Abstract
Cell–cell communication is critical for tissue and organ development. In plants, secretory CLAVATA3/EMBRYO SURROUNDING REGION-related (CLE) peptides function as intercellular signaling molecules in various aspects of tissue development including vascular development. However, little is known about intracellular signaling pathways functioning in vascular development downstream of the CLE ligands. We show that CLE peptides including CLE10, which is preferentially expressed in the root vascular system, inhibit protoxylem vessel formation in Arabidopsis roots. GeneChip analysis displayed that CLE10 peptides repressed specifically the expression of two type-A Arabidopsis Response Regulators (ARRs), ARR5 and ARR6, whose products act as negative regulators of cytokinin signaling. The arr5 arr6 roots exhibited defective protoxylem vessel formation. These results indicate that CLE10 inhibits protoxylem vessel formation by suppressing the expression of type-A ARR genes including ARR5 and ARR6. This was supported by the finding that CLE10 did not suppress protoxylem vessel formation in a background of arr10 arr12, a double mutant of type-B ARR genes. Thus, our results revealed cross-talk between CLE signaling and cytokinin signaling in protoxylem vessel formation in roots. Taken together with the indication that cytokinin signaling functions downstream of the CLV3/WUS signaling pathway in the shoot apical meristem, the cross-talk between CLE and cytokinin signaling pathways may be a common feature in plant development.
Keywords: CLV3/ESR-related (CLE), cytokinin, signal transduction, protoxylem, Arabidopsis thaliana
Microarray data accession number GSE19978.
Introduction
Plant vascular systems are composed of xylem, phloem and procambium/cambium, which is the vascular stem cell giving rise to xylem and phloem cells. Continuous xylem strands provide pathways for water, nutrients and signaling molecules and also function in mechanical support of the plant body. Xylem vessels are classified into two types, protoxylem vessels with a spiral thickening, and metaxylem vessels with a reticulate and pitted thickening pattern. In Arabidopsis roots, two protoxylem vessels are always located at the outer position of two to four metaxylem vessels. Thus, xylem differentiation is strictly controlled to produce and maintain the correct spatial arrangement of two different vessels in the root vascular system and therefore has provided a paradigm of spatial regulation of plant development (Fukuda 2004, Mähönen et al. 2006).
Plant hormones such as auxins, brassinosteroids and cytokinins are crucial intercellular signaling molecules that control the differentiation and proliferation of vascular cells (Fukuda 2004). Of the plant hormones, cytokinins play crucial and pleiotropic roles in vascular development including enhancement of procambial/cambial proliferation and suppression of protoxylem vessel formation (Mähönen et al. 2006, Yokoyama et al. 2007, Matsumoto-Kitano et al. 2008, Nieminen et al. 2008, Hejátko et al. 2009). It is also known that the regulation of vascular cell fate involves the function of specific components of the two-component system, wherein cytokinin receptors transduce the signal through Arabidopsis histidine phosphotransfer proteins (AHPs) to Arabidopsis response regulators (ARRs) via phosphorylation (Mähönen et al. 2000, Hutchison et al. 2002, Mähönen et al. 2006, Yokoyama et al. 2007).
Screening of extracellular factors using Zinnia cultured cells has revealed a new player, tracheary element differentiation inhibitory factor (TDIF), a dodeca-amino acid secretory peptide (Ito et al. 2006). TDIF acts in local cell–cell communication during vascular development (Hirakawa et al. 2008). TDIF belongs to the CLAVATA3/EMBRYO SURROUNDING REGION-related (CLE) family, which contains 32 members in Arabidopsis (Jun et al. 2008). Indeed, it has been reported that CLE19 functions in early xylem development (Fiers et al. 2004). Furthermore, Whitford et al. (2008) reported that overexpression of some CLE genes (CLE6, CLV3 and CLE19) could cooperate with TDIF to enhance vascular proliferation. Therefore, not only TDIF but also other CLE peptides may play some roles in cell communication involved in vascular development.
