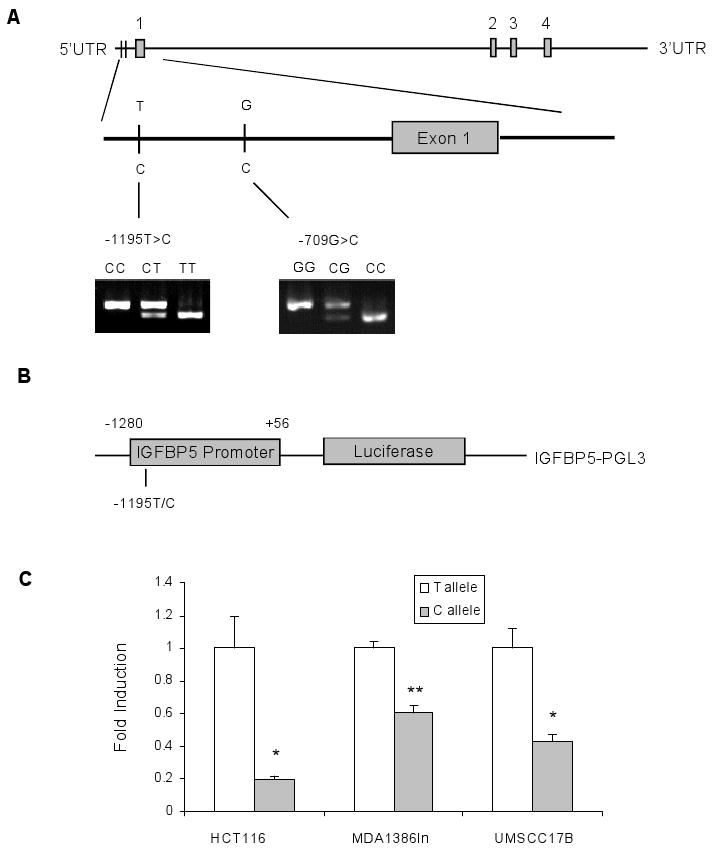
IGFBP5 gene structure, reporter gene constructs for the IGFBP5 promoter, and luciferase expression of the constructed promoter in different cell lines. A, Genomic structure, locations, and genotypes of two selected IGFBP5 SNPs. B, Schematic drawing of the reporter gene constructs containing a 1,337-bp IGFBP5 promoter region; the only difference between the two constructs was a T or C at the -1195 polymorphic site. C, Luciferase activity of the two IGFBP5 promoter constructs in colon cancer cell line HCT116 and two head and neck cancer cell lines: MDA1386Ln and UMSCC-17B. All constructs were co-transfected with pTK-Renilla plasmid as internal control. For each cell line, fold induction of relative luciferase activity was measured by setting reporter gene activity of construct with T allele as 1, using the data (mean ± SD) from three independent transfection experiments. * denotes P < 0.05 for the difference between alleles T and C, ** denotes P < 0.01.
Table 2.
Genotypes, haplotypes and diplotypes distribution of IGFBP5 polymorphisms in SCCHN cases and cancer-free controls
| SNP/Haplotype/Diplotype | Cases No. (%) |
Controls No. (%) |
P value | Crude OR (95% CI) |
Adjusted OR (95% CI)* |
|---|---|---|---|---|---|
| All subjects | 1082 (100) | 1120 (100) | |||
| Genotypes | |||||
| -1195T>C (rs1978346) | 0.797† | ||||
| TT | 461 (42.6) | 466 (41.6) | 1.00 | 1.00 | |
| CT | 486 (44.9) | 519 (46.3) | 0.95 (0.79-1.13) | 0.98 (0.81-1.18) | |
| CC | 135 (12.5) | 135 (12.1) | 1.01 (0.77-1.33) | 1.11 (0.84-1.47) | |
| CT+CC | 621 (57.4) | 654 (58.4) | 0.635‡ | 0.96 (0.81-1.14) | 1.01 (0.84-1.20) |
| -709G>C (rs4480966) | 0.199† | ||||
| GG | 828 (76.5) | 862 (77.0) | 1.00 | 1.00 | |
| CG | 239 (22.1) | 251 (22.4) | 0.99 (0.81-1.21) | 1.05 (0.85-1.29) | |
| CC | 15 (1.4) | 7 (0.6) | 2.23 (0.91-5.50) | 2.10 (0.83-5.31) | |
| CG+CC | 254 (23.5) | 258 (23.0) | 0.807‡ | 1.03 (0.84-1.25) | 1.08 (0.88-1.33) |
| Haplotypes§ | 0.835# | ||||
| T-G | 1405 (64.9) | 1449 (64.7) | 1.00 | 1.00 | |
| C-G | 490 (22.6) | 526 (23.5) | 0.96 (0.84-1.11) | 1.00 (0.86-1.16) | |
| C-C | 266 (12.3) | 263 (11.7) | 1.05 (0.86-1.26) | 1.10 (0.90-1.34) | |
| T-C | 3 (0.1) | 2 (0.1) | 1.50 (0.25-9.00) | 1.64 (0.24-11.28) | |
| Diplotypes | 0.581# | ||||
| T-G/T-G | 458 (42.3) | 464 (41.4) | 1.00 | 1.00 | |
| C-G/T-G | 308 (28.5) | 330 (29.5) | 0.95 (0.77-1.16) | 0.97 (0.79-1.20) | |
| C-C/T-G | 178 (16.5) | 189 (16.9) | 0.95 (0.77-1.22) | 1.00 (0.78-1.29) | |
| C-G/C-G | 62 (5.7) | 68 (6.1) | 0.92 (0.64-1.33) | 0.99 (0.68-1.46) | |
| C-C/C-G | 58 (5.4) | 60 (5.4) | 0.98 (0.67-1.44) | 1.13 (0.76-1.69) | |
| Others‖ | 18 (1.7) | 9 (0.8) | 2.03 (0.90-4.56) | 1.99 (0.86-4.61) |
Adjusted for age, sex, smoking and alcohol status in the logistic regression model.
