Abstract
A universal photochemical method has been established for the immobilization of intact carbohydrates and their analogues, and for the fabrication of carbohydrate microarrays. The method features the use of perfluorophenyl azide (PFPA)-modified substrates and the photochemical reaction of surface azido groups with printed carbohydrates. Various aldoses, ketoses, non-reducing sugars such as alditols and their derivatives can be directly arrayed on the PFPA-modified chips. The lectin-recognition ability of arrayed mannose, glucose and their oligo- and polysaccharides were confirmed using surface plasmon resonance imaging and laser-induced fluorescence imaging.
INTRODUCTION
Carbohydrate microarray chips (CMCs) as a new sensor platform feature high throughput which is highly desirable in the studies of biological recognition, allowing simultaneous observation of multiple events at low cost and low sample consumption (1–4). CMCs have been used increasingly in various biosensing assays in conjunction with fluorescence imaging or surface plasmon resonance imaging (5, 6), and can be fabricated through two strategies: physisorption and covalent immobilization. The physisorption method was first introduced in 2002 for the fabrication of CMCs by Wang et al. (7) and Willats et al. (8). Wang and coworkers used nitrocellulose-coated glass slides to physically deposit polysaccharides or glycoproteins (7) while Willats et al. developed black polystyrene-coated glass slides for the adsorption of polysaccharides and their derivatives (8). Similarly, fluorous-tagged carbohydrates could be strongly adsorbed onto fluorous-derivatized glass slides (9). Biotinylated glycans could be specifically attached onto streptavidin-coated surfaces (10), and the method can be considered as a bio-adsorption process. The physisorption approaches are generally convenient but have limitations of either size-dependence (small sugars have weak van de Waals interaction forces) or requiring functional group derivatization.
To overcome the size-dependent limitation, the second strategy of covalent immobilization has been explored mostly by utilizing various types of functional groups (5). For example, Park et al. synthesized maleimide-derivatized carbohydrates and spotted them onto thiol-modified glass surface (11). Alternatively, Ratner et al. made use of the reaction between thiol-derivatized carbohydrates and maleimide modified surface (12). Houseman and Mrksich utilized the Diels-Alder reaction between cyclopentadiene-derivatized carbohydrates and benzoquinone-functionalized surface (13). Recently, Mercey et al. immobilized pyrrole-derivatized oligosaccharides on pyrrole-modified surfaces by electro-copolymerization (14). Other functionalized carbohydrates such as those derivatized with amine-, aldehyde-, hydrazide-, azide-groups have also been used to construct CMCs (5, 15–18).
However, these covalent approaches frequently require pre-treatment of the carbohydrates, generally not applicable to the direct immobilization of intact carbohydrates on a solid substrate. Direct conjugation of reducing sugars to hydrazide- or aminooxyacetyl- functionalized substrates has been demonstrated (19, 20). However, it remains a great challenge to directly immobilize non-reducing carbohydrates which are more difficult to be derivatized. It is thus highly desirable to develop a universal method that is applicable to the preparation of CMCs from various types of carbohydrates or analogues, such as reducing and non-reducing ketoses and even alditols. In this study, a photochemical approach is employed for the fabrication of CMCs from intact carbohydrates. Photochemical reactions are generally fast, efficient, and can be readily integrated with conventional microfabrication processes. A few examples were reported for the preparation of CMCs where the photoactive reagents of aryltrifluoromethyldiazirines or phthalimide were used (21–23). We have focused our efforts on using perfluorophenyl azides (PFPAs) as the photocoupling agents. PFPAs are highly efficient photoaffinity labelling reagents with significantly enhanced C-H and N-H insertion yields, and have been successfully applied to the covalent immobilization of macromolecules, small molecules and graphene on different substrate materials (24–28). PFPA derivatives can be readily prepared from commercially available starting materials, and they are stable when protected from direct or prolonged light exposure. We have recently demonstrated that PFPAs can be used to couple unmodified carbohydrates onto gold (29, 30) and iron oxide nanoparticles (31, 32). Ramstrom et al. have also shown that PFPA-derivatized sugars could readily be immobilized onto poly(ethylene oxide)-modified surfaces by a fast photoativation (33, 34). However, the derivatization of sugars with a PFPA is not yet universally applicable.
In order to broaden the application of PFPA photocoupling chemistry, we developed a general method for the fabrication of CMCs. The idea is to modify the substrate with PFPAs, which is then used to attach intact carbohydrates and their analogues by light activation. Covalent bond formation has been characterized by various methods. Successful preparation of various CMCs has been demonstrated using different types of carbohydrates including aldoses, ketoses, alditols and their derivatives. The bioactivities of the immobilized carbohydrates on CMCs were well maintained with respect to their recognition with lectins, as demonstrated by surface plasmon resonance imaging (SPRi) and laser-induced fluorescence imaging (LIFi).
EXPERIMENTAL SECTION
Materials and Reagents
D-Mannosamine, 3-mercaptopropyltrimethoxysilane (MPTMS), 3-aminopropyltrimethoxysilane (APTMS), concanavalin A (Con A), peanut agglutinin (PNA), fluorescein isothiocyanate (FITC), methylpentafluorobenzoate, 1-ethyl-3-(3-dimethyllaminopropyl)carbodiimide (EDC) hydrochloride, and poly(L-lysine) (PLL) hydrobromide with the molecule weight of 70–150 kD were purchased from Sigma (St. Louis, Mo, USA). 11-Mercaptoundecanoic acid (MUA) was obtained from Aldrich (Milwaukee, WI, USA). N-Hydroxysuccinimide (NHS) was purchased from Acros (New Jersey, USA). N, N′-Dicyclohexylcarbodiimide (DCC), sodium azide, KH2PO4, NaOH, NaCl, CaCl2, MnCl2, absolute ethanol, ethanolamine (EOA), ethylenediamine (EDA) and tris(hydroxylmethyl)aminomethane hydrochloride (Tris-HCl) were of analytical reagent grade and were obtained from Beijing Chemical Works (Beijing, China). All other carbohydrates were of biochemical reagent grade from Beijing Reagent Work (Beijing, China). All reagents and solvents were used as received. Triply distilled water was used for the preparation of all solutions and rinsing.