In this report, we found a novel function of CLE peptides in vascular development in Arabidopsis roots. Comprehensive analyses of CLE peptides revealed that some CLE peptides, including CLE10, suppressed protoxylem vessel formation, similarly to cytokinin. Microarray analysis and genetic analyses indicated that this suppression by the CLE peptides occurred through activation of cytokinin signaling. These results suggest that there is cross-talk between CLE peptide signaling and cytokinin signaling in protoxylem vessel formation.
Results
CLE peptides causing inhibition of protoxylem development in roots
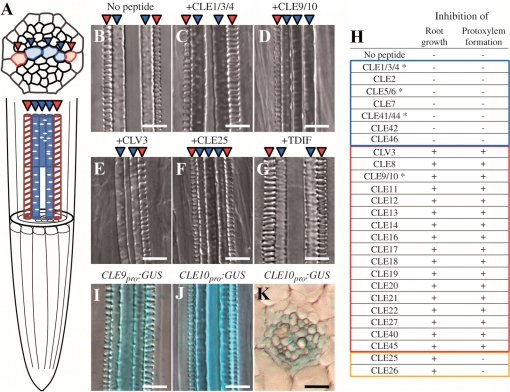
To understand the function of Arabidopsis CLE peptides in vascular development, we examined the effect of exogenously supplied 26 dodeca-peptides on vascular development in the primary root and lateral roots. Xylem in roots contains two different vessels: metaxylem and protoxylem (Fig. 1A). Treatment with some CLE peptides at 1 μM inhibited formation of protoxylem vessels but not of metaxylem vessels, in both the primary root (Fig. 1B–H) and lateral roots (Supplementary Fig. S1). In a series of experiments, defects of vessel formation occurred similarly both in the primary root and lateral roots, although protoxylem vessel formation in lateral roots seems to be affected by a lower concentration of CLE10 than that in the primary roots (e.g. Fig. S1C compared with D). Because of easier and more accurate quantification of vascular defects in the primary root than in lateral roots, we usually used the primary root for evaluating vascular defects. Previous studies have revealed that 19 of 26 Arabidopsis CLE dodeca-peptides inhibit root growth (Ito et al. 2006, Kinoshita et al. 2007). We found that 17 of these 19 CLE peptides inhibited protoxylem vessel formation without affecting metaxylem vessel formation (Figs. 1D, E, H, 2A, B). Interestingly, CLE25 and CLE26, which most severely inhibit root growth (Kinoshita et al. 2007), were much less effective against protoxylem vessel formation (Fig. 1F). These defects were enhanced in a dose-dependent manner as shown by the experiment with CLE9/10 peptide (hereafter referred to as CLE10 peptide), although a control peptide (CLE25) treatment had little effect on protoxylem vessel formation even at the highest concentration (Fig. 3A–D). These results suggest that CLE peptides function in protoxylem formation directly, but not indirectly through a root growth defect. CLE-induced vascular defect occurred in one or both of the two protoxylem vessel cell files. In most cases, one file was almost completely lost (Fig. 1D, E), and the other was partially disrupted (Fig. 2C–E). Interestingly, the seven CLE peptides that do not inhibit root growth, including TDIF (CLE41/44), did not cause defects in protoxylem vessel formation (Fig. 1G, H).
Fig. 1.