Distribution of three genotypes
Distribution of combined genotypes
The number for cases and controls in Haplotype analysis is chromosome number.
Includes 2 minor diplotypes, each with a frequency < 0.05.
χ2 tests for the difference of haplotypes and diplotypes frequencies distributions between cases and controls
Table 3.
Stratification analysis of the association between IGFBP5 SNPs and SCCHN risk
| Variables | -1195T>C | P, Interaction* | -709G>C | P, Interaction* | |||
|---|---|---|---|---|---|---|---|
| TT | CT+CC | GG | CG+CC | ||||
| Age | 0.321 | 0.261 | |||||
| < 45 years | Cases/Controls | 62/57 | 72/97 | 106/115 | 28/39 | ||
| Adjusted OR (95% CI)† | 1.00 | 0.70 (0.43-1.14) | 1.00 | 0.77 (0.44-1.36) | |||
| ≥ 45 years | Cases/Controls | 399/409 | 549/557 | 722/747 | 226/219 | ||
| Adjusted OR (95% CI)† | 1.00 | 1.06 (0.88-1.28) | 1.00 | 1.14 (0.91-1.43) | |||
| Sex | 0.409 | 0.955 | |||||
| Female | Cases/Controls | 120/108 | 147/154 | 210/208 | 57/54 | ||
| Adjusted OR (95% CI)† | 1.00 | 0.89 (0.61-1.28) | 1.00 | 1.09 (0.70-1.70) | |||
| Male | Cases/Controls | 341/358 | 474/500 | 618/654 | 197/204 | ||
| Adjusted OR (95% CI)† | 1.00 | 1.05 (0.85-1.28) | 1.00 | 1.08 (0.85-1.36) | |||
| Smoking Status | 0.222 | 0.206 | |||||
| Never | Cases/Controls | 124/216 | 181/329 | 224/413 | 81/132 | ||
| Adjusted OR (95% CI)† | 1.00 | 0.96 (0.72-1.28) | 1.00 | 1.14 (0.83-1.58) | |||
| Ever | Cases/Controls | 337/250 | 440/325 | 604/449 | 173/126 | ||
| Adjusted OR (95% CI)† | 1.00 | 1.01 (0.81-1.27) | 1.00 | 1.02 (0.78-1.33) | |||
| Drinking Status | 0.131 | 0.508 | |||||
| Never | Cases/Controls | 131/202 | 168/287 | 231/376 | 68/113 | ||
| Adjusted OR (95% CI)† | 1.00 | 0.92 (0.68-1.23) | 1.00 | 1.03 (0.73-1.46) | |||
| Ever | Cases/Controls | 330/264 | 453/367 | 597/486 | 186/145 | ||
| Adjusted OR (95% CI)† | 1.00 | 1.06 (0.85-1.33) | 1.00 | 1.11 (0.86-1.44) | |||
| Education Level | 0.024 | 0.062 | |||||
| ≤ High School | Cases/Controls | 166/140 | 213/159 | 292/239 | 87/60 | ||
| Adjusted OR (95% CI)† | 1.00 | 1.20 (0.88-1.65) | 1.00 | 1.29 (0.88-1.90) | |||
| > High School | Cases/Controls | 295/326 | 408/495 | 536/623 | 167/198 | ||
| Adjusted OR (95% CI)† | 1.00 | 0.94 (0.76-1.16) | 1.00 | 1.01 (0.79-1.29) | |||
| Income Level | 0.039 | 0.327 | |||||
| <$50,000 | Cases/Controls | 164/149 | 235/179 | 304/255 | 95/73 | ||
| Adjusted OR (95% CI)† | 1.00 | 1.23 (0.90-1.67) | 1.00 | 1.14 (0.79-1.64) | |||
| ≥ $50,000 | Cases/Controls | 297/317 | 386/475 | 524/607 | 159/185 | ||
| Adjusted OR (95% CI)† | 1.00 | 0.90 (0.73-1.12) | 1.00 | 1.05 (0.82-1.35) | |||
| Tumor Site | |||||||
| Oropharynx | Cases/Controls | 215/466 | 336/654 | 412/862 | 139/258 | ||
| Adjusted OR (95% CI)† | 1.00 | 1.16 (0.93-1.43) | 1.00 | 1.15 (0.90-1.47) | |||
| Non-Oropharynx | Cases/Controls | 246/466 | 285/654 | 416/862 | 115/258 | ||
| Adjusted OR (95% CI)† | 1.00 | 0.87 (0.69-1.10) | 1.00 | 0.98 (0.75-1.29) | |||
| TNM Stage | 0.001‡ | 0.019‡ | |||||
| I | Cases/Controls | 61/466 | 56/654 | 98/862 | 19/258 | ||
| Adjusted OR (95% CI)† | 1.00 | 0.68 (0.46-1.00) | 1.00 | 0.67 (0.40-1.13) | |||
| II | Cases/Controls | 74/466 | 82/654 | 117/862 | 39/258 | ||
| Adjusted OR (95% CI)† | 1.00 | 0.86 (0.61-1.22) | 1.00 | 1.19 (0.80-1.78) | |||
| III | Cases/Controls | 81/466 | 101/654 | 144/862 | 38/258 | ||
| Adjusted OR (95% CI)† | 1.00 | 0.96 (0.69-1.33) | 1.00 | 0.96 (0.65-1.42) | |||
| IV | Cases/Controls | 245/466 | 381/654 | 469/862 | 157/258 | ||
| Adjusted OR (95% CI)† | 1.00 | 1.19 (0.97-1.47) | 1.00 | 1.17 (0.92-1.48) | |||
The interaction p value was the p value of chi-square test for the product of two variables, and was adjusted by age, sex, smoking and alcohol status in the logistic regression model.