Synthesis of NHS-PFPA
The photoactive reagent of NHS-PFPA was synthesized according to a previously reported procedure (24). Briefly, the starting materials, sodium azide and methyl pentafluorobenzoate, were mixed and refluxed for 8 h in a mixed solvent of acetone and water. The resulting mixture was extracted with ether and dried in vacuum. The residue, methyl 4-azidotetrafluorobenzoate, was redissolved in CH3OH containing 1% NaOH and was stirred overnight. After acidification of the solution to pH<1 with 2 M HCl, the solution was extracted with CHCl3 and 4-azidotetrafluorobenzoic acid was obtained from the CHCl3 layer after evaporation. The acid was then allowed to react with DCC and NHS in CH2Cl2 under stirring for 8 h. The mixture was filtered and the filtrate was evaporated to leave NHS-PFPA. At last, the resulting NHS-PFPA was recrystallized using CHC13/hexane.
Preparation of NHS-PFPA-based photoactive surfaces
Two types of substrates, glass slides and gold-coated glass slides, were first functionalized with amine and were then reacted with NHS-PFPA in CH2Cl2 (2 mg/mL) at room temperature for 12 h. They were rinsed with CH2Cl2 and dried with N2 gas. The procedures for the amination of the two substrates are described as below.
Silanization of glass slides
Glass slides (24 mm × 24 mm, Shitai Co. Ltd., China) were cleaned in the piranha solution (7: 3 v/v concentrated H2SO4-35wt% H2O2. Warning: this solution is highly oxidative and should be used under protection!) at 70°C for 40 min, washed thoroughly with water and methanol by sonicating in the corresponding solvent for 5 times each. They were immediately immersed in a freshly prepared APTMS solution (3% in methanol) for 12 h. The silanized glass slides were sonicated in methanol three times and dried under N2 stream.
Modification of gold slides with EDA or PLL
MPTMS- functionalized glass slides (20 mm × 20 mm) were deposited with 50 nm thick gold films by vapor-deposition in a JEE-4X vacuum evaporator (JEOL Ltd., Tokyo, Japan). The gold chips were immediately immersed in a solution of MUA in ethanol (1 mM) for at least 16 h, rinsed with ethanol, and were then immersed for 30 min in an aqueous solution of EDC (75 mM) and NHS (15 mM). The resulting chips were treated with either an aqueous solution of EDA (1 M, adjusted with 5 M HCl to pH 8.6) for 1 h, or a solution of PLL in the pH 8.0 buffer of TEA (0.05 M) and NaCl (0.25 M) (1 mg/mL) for 1 h (35). Finally, the chips were soaked in NaOH (pH 12.2) for about 15 min to remove the unbound EDA or PLL, washed with ethanol three times and dried with N2 gas.
Preparation of CMCs
An aqueous carbohydrate solution was spotted on the NHS-PFPA functionalized chip by a laboratory-fabricated microprinting system which allows the delivery of 15-nL solution onto the substrate forming a spot of ~250 μm in diameter. After spotting all carbohydrate samples, the chips were dried in air. The dried chips were exposed for 5 min with a 450-W medium pressure Hg lamp (Blue Sky Special Lamps Development Co. Ltd., China) where a 300-nm optical filter was placed above the chips to filter out the short wavelength UV which may oxidize the sulfur-gold bond (36) The UV-exposed chips were rinsed successively with water, ethanol and water, and dried under an N2 gas stream. These CMCs were used directly or stored at −20 °C for at least two weeks before use. For biological tests, the chips were further blocked by glutaraldehyde (10% in 20 mM phosphate buffer, pH 7.4) for 1 h and then ethanolamine aqueous solution (1 M, pH 8.6 adjusted with HCl) for 1 h.
Determination of coupling efficiency of carbohydrates on PFPA surfaces
The coupling efficiency of carbohydrates on PFPA-modified surfaces was measured based on specific reaction of carbohydrates with anthrone reagent that is composed of 0.1% anthrone in 98% concentrated sulfuric acid (Note: the anthrone reagent should be stored in dark and used within 12 h) (37). Briefly, a PFPA-modified chip was first coated with 100 μL carbohydrate solution (for example, 5 mg/mL mannose in water), dried in vacuum at 0.1 MPa for 5 min and exposed to UV light for 5 min. The chip was then immersed in 1.9 mL water and sonicated for about 5 min at room temperature to remove the un-reacted carbohydrate off the chip. The aqueous solution (1 mL) was transferred to a 25-mL borosilicate glass vial in an ice bath and the anthrone reagent (4 mL) was added dropwise. The solution was then heated in boiling water for 10 min and cooled in tap water. The solution was finally subjected to spectrometric measurement on a TU 1900 UV-Vis spectrophotometer (Beijing Purkinje General Instrument Co., Beijing, China). Depending on the type of carbohydrates, maximal absorbance was measured and was compared to the corresponding calibration curve, which was prepared by plotting the maximal absorbance at 624 nm against the carbohydrate concentration in the range of 31.5–521.9 μM.
Determination of amine density by UV-vis spectrometry
The free amino groups can be measured by reacting the amine chips with 4-nitrobenzaldehyde, which is then released by hydrolyzing the formed imine groups under acidic conditions (38). The concentration of freed 4-nitrobenzaldehyde is subsequently determined by UV-Vis spectroscopy at 267 nm, and the result correlated to the density of amino groups on the chip. Briefly, amine-modified chip were immersed in anhydrous ethanol (5 mL) containing 4-nitrobenzaldehyde (2 mg) and acetic acid (4 μL), and was then incubated in water bath at 50 °C for 3 h. The chips were washed with absolute ethanol by sonication three times for 1 min each, and dried with N2 gas. The resulting imine substrates were hydrolyzed in water (500 μL) containing acetic acid (1 μL) at 30 °C for 1 h, after which, the hydrolysate (400 μL) was subjected to spectrometric analysis on a TU 1900 UV-Vis spectrophotometer. Both samples and standards were measured at 200–350 nm. The calibration curve was generated by plotting the maximal absorbance at 267 nm against 4-nitrobenzaldehyde concentration in the range of 0–100 μM.