Effects of CLE peptides on protoxylem vessel formation and growth in the primary root. (A) A schematic vascular structure in Arabidopsis roots. Red and blue arrowheads represent protoxylem and metaxylem cell files, respectively. (B–H) Arabidopsis seeds were germinated in liquid medium with one of 26 dodeca-amino acid CLE peptides at 1 μM. Seven-day-old seedlings were collected to observe root growth and protoxylem vessel formation in roots. (B–G) Effects on protoxylem vessel formation of no peptide (B), CLE1/3/4 (C), CLE9/10 (D), CLV3 (E), CLE25 (F) and TDIF (G). (H) Twenty-six CLE peptides were classified into three groups (blue, red and orange frames) depending on their effects on the suppression of root growth and protoxylem vessel formation. Asterisks indicate synonymous names for the same peptide. For example, CLE1/3/4 means the same peptide derived from three different CLE1, CLE3 and CLE4 genes. Root growth and protoxylem formation were represented as inhibited (+) or not inhibited (−). (I–K) Vascular-specific expression of CLE9 and CLE10 in the primary root. GUS activity was observed in the root of Arabidopsis seedlings harboring a chimeric gene of the promoter of a CLE gene and the GUS gene. (I) CLE9pro:GUS. (J) CLE10pro:GUS. (K) is a cross-section of (J). Scale bars are 25 μm.
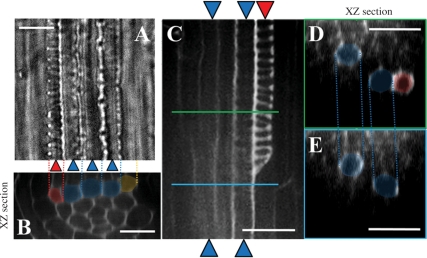
Fig. 2.
Confocal sections of the root vasculature. Confocal images of the protoxylem vessels and metaxylem vessels were taken from the mPS–PI-stained primary root of seedlings treated with 1 μM CLE10. (A) A bright field image. (B) A radial optical section of (A), which was constructed from Z-stack images by using image J (http://rsbweb.nih.gov/ij/). (C) A longitudinal image of a partially disrupted protoxlem vessel. (D, E) Radial optical sections of (C) at the green line position (D) and at the blue line position (E). Red and blue indicate protoxylem and metaxylem vessels, respectively. Orange indicates a cell where differentiation into protoxylem vessels has been inhibited. Scale bars are 25 μm.
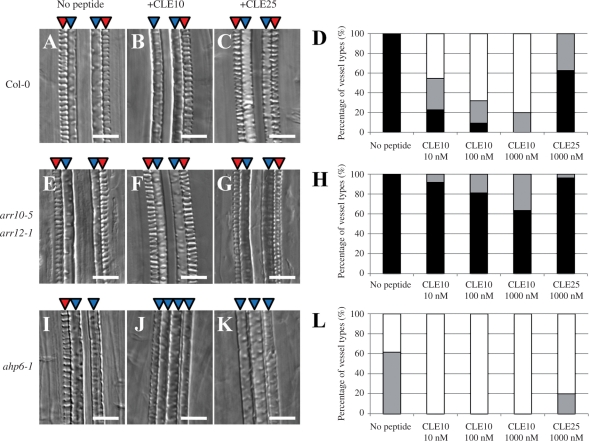
Fig. 3.
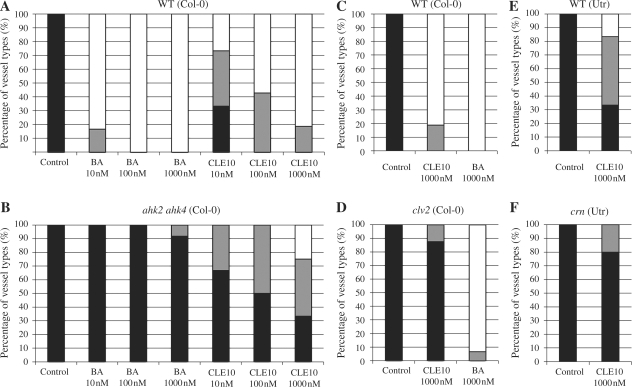
CLE10 peptide-dependent suppression of protoxylem vessel formation in mutants defective in cytokinin signaling. (A–C, E–G, I–K) The WT, arr10-5 arr12-1 and ahp6-1 seedlings were grown for 7 d on agar plates (with 0.8% agar) containing no peptide, 1 μM CLE10 and 1 μM CLE25. (D, H, L) Dose-dependent suppression of protoxylem vessel formation by CLE10. The WT (D), arr10-5 arr12-1 (H) and ahp6-1 (L) seedlings were grown for 7 d on plates with no peptide, 10–1000 nM CLE10 and 1000 nM CLE25, and classified into three types depending on the phenotype of protoxylem vessels in the primary root. Black represents normal protoxylem vessel formation where two protoxylem cell files differentiated correctly. Gray represents a weak abnormal phenotype showing that one of the two protoxylem cell files was missing or interrupted. White represents a severe abnormal phenotype showing that both protoxylem cell files were missing or interrupted. (N = 19–30). Scale bars are 25 μm.