Adjusted for age, sex, smoking and alcohol status in the logistic regression model.
P values for trend test.
Association between IGFBP5 polymorphisms and risk of late-stage SCCHN
Compared with the -1195TT genotype, CT+CC genotypes were associated with increasing risk of SCCHN in a TNM stage-dose response manner (adjusted OR 0.68, 0.86, 0.96, and 1.19 for TNM stages I, II, III, and IV, respectively, Ptrend = 0.001), even after Bonferroni correction for multiple testing, where there was a borderline protective effect for the stage I disease, but this was reversed to an increased risk for the stage IV disease (Table 3). This dose-response effect appeared to be present only for HPV-unrelated sites (P for trend < 0.001) (Table 4), in which similar trends were seen even when data were stratified by the extent of the primary tumor (T stage, low vs. high) and nodal involvement (N stage, none vs. involved) (Table 4). Additional case-only analysis revealed that the -1195 C variant genotypes were significantly associated with late stages (III and IV vs. I and II) at the presentation of SCCHN at HPV-unrelated sites (adjusted OR, 2.21; 95% CI, 1.19-4.11 for CC; and adjusted OR, 1.42; 95% CI, 0.99-2.04 for CT+CC) with a trend of the C allele dose-response (P for trend = 0.014) (Table 5); furthermore, haplotypes and diplotypes of -1195T>C and -709G>C were also significantly associated with advanced stage SCCHN at HPV-unrelated sites (adjusted OR, 1.48; 95% CI, 1.06-2.07 for C-G haplotype and adjusted OR, 2.90; 95% CI, 1.10-7.64 for C-G/C-G diplotype), comparing with the most frequent haplotype (T-G) and diplotype (T-G/T-G) (Table 5). All statistically significant P values remained significant after Bonferroni correction in the case-only analysis. All above-mentioned associations or trends were not observed for oropharyngeal cancer, or for the -709C variant genotypes.
Table 4.
Association between IGFBP5 SNPs and SCCHN risk stratified by TNM stage and tumor site
| Variables | -1195T>C | P* | -709G>C | P* | |||
|---|---|---|---|---|---|---|---|
| TT | CT+CC | GG | CG+CC | ||||
| Oropharynx | |||||||
| Stage I | Cases/Controls | 4/466 | 5/654 | 0.863 | 7/862 | 2/258 | 0.954 |
| Adjusted OR (95% CI)† | 1.00 | 0.94 (0.25-3.56) | 1.00 | 1.10 (0.22-5.41) | |||
| Stage II | Cases/Controls | 13/466 | 25/654 | 0.363 | 27/862 | 11/258 | 0.396 |
| Adjusted OR (95% CI)† | 1.00 | 1.43 (0.72-2.83) | 1.00 | 1.39 (0.68-2.86) | |||
| Stage III | Cases/Controls | 42/466 | 53/654 | 0.621 | 71/862 | 24/258 | 0.622 |
| Adjusted OR (95% CI)† | 1.00 | 0.92 (0.60-1.41) | 1.00 | 1.15 (0.71-1.87) | |||
| Stage IV | Cases/Controls | 156/466 | 253/654 | 0.222 | 307/862 | 102/258 | 0.438 |
| Adjusted OR (95% CI)† | 1.00 | 1.22 (0.96-1.55) | 1.00 | 1.13 (0.86-1.48) | |||
| P, Trend | 0.578 | 0.830 | |||||
| T-stage 1 or 2 | Cases/Controls | 140/466 | 221/654 | 0.342 | 270/862 | 91/258 | 0398 |
| Adjusted OR (95% CI)† | 1.00 | 1.17 (0.91-1.50) | 1.00 | 1.14 (0.86-1.51) | |||
| T-stage 3 or 4 | Cases/Controls | 75/466 | 115/654 | 0.581 | 142/862 | 48/258 | 0.502 |
| Adjusted OR (95% CI)† | 1.00 | 1.14 (083-1.57) | 1.00 | 1.15 (0.80-1.65) | |||
| N-stage 0 | Cases/Controls | 35/466 | 52/654 | 0.802 | 64/862 | 23/258 | 0.470 |
| Adjusted OR (95% CI)† | 1.00 | 1.10 (0.70-1.72) | 1.00 | 1.23 (0.74-2.03) | |||
| N-stage 1-3 | Cases/Controls | 180/466 | 284/654 | 0.300 | 348/862 | 116/258 | 0.402 |
| Adjusted OR (95% CI)† | 1.00 | 1.17 (0.93-1.47) | 1.00 | 1.14 (0.88-1.47) | |||