Control experiments were conducted at the same time using the same procedure using MUA- or amine-modified chips. The chips were reacted with glutaraldehyde (10% in 20 mM phosphate buffer, pH 7.4) for 1 h, washed with absolute ethanol by sonication for 1 min, three times total, and dried with N2 gas.
Vapor condensation imaging (VCI)
VCI was conducted under a stereo microscope model XTL-500 from Guilin Optical Instrument Factory (Guilin, China) with a charge-coupled device (CCD) camera. Images were recorded after a gentle breath from the operator. The recorded images were subsequently saved and analyzed.
Surface Plasmon Resonance Imaging
CMCs prepared on gold slides were studied using our laboratory-built SPR imager, model ICCAS Most I. Briefly, a collimated beam of p-polarized light was directed, at a fixed angle, toward a prism on which a CMC chip was assembled via a thin layer of index matching fluid and sealed in a flow cell. The reflected light beam filtered through a narrow bandpass filter (λ645 nm) was collected by a CCD camera (WAT-902B, Watec Co., Ltd., Japan) and the images were saved and analyzed with a laboratory-edited imaging workstation. The images were recorded against pH 7.6 Tris buffer (25 mM) containing 1 mM CaCl2, 1 mM MnCl2 and 0.1% Tween 20, which was pneumatically pumped through the cell at a flow rate of 50 μL/min. A Con A solution (pH 7.6 Tris buffer) was introduced into the cell at a flow rate of 50 μL/min. The experiments were carried out at room temperature (about 25 °C).
Preparation of FITC-labeled Con A
FITC-labeled Con A was synthesized according to a previously reported procedure with minor changes (39). Briefly, a solution of Con A in pH 9.5 Tris buffer (50 mM) (5 mg/mL) was mixed with FITC (final concentration: 100 μg/mL) at 20°C for 20 h. The resulting product was dialyzed against pH 7.0 Tris buffer (0.1 M) 3 times for 8 h each. The dialyzed solution was diluted with the pH 7.6 Tris buffer (25 mM) to the appropriate concentration and was used immediately for the subsequent experiments with CMCs.
Laser-induced fluorescence imaging
A CMC prepared on a gold slide was allowed to react with a solution of FITC-labeled Con A in pH 7.6 Tris buffer (20 μg/mL) for 1 h. The chip was washed with the Tris buffer by vortexing for three times, about 1 min each. After drying under a nitrogen stream, fluorescent images were recorded using a Typhoon Trio Variable Mode Imager (Amersham, Biosciences/GE Healthcare) at a pixel size of 25 μm. The fluorescence was excited by 488 nm laser and collected through a 520 nm filter (40 nm band pass).
RESULTS AND DISCUSSION
Coupling intact carbohydrates on PFPA surface
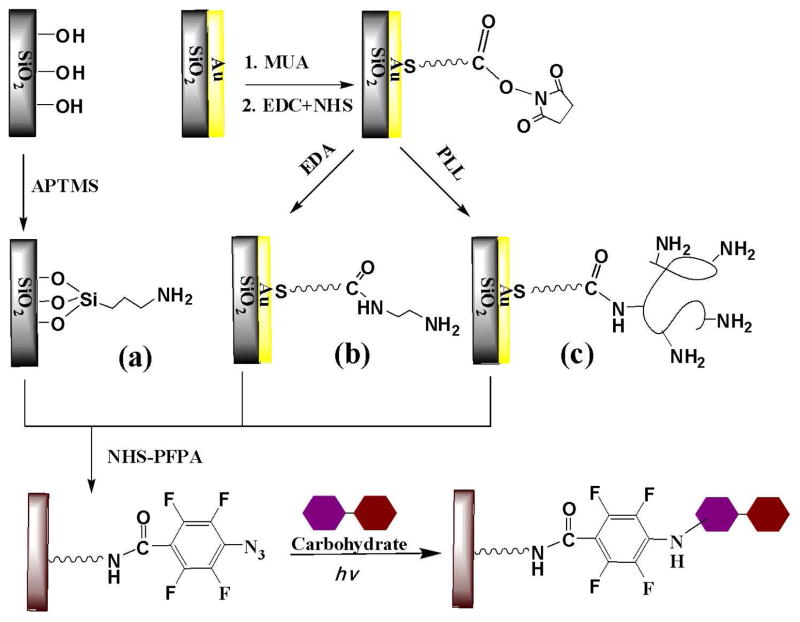
PFPA-functionalized surfaces were generated for the covalent immobilization of intact carbohydrates. A well-established amine-NHS chemistry was adopted (40) whereby amine-based matrices were treated with NHS-PFPA, thus introducing PFPA on the substrate surface. A versatile strategy (Figure 1) was designed and tested, and it could be conveniently adapted to prepare carbohydrate microarrays. For the demonstration of principles, three types of amine matrices were prepared on which PFPA was assembled for further immobilization of intact carbohydrates. The first was APTMS-silanized glass slides, and the second and third were gold films modified with EDA and PLL, respectively.
Figure 1.
Schematic representation of functionalizing three types of amine-based chips with photoactive NHS-PFPA and the subsequent immobilization of intact carbohydrates by UV activation: (a) is APTMS-silanized glass slides. (b) and (c) are gold films modified with EDA and PLL, respectively.
The successful coupling of NHS-PFPA on the surfaces was confirmed by XPS. Since F atoms in PFPA is unique, the F 1s and C 1s from C-F bonds in XPS spectra can serve as the markers to prove the presence of PFPA. Two surfaces, PFPA/EDA/MUA/Au and PFPA/PLL/MUA/Au, were analyzed by XPS. F1s peaks were clearly observed and the concentrations of F atoms on the surface of the two substrates were 2.43% and 1.66%, respectively (for detailed XPS data, see Figure 2S and Table 2S in Supporting Information).