| Non-Oropharynx | |||||||
| Stage I | Cases/Controls | 57/466 | 51/654 | 0.025 | 91/862 | 17/258 | 0.082 |
| Adjusted OR (95% CI)† | 1.00 | 0.67 (0.44-1.00) | 1.00 | 0.67 (0.39-1.15) | |||
| Stage II | Cases/Controls | 61/466 | 57/654 | 0.035 | 90/862 | 28/258 | 0.865 |
| Adjusted OR (95% CI)† | 1.00 | 0.74 (0.50-1.10) | 1.00 | 1.20 (0.75-1.92) | |||
| Stage III | Cases/Controls | 39/466 | 48/654 | 0.558 | 73/862 | 14/258 | 0.135 |
| Adjusted OR (95% CI)† | 1.00 | 0.98 (0.62-1.57) | 1.00 | 0.73 (0.39-1.36) | |||
| Stage IV | Cases/Controls | 89/466 | 128/654 | 0.871 | 162/862 | 55/258 | 0.462 |
| Adjusted OR (95% CI)† | 1.00 | 1.17 (0.85-1.61) | 1.00 | 1.26 (0.87-1.82) | |||
| P, Trend | <0.001 | 0.013 | |||||
| T-stage 1 or 2 | Cases/Controls | 153/466 | 157/654 | 0.015 | 250/862 | 60/258 | 0.167 |
| Adjusted OR (95% CI)† | 1.00 | 0.78 (0.60-1.02) | 1.00 | 0.87 (0.62-1.21) | |||
| T-stage 3 or 4 | Cases/Controls | 93/466 | 127/654 | 0.855 | 166/862 | 54/258 | 0.628 |
| Adjusted OR (95% CI)† | 1.00 | 1.09 (078-1.50) | 1.00 | 1.22 (0.84-1.78) | |||
| N-stage 0 | Cases/Controls | 152/466 | 159/654 | 0.022 | 247/862 | 64/258 | 0.359 |
| Adjusted OR (95% CI)† | 1.00 | 0.81 (0.61-1.05) | 1.00 | 0.95 (0.69-1.32) | |||
| N-stage 1-3 | Cases/Controls | 94/466 | 125/654 | 0.718 | 169/862 | 50/258 | 0.948 |
| Adjusted OR (95% CI)† | 1.00 | 1.03 (0.75-1.42) | 1.00 | 1.07 (0.74-1.56) | |||
P value for detecting the difference of combined genotype distribution between cases and controls.
Adjusted for age, sex, smoking and alcohol status in the logistic regression model.
Table 5.
Genotypes, haplotypes and diplotypes distribution of IGFBP5 polymorphisms associated with high TNM stage among patients with SCCHN at HPV-unrelated sites (oral cavity and larynx/hypopharynx only)
| SNP position | Stages I,II No. (%) |
Stages III, IV No. (%) |
P value | Crude OR (95% CI) |
Adjusted OR (95% CI)* |
|---|---|---|---|---|---|
| All subjects | 226 (100%) | 304 (100%) | |||
| Genotypes | |||||
| -1195T>C (rs1978346) | 0.018† | ||||
| TT | 118 (52.2) | 128 (42.1) | 1.00 | 1.00 | |
| CT | 89 (39.4) | 130 (42.8) | 1.35 (0.93-1.95) | 1.25 (0.85-1.85) | |
| CC | 19 (8.4) | 46 (15.1) | 2.23 (1.24-4.03) | 2.21 (1.19-4.11) | |
| P, Trend | 0.014§ | ||||
| CT+CC | 128 (42.1) | 176 (57.9) | 0.021‡ | 1.50 (1.06-2.12) | 1.42 (0.99-2.04) |
| -709G>C (rs4480966) | 0.689† | ||||
| GG | 181 (80.1) | 235 (77.3) | 1.00 | 1.00 | |
| CG | 42 (18.6) | 63 (20.7) | 1.16 (0.75-1.79) | 1.09 (0.69-1.72) | |
| CC | 3 (1.3) | 6 (2.0) | 1.54 (0.38-6.24) | 1.40 (0.34-5.80) | |
| P, Trend | 0.592§ | ||||
| CG+CC | 45 (19.9) | 69 (22.7) | 0.440‡ | 1.18 (0.77-1.80) | 1.11 (0.71-1.73) |
| Haplotypes‖ | 0.020†† | ||||
| T-G | 324 (71.7) | 386 (63.5) | 1.00 | 1.00 | |
| C-G | 80 (17.7) | 147 (24.2) | 1.50 (1.09-2.05) | 1.48 (1.06-2.07) | |
| C-C | 47 (10.4) | 75 (12.3) | 1.37 (0.80-2.35) | 1.31 (0.75-2.28) | |
| T-C | 1 (0.2) | 0 (0) | NA | NA | |
| Diplotypes | 0.163†† | ||||
| T-G/T-G | 117 (51.8) | 128 (42.1) | 1.00 | 1.00 | |
| C-G/T-G | 57 (25.2) | 87 (28.6) | 1.40 (0.92-2.12) | 1.31 (0.84-2.04) | |
| C-C/T-G | 32 (14.2) | 43 (14.1) | 1.23 (0.73-2.08) | 1.13 (0.65-1.95) | |
| C-C/C-G | 9 (4.0) | 20 (6.6) | 2.03 (0.89-4.64) | 1.91 (0.81-4.51) | |
| C-G/C-G | 7 (3.1) | 20 (6.6) | 2.61 (1.07-6.40) | 2.90 (1.10-7.64) | |
| Others** | 4 (1.8) | 6 (2.0) | 1.37 (0.38-4.98) | 1.29 (0.35-4.78) |
Adjusted for age, sex, smoking and alcohol status, education level, and income level in the logistic regression model.
Distribution of three genotypes
Distribution of combined genotypes
P trend was tested according to the distribution of tumor stages over 3 genotypes
The number for cases and controls in Haplotype analysis is chromosome number.
Includes 2 minor diplotypes, each with a frequency < 0.05.
χ2 tests for the difference of haplotypes and diplotypes frequencies distributions between cases and controls
The -1195T>C allele change resulted in altered promoter activity regulated byAP-1
To determine allele-specific effects of the -1195T>C SNP on the IGFBP5 promoter activity, we generated two luciferase reporter gene constructs with 1,347 bp of the IGFBP5 promoter region (from -1280 to +56), containing either T or C at -1195 position (Fig. 1B). As expected, the IGFBP5 promoter containing -1195C exhibited a significantly lower reporter gene expression, compared with the -1195T in two human head and neck carcinoma cell lines MDA-1386Ln (P < 0.01) and UMSCC-17B (P < 0.05) and a human colon cancer cell line HCT116 (P < 0.05) (Fig. 1C), suggesting that T to C allele change at IGFBP5 -1195 may decrease the IGFBP5 promoter activity in a non-tissue specific manner.
To determine the mechanism underlying differential regulation of the IGFBP5 promoter activity, the potential transcription binding site change at -1195T>C was predicted with TFSearch (http://www.cbrc.jp/research/db/TFSEARCH.html) and transcription element search software (TESS) (http://www.cbil.upenn.edu/cgi-bin/tess/tess). Both software tools predicted the activator protein 1 (AP-1) as the potential transcription factor to bind to T and C alleles differentially. A schematic diagram of the predicted AP-1 binding site by TESS is shown in Fig. 2A. We then conducted an EMSA to test whether AP-1 differentially binds to the T and C alleles at the -1195 site of IGFBP5. As shown in Fig. 2B, the biotin-labeled oligonucleotide probe with the T allele had a 20-30% stronger binding than that with the C allele (lane 1 comparing to lane 6), which is consistent with luciferase reporter gene assay results. Moreover, a 50-fold excess of unlabeled oligonucleotide with the T or C allele competed for this binding activity (lanes 2, 3, 7, and 8). The specificity of the protein-DNA complex was further confirmed by the gel supershift assay, because the antibody specifically against c-fos, a subunit of AP-1, abolished the AP-1 DNA binding activity (lanes 4 and 9), and the non-specific IgG failed to do so (lanes 5 and 10). The in vivo binding of AP-1 to the IGFBP5 promoter was also confirmed by the chromatin immunoprecipitation assay in UMSCC-17B cells, as demonstrated by the specific binding of c-fos, but not IgG, to the DNA sequence (Fig. 2C).
Figure 2.
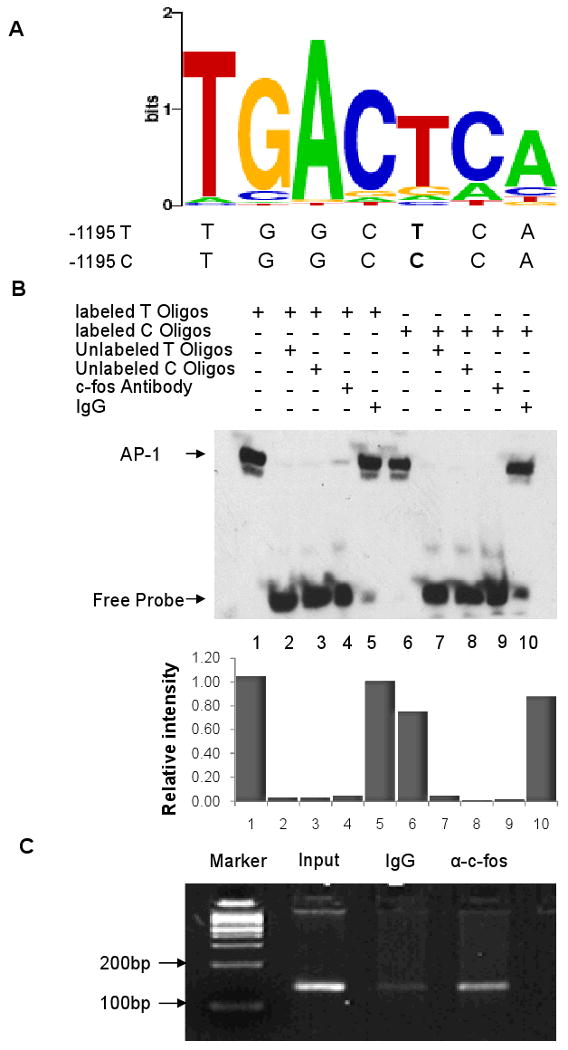
Detection of AP-1 binding to IGFBP5 promoter at the -1195 T>C. A, Prediction of AP-1 binding site at IGFBP5 promoter -1195 position by TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess); B, electrophoretic mobility-shift assay with biotin-labeled -1195 T (lanes 1 to 5) or C probes (lanes 6 to 10) and UMSCC-17B nuclear extract. Lanes 1 and 6, probe and nuclear extracts; lanes 2, 7 and 3, 8, probe and nuclear extracts plus 50× unlabeled -1195 T (lanes 2 and 7), or C (lanes 3 and 8) probes; lanes 4 and 9, probe and nuclear extracts and c-fos antibody; lanes 5 and 10, probe and nuclear extracts and rabbit IgG, a relative densitometric assessment of the gel was shown in the bottom panel, with the density of lane 5 setting as 1. The EMSA experiments were repeated once to ensure the consistency; C, ChIP assay using USMCC-17B cells. The binding of IGFBP5 promoter with AP-1 was verified by PCR.