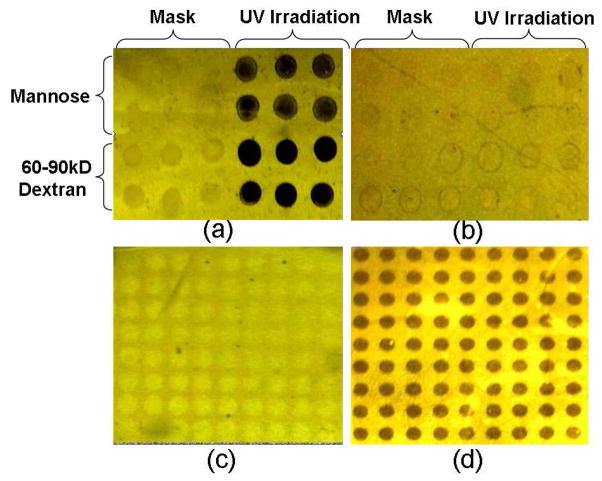
No matter on which type of the amine matrices the PFPA was assembled, the resulting surfaces were all shown to be suitable for photocoupling the intact carbohydrates tested. A simple vapor condensation imaging (VCI) method (41, 42) was used to confirm the immobilization of carbohydrates. VCI is especially useful for testing the presence of carbohydrates as they are highly hydrophilic. Figure 2a shows the VCI image of a CMC prepared by spotting carbohydrates on PFPA/PLL surface followed by irradiating with half of the sample. As expected, spots that were irradiated were highly visible, and in the area that was protected from the irradiation, no obvious spots were observed. To further exclude the possibility that carbohydrates adhered to surfaces by physical adsorption rather than covalent attachment, control CMCs were prepared following the same procedure except that the carbohydrates were spotted directly on the PLL surface without the PFPA layer. In this case, no recognizable carbohydrate spots were obtained in the VCI image (Figure 2b), indicating that the physical or non-specific adsorption is very weak or negligible. An additional control experiment was carried out where the PLL/PFPA surface was irradiated through a photomask without applying the carbohydrates. No obvious patterns were obtained in this case (Figure 2c). However, when mannose was coated on the PLL/PFPA surface before irradiation, dark and clear patterns were observed (Figure 2d), demonstrating that the carbohydrate was responsible for the visible spots shown in the VCI image. These results provided strong evidences that the carbohydrates were indeed covalently immobilized on the substrate, and PFPA was responsible for their strong attachment to the surface.
Figure 2.
Vapor condensation images of carbohydrate microarrays showing the mannose and dextran spots printed on surfaces modified with (a) PLL/PFPA, (b) PLL only. In both cases, the left half of the samples was covered during the irradiation. Irradiation of PFPA/PLL surface in the presence of a photomask without (c) and with (d) mannose.
The most striking advantage of this new method is that ketoses and even alditols, such as sucrose and mannitol, can be readily conjugated to PFPA-functionalized surfaces without prior derivatization of the carbohydrate structures (Figure 3). In addition, small molecules such as glucose, fructose, and lactose can also be efficiently immobilized on the PFPA surface (Figure 3), which is not possible by the physical adsorption method. These results show that the new method is indeed universally applicable, and it is especially appealing for non-reducing carbohydrates where the conventional chemical coupling methods involving pre-derivatization can be highly challenging. Furthermore, the process is fast and efficient. The reaction between surface azido groups and the target molecules occurred in less than 5 minutes of UV illumination with the power intensity of 8 mW/cm2 at 365 nm. Compared with the conventional solid-liquid reactions that take at least several hours to complete (13, 19), the present method is among the fastest in the preparation of carbohydrate array chips.
Figure 3.
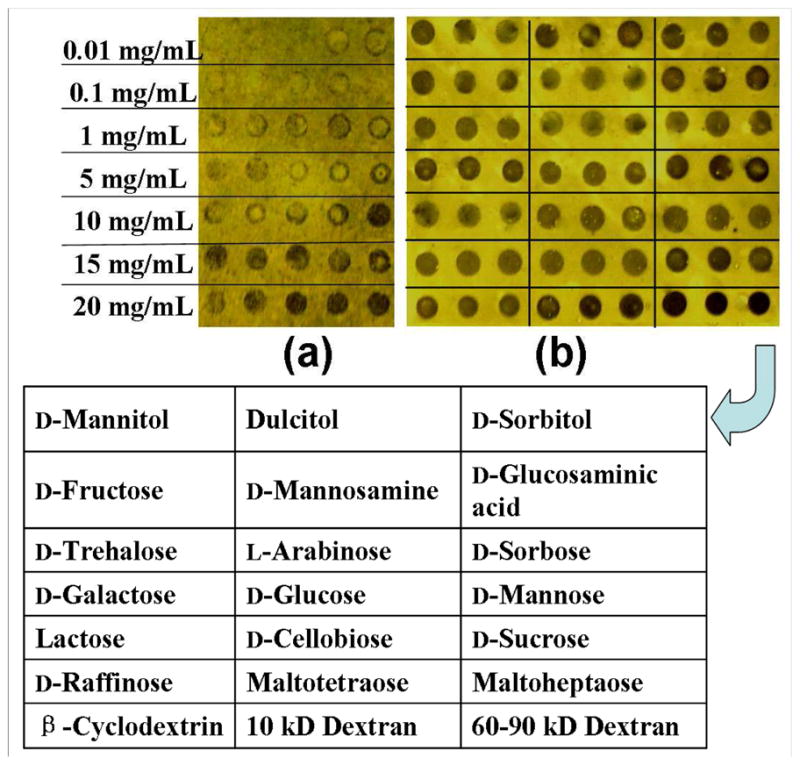
Vapor condensation images of carbohydrate microarray generated on PFPA-functionalized photoactive surface assembled on (a) EDA-based, and (b) EDA/EOA (1:9)-based matrices. The Chip in (a) was printed with a series of maltoheptaose solutions at concentrations of 0.01–20 mg/mL. The chip in (b) contains 21 different carbohydrate samples, each was printed at 5 mg/mL in 3 replicates and the composition of which is shown in the table.
Coupling efficiency
The photocoupling efficiency was fairly high, as demonstrated by the number of immobilized monosaccharide molecules of up to 27.2/nm2 on the PLL-based PFPA surface determined by the anthrone-sulfuric acid method (37). Such a high density of carbohydrates molecules has exceeded the capacity of a two-dimensional surface, likely due to the increased functional group density on the PLL polymer. The amino group density of these two amine matrixes, EDA and PLL, were subsequently determined by UV-vis spectrometry using 4-nitrobenzaldehyde (38). The EDA surface gave an amine density of 8.1/nm2, whereas that of the PLL surface was 4.6 times higher at 37.3/nm2. A higher amine density would attach a larger number of PFPA, which in turn could result in increased number of carbohydrates immobilized.
To determine the immobilization efficiency, maltoheptaose was selected as an example and was spotted on the PFPA-modified surfaces at different concentrations. Figure 3a shows that the density of maltoheptaose spots increased with their printing concentration in the range from 0.01 to 20 mg/mL. However, the spots were not uniform and showed the “ring effect”, likely due to the hydrophobic PFPA surface. EOA-doped matrices were then used because EOA is hydrophilic and should improve the affinity of the resulting surface to carbohydrates. Indeed, the “ring effect” was mostly diminished with the addition of EOA to EDA (Figure 3b). Considering that the arrays were generated on a home-made spotting apparatus, we believe that the spot uniformity can be further improved by using an automated microarrayer. The immobilization efficiency was also enhanced when using EOA-doped matrices. Results in Figure 3b illustrates that EDA/EOA (1:9)-based PFPA surfaces were well-suited for coupling high quality of different carbohydrates, up to at least 21 types tested. In general, a concentration of 5 mg/mL (ca. 28 mM for monosaccharides) is optimal for printing the carbohydrates, which is similar to the value of hydrazide-aldose coupling method where a concentration of 30 mM was found to be the most suitable (19)
Activity and specificity
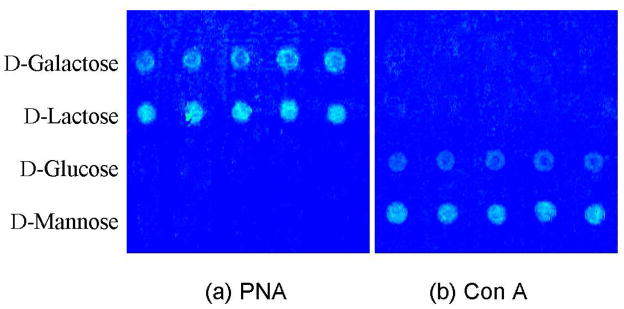
The binding affinity and specificity of the CMCs were next investigated. Because of the non-specific reaction of the azide, possibility exists that the binding sites on the carbohydrates may be altered after the azide insertion and thus impacting their interaction with the binding partners. To investigate whether the immobilized ligands retain their binding affinity and selectivity, SPR chips spotted with PNA-binding carbohydrates, galactose and lactose, and Con A-binding carbohydrates, glucose and mannose, were incubated with PNA or Con A, respectively. As expected, both the immobilized galactose and lactose show clear images after reaction with PNA (Figure 4a, the top two lines), whereas glucose and mannose have no interaction signals at all (Figure 4a, the lower two lines). After the chip was regenerated by phosphoric acid and subjected to Con A treatment, glucose and mannose spots became visible whereas the galactose and lactose spots were not observed. Not only the recognition ability was retained for the immobilized carbohydrates, more importantly, the relative strength of the binding affinity was also preserved. In the SPRi images shown in Figure 4b, mannose exhibited higher intensity than glucose when interacting with Con A. This is consistent with the affinity ranking observed in solution as well as on surfaces reported by others. (43, 44) Similar result was obtained on galactose and lactose when interacting with PNA (Figure 4a) (33).
Figure 4.
SPRi of a carbohydrate microarray after interaction with (a) PNA and (b) Con A. The chip was regenerated with 0.1 M phosphoric acid before treating with the second lectin.
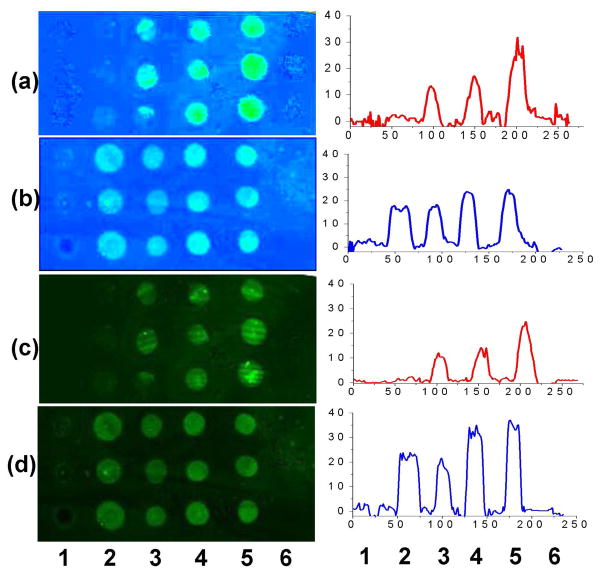
Additional studies were conducted to further confirm that the photocoupling chemistry preserves the affinity differences of the immobilized carbohydrates. A SPR chip containing six intact carbohydrates, monosaccarides (D-galactose and D-mannose), oligosaccharides (maltoheptaose and β-cyclodextrin (β-CD)), and polysaccharides (dextran with molecule weight 10 kD and 60–90 kD, respectively), was prepared. It was then treated with Con A and SPRi images recorded. Results showed that all D-mannose spots immobilized on the PLL-based PFPA surface had strong interaction signals (Figures 5b and 5d, spot 2) whereas the D-galactose spots exhibited negligible signals (Figure 5a–5d, spot 1), which agrees well with well-established literature reports (45). As expected again, the glucose-containing carbohydrates also showed recognition signals that increased with the molecular weight in the order of maltoheptaose, 10 kD and 60–90 kD dextrans, respectively. β-CD had, however, no interaction signal, which agrees with the reported observation of the free β-CD with Con A (46). These data clearly demonstrate that, even though the azide chemistry is not functional group specific, the recognition sites on the carbohydrates are well preserved in terms of both binding affinity and selectivity. Furthermore, the chip can be regenerated without affecting the recognition ability of the immobilized carbohydrates, indicating that this photocoupling chemistry is robust and can benefit their practical uses in the future.
Figure 5.