Discussion
To the best of our knowledge, this study represents the largest study on susceptibility to SCCHN risk associated with potentially functional IGFBP5 polymorphisms. In this study of 1,082 SCCHN patients and 1,120 cancer-free controls of non-Hispanic whites, we did not find evidence of a main effect of IGFBP5 -1195T>C and -709G>C on SCCHN risk in the case-control analysis. However, the -1195C variant genotypes were associated with late-stage SCCHN at HPV-unrelated sites. Further case-only analysis revealed that -1195CC and CT+CC genotypes were significantly associated with the late-stage tumors (stage III and IV) at HPV-unrelated sites. In addition, we found that the change of -1195T allele to C allele resulted in the decreased promoter activity of IGFBP5, which may be regulated by the AP-1 transcription factor.
Very few association studies have been conducted to evaluate the roles of IGFBP5 polymorphisms in cancer risk. In the only published population-based case-control study of 460 African-American women with breast cancer and 279 matched controls as well as 461 Nigerian women with breast cancer and 163 matched controls, a 50-kb DNA sequence covering three exons in the 3′ end of IGFBP2 and three exons in the 3′ end of IGFBP5 was significantly associated with breast cancer risk (20), but this study did not include any functional SNPs, nor the promoter polymorphisms that were studied in our current study of non-Hispanic whites. Although these two studies did not provide relevant data for the comparisons, our larger sample size provided adequate statistical power for detecting a meaningful main effect of IGFBP5 variant genotypes on SCCHN risk and tumor progression in non-Hispanic whites.
In our study, the genotype distribution of -709G>C in controls did not follow HWE. This could have been caused by genotyping error of 1% in our study, but this is unlikely to play a major role; selection bias could not be fully ruled out, because our control subjects were recruited from the cancer-free visitors to MD Anderson Cancer Center, who may not represent the general population; another explanation is that the adverse effect of the rare homozygous CC may be lethal during the embryonic development, because only 22 out of 2202 study subjects had the CC genotype.
IGFBP-5 has been reported to be involved in tumorigenesis and metastasis of various cancers, including SCCHN. Although we did not find any main effect on overall risk of SCCHN, we did find that the -1195C allele was significantly associated with late TNM stages of SCCHN at HPV-unrelated sites. It is possible that the very limited sample size of patients with early-stage I and II oropharyngeal cancer (9 stage I and 38 stage II) did not provide enough power to detect such an association in this HPV-related site. Because a direct association between HPV-infection and the -1195 SNP was not established here, larger studies with more early-stage I and II oropharyngeal cancer patients are needed to confirm this observed association.
Nevertheless, our findings are biologically plausible, because several studies have shown that IGFBP-5 regulate cell migration, invasion and angiogenesis through interactions with ECM proteins. For example, IGFBP-5 has been shown to function as a tumor suppressor by inhibiting tumor vascularity of human ovarian cancer in a xenograft model (21). An important ECM protein, matrix metalloproteinase 7 (MMP-7) has been shown to cleave IGFBP-5 and other IGFBPs, hence to affect the bioavailability of IGFs and to regulate tumor cell growth during invasion and metastasis (22). Another study showed that IGFBPs, including IGFBP-5, bound to vitronectin and enhanced keratinocyte cell migration (23). Indeed, the expression levels of IGFBP-5 in multiple cancers, including breast cancer, ovarian cancer, cervical cancer, mesothelioma, and SCCHN, were associated with advanced TNM stages (15, 21, 24-26). In SCCHN, it has been shown that the down-regulation of IGFBP-5 was associated with a higher tendency for nodal metastasis (15). Therefore, our findings may provide a useful biomarker for the association between IGFBP5 polymorphisms and SCCHN risk, particularly for late-stage tumors, if validated by others.
To provide additional mechanistic insight into our findings, we also showed that the transcription factor AP-1 differentially bound to the IGFBP5 promoter region containing either T or C allele, which may lead to different expression levels of IGFBP-5. AP-1 is a dimeric transcription factor that consists of members of Fos (c-Fos, FosB, Fra1, and Fra2) and Jun (c-Jun, JunB, and JunD) gene families, and Fos/Jun heterodimers are the predominant forms of AP-1 in most tissues (27). A number of extracellular signals, including growth factors, cytokines and ECM proteins can activate AP-1. It is reported that AP-1 may play critical roles in tumor invasion and metastasis through regulating the expression of genes, MMPs, involved in cell migration and invasion (28).
In an early study, multiple AP-1 and AP-2 transcription factor binding sites were found to be present in the rat IGFBP5 promoter region (29), suggesting that human IGFBP5 may also be regulated by AP-1 at the highly conserved promoter region. Although a direct interaction between AP-1 and IGFBP-5 has not been fully studied, the activation of the mitogen activated protein kinase (MAPK) pathway can increase the expression of IGFBP-5. Indeed, IGFBP-5 induce lung fibroblast and mononuclear cell migration through the activation of the MAPK signaling pathway, as blocking MEK1/2, the key mediator of the MAPK pathway, inhibited the IGFBP-5 induced cell migration (30). IGF-I has been reported to regulate the expression of IGFBP-5 (31), and the MAPK pathway played critical roles in IGF-I induced expression of IGFBP-5 in intestinal smooth muscle cells (32). More recently, it was shown that inhibition of MEK1/2 abolished IGFBP-5 mediated cell survival in pancreatic cancer cells (33).