SPRi (a, b) and LIFi (c, d) of carbohydrates microarray after treating with Con A and FITC-labeled Con A, respectively. The SPRi experiments were conducted after LIFi by washing the sensor chip with 0.1 M phosphoric acid to regenerate the surfaces. Chip (a) is identical to (c) with carbohydrates immobilized on EDA-PFPA surfaces, and (b) and (d) are also identical but with carbohydrates immobilized on PLL-PFPA photoactive surfaces. Numbers 1–6 are D-galactose, D-mannose, maltoheptaose, 10 kD dextran, 60–90 kD dextran and β-cyclodextrin, respectively. The line profiles represent the relative signal intensities of the corresponding sensor chips.
The interaction signal was dependent of lectin concentration (0.1–50 μg/mL) as shown in Figure 6. Although the signals are difficult to differentiate from the background at the concentration lower than 1μg/mL Con A, distinct increases were clearly observed for different sugars, with galactose as a control, as the concentration of Con A increased from 2 μg/mL to 30 μg/mL. The curves gradually plateured after 30–50 μg/mL Con A. These data demonstrated, in another aspect, the preservation of the binding affinity of the immobilized carbohydrates.
Figure 6.
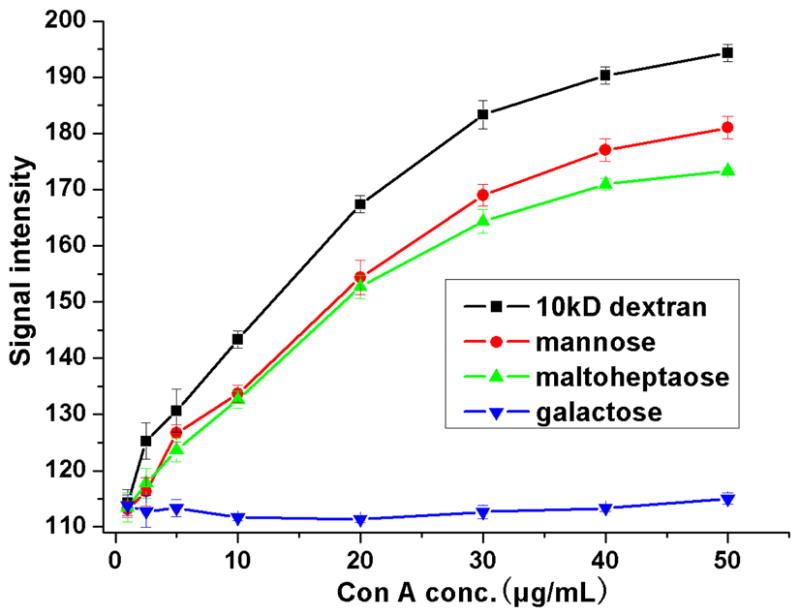
Maximal interaction intensity obtained from SPRi as a function of Con A concentration (0.1–50 μg/mL) on different carbohydrates spots immobilized on PLL-based PFPA sensor chip. Each data was obtained from the average value of 3 replicate spots.
Sensitivity enhancement
Detection sensitivity is another key parameter in the evaluation of sensor performance. The detection sensitivity is dictated by a number of factors, of which the most important are the coupling efficiency of analytes to chip matrices, strength of recognition, and method of detection. Since the coupling efficiency was high by the current method and the recognition system was kept the same (i.e., carbohydrate-Con A interaction) in our studies, the detection sensitivity should be depended on the detection method selected. With maltoheptaose as a test sample, the limit of detection (LOD) was estimated to be 10 μg/mL of the printing concentration for the direct vapor imaging method (Figure 3a). At a maltoheptaose printing concentration of 5 mg/mL, the LOD was determined to be 10 μg/mL Con A for SPRi, and 4 μg/mL FITC-labeled Con A when using LIFi as the detection method. Clearly, LIFi is more than two times more sensitive than SPRi. However, LIFi requires a fluorescently-labeled reagent which usually takes extra effort to prepare. On the other hand, SPRi is a label-free method that uses native proteins without additional chemical derivatization. SPRi is hence a highly attractive sensing technique especially suited for studying biorecognition events.
During the study on the selectivity of newly prepared CMCs, an additional observation was noted, that is, the mannose microdots showed much stronger signals on PLL-based PFPA surfaces (Figures 5b and 5d, No. 2) than on EDA-based PFPA surfaces which showed almost no recognizable signals (Figures 5a and 5c, No. 2). Carroll et al. have also observed that monosaccharides immobilized on phthalimide-based glass substrates could not be recognized by Con A (22). Clearly, the matrix composition and configuration play important role affecting the bioactivity of the resulting carbohydrate ligands. The EDA-modified surface yields a monolayer of amine and subsequently a monolayer of PFPA, and hence the carbohydrates were in turn immobilized as a monolayer. Furthermore, the shorter linkage of EDA creates high steric hindrance preventing the effective interaction of approaching Con A molecules with the surface ligands, and thus producing much weaker signal. PLL, on the other hand, offers multiple layers of amines for anchoring PFPA, and subsequently leads to higher density of the immobilized mannose on PLL than on EDA over the same surface area. In addition, the longer and more flexible PLL reduces steric hindrance when Con A approaches the surface, resulting in increased detection sensitivity. Therefore, surface modification by using flexible polymer or longer spacer is necessary in enhancing the detection limit and improving the signal-to-noise ratio. We expect that the extent of sensitivity enhancement will vary depending on the molecular weight of PLL, which is a subject that warrants further investigation. This amplification effect by PLL was however not observed for polysaccharides which are themselves larger and more flexible for favorable interactions with Con A.
CONCLUSIONS
In summary, a universal method has been established for immobilizing various intact carbohydrates and has demonstrated to be suitable for the fabrication of carbohydrate chips with well-preserved recognition abilities. The key feature of the method is the use of PFPA-modified photoactive surfaces, and the immobilization takes the advantage of solid phase reaction rather than solution reaction. Notably, although the attachment chemistry is not site-specific, the binding assays carried out using both SPRi and LIFi showed that the attached carbohydrates on the arrays retained their binding affinity and specificity. The techniques could be used for both qualitative and quantitative analyses, and the detection sensitivity was comparable with other reported methods. Additionally, by using polymer-based photoactive matrices, interactions between CMCs and their receptors were greatly enhanced, especially for monosaccharides which could not be detected with the monolayer surface. This new platform can be readily applied to a wide range of carbohydrate structures in the construction of CMCs for sensing, ligand screening, and other applications involving carbohydrate recognitions.