It is well known that AP-1 can be activated through the induction of the MAPK pathway by growth factors, including IGF-I (34). Therefore, it is possible that IGF-I or other growth factors may activate the MAPK pathway and then induce AP-1, and the activated AP-1 then binds to the IGFBP5 promoter and regulates its expression. Our data suggested that IGFBP-5 with the C allele at the -1195T>C site may have a weaker binding to AP-1, compared with that with the T allele. Consequently, lower expression of IGFBP-5 in tissues of C allele carriers may explain the higher TNM stage. These results are consistent with the previous work showing that decreased expression of IGFBP-5 was related to the higher propensity for metastasis in SCCHN (15). However, more detailed molecular studies are needed to provide direct evidences that the differential binding of AP-1 to the IGFBP5 promoter alters the expression level of IGFBP-5.
In summary, our study showed that carriers of the -1195C allele were likely to have late-stage or invasive SCCHN and that T and C alleles at -1195T>C resulted in different promoter activities, which may be regulated by the AP-1 transcription factor. These results provide evidence of a possibly novel mechanism by which the IGFBP-5 regulates SCCHN metastasis. Although our large study provides adequate statistical power and reliable association estimates, an uncontrolled selection bias may exist due to the nature of a hospital-based case-control study. Larger, population-based, preferably prospective studies should be performed to further validate our findings.
Acknowledgments
This work was partly supported by the National Institute of Health grants R01 CA131274 and R01 ES011740 (Q. Wei), P50 CA097007 (S. Lippman), and P30 CA016672 (The University of Texas M. D. Anderson Cancer Center) and a cancer prevention fellowship supported by R25T CA057730 (to R M. Chamberlain. and S. Chang.).
Abbreviations
- AP-1
Activator protein 1
- ChIP
chromatin immunoprecipitation
- CI
confidence interval
- ECM
extracellular matrix
- EMSA
electrophoretic mobility shift assay
- HPV
human papillomavirus
- IGF
insulin-like growth factor
- IGFBP5
insulin-like growth factor binding protein 5
- MAF
minor allele frequency
- OR
odds ratio
- PCR
polymerase chain reaction
- SCCHN
Squamous cell carcinoma of the head and neck
- SNP
single nucleotide polymorphism
- HWE
Hardy-Weinberg equilibrium
- TNM
tumor-node-metastasis
Footnotes
Disclosure of Potential Conflicts of Interest. No potential conflicts of interest were disclosed.
References
- 1.Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers? Cancer. 2007;110(7):1429–35. doi: 10.1002/cncr.22963. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Hashibe M, Brennan P, Chuang SC, et al. Interaction between tobacco and alcohol use and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol Biomarkers Prev. 2009;18(2):541–50. doi: 10.1158/1055-9965.EPI-08-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu TC, Spitz MR, Schantz SP. Mutagen sensitivity: a biological marker of cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1991;1(1):83–9. [PubMed] [Google Scholar]
- 5.Ho T, Wei Q, Sturgis EM. Epidemiology of carcinogen metabolism genes and risk of squamous cell carcinoma of the head and neck. Head Neck. 2007;29(7):682–99. doi: 10.1002/hed.20570. [DOI] [PubMed] [Google Scholar]
- 6.Neumann AS, Sturgis EM, Wei Q. Nucleotide excision repair as a marker for susceptibility to tobacco-related cancers: a review of molecular epidemiological studies. Mol Carcinog. 2005;42(2):65–92. doi: 10.1002/mc.20069. [DOI] [PubMed] [Google Scholar]
- 7.Valentinis B, Baserga R. IGF-I receptor signalling in transformation and differentiation. Mol Pathol. 2001;54(3):133–7. doi: 10.1136/mp.54.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JJ, Accili D. Signalling through IGF-I and insulin receptors: where is the specificity? Growth Horm IGF Res. 2002;12(2):84–90. doi: 10.1054/ghir.2002.0265. [DOI] [PubMed] [Google Scholar]
- 9.Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev. 1999;20(6):761–87. doi: 10.1210/edrv.20.6.0382. [DOI] [PubMed] [Google Scholar]
- 10.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23(6):824–54. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 11.Mohan S, Baylink DJ. IGF-binding proteins are multifunctional and act via IGF-dependent and -independent mechanisms. J Endocrinol. 2002;175(1):19–31. doi: 10.1677/joe.0.1750019. [DOI] [PubMed] [Google Scholar]
- 12.Beattie J, Allan GJ, Lochrie JD, Flint DJ. Insulin-like growth factor-binding protein-5 (IGFBP-5): a critical member of the IGF axis. Biochem J. 2006;395(1):1–19. doi: 10.1042/BJ20060086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lochrie JD, Phillips K, Tonner E, et al. Insulin-like growth factor binding protein (IGFBP)-5 is upregulated during both differentiation and apoptosis in primary cultures of mouse mammary epithelial cells. J Cell Physiol. 2006;207(2):471–9. doi: 10.1002/jcp.20587. [DOI] [PubMed] [Google Scholar]