Supplementary Material
Acknowledgments
We acknowledge the financial support of this work from NSFC (No. 90717120), MOST (No. 2007CB714504) and CAS (No. KJCX2-YW-H11). M. Yan thanks the financial support from the National Institutes of General Medical Science (NIGMS) under NIH Award Numbers R01GM080295 and R15GM066279.
Footnotes
Supporting Information Available: Methods and calculations for determining lectin LOD, surface composition characterization by XPS and surface concentration of amino groups or carbohydrates by UV-Vis spectroscopy. This material is available free of charge via the Internet at http://pubs.acs.org.
LITERATURE CITED
- 1.Fukui S, Feizi T, Galustian C, Lawson AM, Chai W. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- 2.Disney MD, Seeberger PH. The Use of Carbohydrate Microarrays to Study Carbohydrate-Cell Interactions and to Detect Pathogens. Chem Biol. 2004;11:1701–1707. doi: 10.1016/j.chembiol.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-Throughput Carbohydrate Microarray Analysis of 24 Lectins. Angew Chem Int Ed. 2006;45:3607–3610. doi: 10.1002/anie.200600591. [DOI] [PubMed] [Google Scholar]
- 4.Oyelaran O, Gildersleeve JC. Glycan arrays: recent advances and future challenges. Curr Opin Chem Biol. 2009;13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang CH, Wu CY. Glycan array: a powerful tool for glycomics studies. Expert Rev Proteomics. 2009;6:631–645. doi: 10.1586/epr.09.82. [DOI] [PubMed] [Google Scholar]
- 6.Wu CY, Liang PH, Wong CH. New development of glycan arrays. Org Biomol Chem. 2009;7:2247–2254. doi: 10.1039/b902510n. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Liu S, Trummer BJ, Deng C, Wang A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat Biotechnol. 2002;20:275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- 8.Willats WG, Rasmussen SE, Kristensen T, Mikkelsen JD, Knox JP. Sugar-coated microarrays: a novel slide surface for the high-throughput analysis of glycans. Proteomics. 2002;2:1666–1671. doi: 10.1002/1615-9861(200212)2:12<1666::AID-PROT1666>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 9.Ko KS, Jaipuri FA, Pohl NL. Fluorous-based carbohydrate microarrays. J Am Chem Soc. 2005;127:13162–13163. doi: 10.1021/ja054811k. [DOI] [PubMed] [Google Scholar]
- 10.Karamanska R, Clarke J, Blixt O, Macrae JI, Zhang JQ, Crocker PR, Laurent N, Wright A, Flitsch SL, Russell DA, Field RA. Surface plasmon resonance imaging for real-time, label-free analysis of protein interactions with carbohydrate microarrays. Glycoconj J. 2008;25:69–74. doi: 10.1007/s10719-007-9047-y. [DOI] [PubMed] [Google Scholar]
- 11.Park S, Shin I. Fabrication of carbohydrate chips for studying protein-carbohydrate interactions. Angew Chem Int Ed. 2002;41:3180–3182. doi: 10.1002/1521-3773(20020902)41:17<3180::AID-ANIE3180>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 12.Ratner DM, Adams EW, Su J, O’Keefe BR, Mrksich M, Seeberger PH. Probing protein–carbohydrate interactions with microarrays of synthetic oligosaccharides. ChemBioChem. 2004;5:379–382. doi: 10.1002/cbic.200300804. [DOI] [PubMed] [Google Scholar]
- 13.Houseman BT, Mrksich M. Carbohydrate arrays for the evaluation of protein binding and enzymatic modification. Chem Biol. 2002;9:443–454. doi: 10.1016/s1074-5521(02)00124-2. [DOI] [PubMed] [Google Scholar]
- 14.Mercey E, Sadir R, Maillart E, Roget A, Baleux F, Lortat-Jacob H, Livache T. Polypyrrole oligosaccharide array and surface plasmon resonance imaging for the measurement of glycosaminoglycan binding interactions. Anal Chem. 2008;80:3476–3482. doi: 10.1021/ac800226k. [DOI] [PubMed] [Google Scholar]
- 15.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, van Die I, Burton D, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. J C Proc Natl Acad Sci. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Paz JL, Spillmann D, Seeberger PH. Microarrays of heparin oligosaccharides obtained by nitrous acid depolymerization of isolated heparin. Chem Commun. 2006;7:3116–3118. doi: 10.1039/b605318a. [DOI] [PubMed] [Google Scholar]
- 17.Lee MR, Shin I. Fabrication of chemical microarrays by efficient immobilization of hydrazide-linked substances on epoxide-coated glass surfaces. Angew Chem Int Ed. 2005;44:2881–2884. doi: 10.1002/anie.200462720. [DOI] [PubMed] [Google Scholar]
- 18.Sun XL, Stabler CL, Cazalis CS, Chaikof EL. Carbohydrate and protein immobilization onto solid surfaces by sequential diels-alder and azide-alkyne cycloadditions. Bioconjugate Chem. 2006;17:52–57. doi: 10.1021/bc0502311. [DOI] [PubMed] [Google Scholar]
- 19.Lee MR, Shin I. Facile preparation of carbohydrate microarrays by site-specific, covalent immobilization of unmodified carbohydrates on hydrazide-coated glass slides. Org Lett. 2005;15:4269–4272. doi: 10.1021/ol051753z. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, Zhou J. Oligosaccharide microarrays fabricated on aminooxyacetyl functionalized glass surface for characterization of carbohydrate-protein interaction. Biosens Bioelectron. 2006;21:1451–1458. doi: 10.1016/j.bios.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Angeloni S, Ridet JL, Kusy N, Gao H, Crevoisier F, Guinchard S, Kochhar S, Sigrist H, Sprenger N. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology. 2005;15:31–41. doi: 10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- 22.Carroll GT, Wang D, Turro NJ, Koberstein JT. Photochemical micropatterning of carbohydrates on a surface. Langmuir. 2006;22:2899–2905. doi: 10.1021/la0531042. [DOI] [PubMed] [Google Scholar]