- 14.Chang KW, Hung PS, Lin IY, et al. Curcumin up-regulates insulin-like growth factor binding protein-5 (IGFBP-5) and C/EBPalpha during oral cancer suppression. Int J Cancer. doi: 10.1002/ijc.25220. [DOI] [PubMed] [Google Scholar]
- 15.Hung PS, Kao SY, Shih YH, et al. Insulin-like growth factor binding protein-5 (IGFBP-5) suppresses the tumourigenesis of head and neck squamous cell carcinoma. J Pathol. 2008;214(3):368–76. doi: 10.1002/path.2280. [DOI] [PubMed] [Google Scholar]
- 16.Lin SC, Wang CP, Chen YM, et al. Regulation of IGFBP-5 expression during tumourigenesis and differentiation of oral keratinocytes. J Pathol. 2002;198(3):317–25. doi: 10.1002/path.1220. [DOI] [PubMed] [Google Scholar]
- 17.Patel SG, Shah JP. TNM staging of cancers of the head and neck: striving for uniformity among diversity. CA Cancer J Clin. 2005;55(4):242–58. doi: 10.3322/canjclin.55.4.242. quiz 261-2, 264. [DOI] [PubMed] [Google Scholar]
- 18.Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19(9):2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong Y, Miao X, Zhang X, et al. The role of P53 and MDM2 polymorphisms in the risk of esophageal squamous cell carcinoma. Cancer Res. 2005;65(20):9582–7. doi: 10.1158/0008-5472.CAN-05-1460. [DOI] [PubMed] [Google Scholar]
- 20.Garner CP, Ding YC, John EM, et al. Genetic variation in IGFBP2 and IGFBP5 is associated with breast cancer in populations of African descent. Hum Genet. 2008;123(3):247–55. doi: 10.1007/s00439-008-0468-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rho SB, Dong SM, Kang S, et al. Insulin-like growth factor-binding protein-5 (IGFBP-5) acts as a tumor suppressor by inhibiting angiogenesis. Carcinogenesis. 2008;29(11):2106–11. doi: 10.1093/carcin/bgn206. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura M, Miyamoto S, Maeda H, et al. Matrix metalloproteinase-7 degrades all insulin-like growth factor binding proteins and facilitates insulin-like growth factor bioavailability. Biochem Biophys Res Commun. 2005;333(3):1011–6. doi: 10.1016/j.bbrc.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Hyde C, Hollier B, Anderson A, Harkin D, Upton Z. Insulin-like growth factors (IGF) and IGF-binding proteins bound to vitronectin enhance keratinocyte protein synthesis and migration. J Invest Dermatol. 2004;122(5):1198–206. doi: 10.1111/j.0022-202X.2004.22527.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Arun BK, Fuller GN, Zhang W, Middleton LP, Sahin AA. IGFBP2 and IGFBP5 overexpression correlates with the lymph node metastasis in T1 breast carcinomas. Breast J. 2008;14(3):261–7. doi: 10.1111/j.1524-4741.2008.00572.x. [DOI] [PubMed] [Google Scholar]
- 25.Hou XJ, Zhang YZ, Liu X, Meng LH, Qiao YB. Expressions of IGFBP-5, cFLIP in cervical intraepithelial neoplasia, cervical carcinoma and their clinical significances: a molecular pathology. J Exp Clin Cancer Res. 2009;28:70. doi: 10.1186/1756-9966-28-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoang CD, D'Cunha J, Kratzke MG, et al. Gene expression profiling identifies matriptase overexpression in malignant mesothelioma. Chest. 2004;125(5):1843–52. doi: 10.1378/chest.125.5.1843. [DOI] [PubMed] [Google Scholar]
- 27.Curran T, Franza BR., Jr Fos and Jun: the AP-1 connection. Cell. 1988;55(3):395–7. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- 28.Ozanne BW, Spence HJ, McGarry LC, Hennigan RF. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007;26(1):1–10. doi: 10.1038/sj.onc.1209759. [DOI] [PubMed] [Google Scholar]
- 29.Zhu X, Ling N, Shimasaki S. Cloning of the rat insulin- like growth factor binding protein-5 gene and DNA sequence analysis of its promoter region. Biochem Biophys Res Commun. 1993;190(3):1045–52. doi: 10.1006/bbrc.1993.1154. [DOI] [PubMed] [Google Scholar]
- 30.Yasuoka H, Yamaguchi Y, Feghali-Bostwick CA. The pro-fibrotic factor IGFBP-5 induces lung fibroblast and mononuclear cell migration. Am J Respir Cell Mol Biol. 2009;41(2):179–88. doi: 10.1165/rcmb.2008-0211OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiepe D, Ciarmatori S, Hoeflich A, Wolf E, Tonshoff B. Insulin-like growth factor (IGF)-I stimulates cell proliferation and induces IGF binding protein (IGFBP)-3 and IGFBP-5 gene expression in cultured growth plate chondrocytes via distinct signaling pathways. Endocrinology. 2005;146(7):3096–104. doi: 10.1210/en.2005-0324. [DOI] [PubMed] [Google Scholar]
- 32.Xin X, Hou YT, Li L, et al. IGF-I increases IGFBP-5 and collagen alpha1(I) mRNAs by the MAPK pathway in rat intestinal smooth muscle cells. Am J Physiol Gastrointest Liver Physiol. 2004;286(5):G777–83. doi: 10.1152/ajpgi.00293.2003. [DOI] [PubMed] [Google Scholar]
- 33.Johnson SK, Haun RS. Insulin-like growth factor binding protein-5 influences pancreatic cancer cell growth. World J Gastroenterol. 2009;15(27):3355–66. doi: 10.3748/wjg.15.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hipskind RA, Buscher D, Nordheim A, Baccarini M. Ras/MAP kinase-dependent and -independent signaling pathways target distinct ternary complex factors. Genes Dev. 1994;8(15):1803–16. doi: 10.1101/gad.8.15.1803. [DOI] [PubMed] [Google Scholar]