- 23.Carroll GT, Wang D, Turro NJ, Koberstein JT. Photons to illuminate the universe of sugar diversity through bioarrays. Glycoconj J. 2008;25:5–10. doi: 10.1007/s10719-007-9052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keana JFW, Cai SX. New reagents for photoaffinity labeling: synthesis and photolysis of functionalized perfluorophenyl azides. J Org Chem. 1990;55:3640–3647. [Google Scholar]
- 25.Bartlett MA, Yan M. Fabrication of polymer thin films and arrays with spatial and topographical controls. Adv Mater. 2001;13:1449–1451. [Google Scholar]
- 26.Liu LH, Yan M. Simple method for the covalent immobilization of graphene. Nano Lett. 2009;9:3375–3378. doi: 10.1021/nl901669h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, Yan M. A general approach to the covalent immobilization of single polymers. Angew Chem Int Ed. 2006;45:6207–6210. doi: 10.1002/anie.200602097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu LH, Yan M. Perfluorophenyl azides: new applications in surface functionalization and nanomaterial synthesis. Acc Chem Res. 2010;43:1434–1443. doi: 10.1021/ar100066t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Ramström O, Yan M. A Photochemically initiated chemistry for coupling underivatized carbohydrates to gold nanoparticles. J Mater Chem. 2009;19:8944–8949. doi: 10.1039/B917900C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Ramström O, Yan M. Glyconanomaterials: synthesis, characterization, and ligand presentation. Adv Mater. 2010;22:1946–1953. doi: 10.1002/adma.200903908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu LH, Dietsch H, Schurtenberger P, Yan M. Photoinitiated coupling of unmodified monosaccharides to iron oxide nanoparticles for sensing proteins and bacteria. Bioconjugate Chem. 2009;20:1349–1355. doi: 10.1021/bc900110x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Liu LH, Ramström O, Yan M. Engineering nanomaterial surfaces for biomedical applications. Exp Biol Med. 2009;234:1128–1139. doi: 10.3181/0904-MR-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pei Z, Yu H, Theurer M, Waldén A, Nilsson P, Yan M, Ramström O. Photogenerated carbohydrate microarrays. Chembiochem. 2007;22:166–168. doi: 10.1002/cbic.200600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norberg O, Deng L, Yan M, Ramström O. Photo-click immobilization of carbohydrates on polymeric surfaces-a quick and effortless method to functionalize surfaces for biomolecular recognition studies. Bioconjugate Chem. 2009;20:2364–2370. doi: 10.1021/bc9003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frey BL, Corn RM. Covalent attachment and derivatization of poly (L-lysine) monolayers on gold surfaces as characterized by polarization-modulation FT-IR spectroscopy. Anal Chem. 1996;68:3187–3193. [Google Scholar]
- 36.Li Y, Huang J, McIver RT, Hemminger JC. Characterization of thiol self-assembled films by laser desorption Fourier transform mass spectrometry. J Am Chem Soc. 1992;114:2428–2432. [Google Scholar]
- 37.Leyva A, Quintana A, Sánchez M, Rodríguez EN, Cremata J, Sánchez JC. Rapid and sensitive anthrone–sulfuric acid assay in microplate format to quantify carbohydrate in biopharmaceutical products: method development and validation. Biologicals. 2008;36:134–141. doi: 10.1016/j.biologicals.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Moon JH, Kim JH, Kim KJ, Kang TH, Kim B, Kim CH, Hahn JH, Park JW. Absolute surface density of the amine group of the aminosilylated thin layers: ultraviolet-visible spectroscopy, second harmonic generation, and synchrotron-radiation photoelectron spectroscopy study. Langmuir. 1997;13:4305–4310. [Google Scholar]
- 39.The TH, Feltkamp TEW. Conjugation of fluorescein isothiocyanate to antibodies. 1 Experiments on the conditions of conjugation. Immunology. 1970;18:865–873. [PMC free article] [PubMed] [Google Scholar]
- 40.Yan M, Cai S, Wybourne MN, Keana JFW. N-hydroxysuccinimide ester functionalized perfluorophenyl azides as novel photoactive heterobifunctional crosslinking reagents. The Covalent Immobilization of Biomolecules to Polymer Surfaces. Bioconjugate Chem. 1994;5:151–157. doi: 10.1021/bc00026a007. [DOI] [PubMed] [Google Scholar]
- 41.Holmgren J, Svennerholm L, Elwing H, Fredman P, Strannegård O. Sendai virus receptor: proposed recognition structure based on binding to plastic-adsorbed gangliosides. Proc Natl Acad Sci U S A. 1980;77:1947–1950. doi: 10.1073/pnas.77.4.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elwing H, Nilsson LA, Ouchterlony O. A simple spot technique for thin layer immunoassays (TIA) on plastic surfaces. J Immunol Methods. 1977;17:131–145. doi: 10.1016/0022-1759(77)90084-9. [DOI] [PubMed] [Google Scholar]
- 43.Mandal DK, Kishore N, Brewer CF. Thermodynamics of lectin-carbohydrate interactions. Titration microcalorimetry measurements of the binding of N-linked carbohydrates and ovalbumin to concanavalin A. Biochemistry. 1994;33:1149–1156. doi: 10.1021/bi00171a014. [DOI] [PubMed] [Google Scholar]
- 44.Park S, Lee MR, Pyo SJ, Shin I. Carbohydrate chips for studying high-throughput carbohydrate-protein interactions. J Am Chem Soc. 2004;126:4812–4819. doi: 10.1021/ja0391661. [DOI] [PubMed] [Google Scholar]
- 45.Smith EA, Thomas WD, Kiessling LL, Corn RM. Surface plasmon resonance imaging studies of protein-carbohydrate interactions. J Am Chem Soc. 2003;125:6140–6148. doi: 10.1021/ja034165u. [DOI] [PubMed] [Google Scholar]
- 46.Voskuhl J, Stuart MCA, Ravoo BJ. Sugar-decorated sugar vesicles: lectin-carbohydrate recognition at the surface of cyclodextrin vesicles. Chem Eur J. 2010;16:2790–2796. doi: 10.1002/chem.200902423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.