Vascular-specific expression of CLE9 and CLE10 genes in roots
To determine which CLE peptides function in protoxylem differentiation in situ, we screened for CLE genes involved in vascular development based on expression data of the Arabidopsis in vitro xylogenic culture system (see expression data in Kubo et al. 2005). We found that transcripts for two CLE genes (CLE10 and CLE26) increased in the early stages of xylem differentiation. Because CLE26 peptide was not involved in protoxylem vessel formation as aforementioned, we focused on the CLE10 gene. We then examined the expression pattern of CLE10 in Arabidopsis seedlings harboring a chimeric gene of its promoter and the β-glucuronidase (GUS) reporter gene (CLE10pro:GUS). We also analyzed the expression of CLE9pro:GUS, because CLE9 encodes the same CLE dodeca-peptide as CLE10 (Ito et al. 2006). Both genes exhibited stele-specific expression in roots (Fig. 1I, J), in particular in the differentiation zone (DZ) and the elongation zone (EZ) near the root basal meristem (RBM) (Supplementary Fig. S2A, B). Transverse sections of roots revealed ubiquitous expression of CLE10 in the stele (Fig. 1K).
CLE10 peptide regulates protoxylem vessel formation
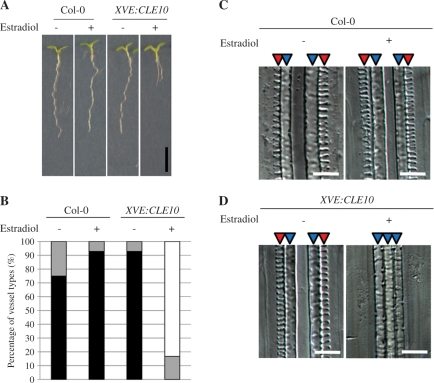
Some CLE peptides take their active form after post-translational glycosylation (Ohyama et al. 2009). Therefore the result of exogenous CLE10 peptide treatment might not reflect the function of endogenous CLE10 peptide. To examine whether the CLE10 gene is capable of inhibiting protoxylem vessel formation in vivo, we overexpressed CLE10 using an estradiol-inducible system (Curtis and Grossniklaus 2003). In the presence of estradiol, these transgenic plants exhibited the same protoxylem vessel defect as plants treated with CLE10 peptide (Fig. 4). These results strongly suggest that chemically synthesized CLE10 dodeca-peptide reflects the function of endogenous CLE10 peptide in the inhibition of protoxylem vessel formation.
Fig. 4.
Effects of CLE10 overexpression on root growth and protoxylem vessel formation. The WT (Col-0) and XVE:CLE10-harboring seedlings were grown on agar plates (with 1.5% agar) for 7 d with (+) or without (−) 5 μM estradiol. (A) Inhibition of root growth by overexpression of CLE10. Scale bar is 5 mm. (B) Inhibition of protoxylem vessel formation by overexpression of CLE10. Black, gray and white bars are explained in Fig. 3 (N = 12–14). (C, D) Images of root vessels in the WT (C) and XVE:CLE10-harboring (D) seedlings, when treated with and without estradiol. Scale bars are 25 μm.
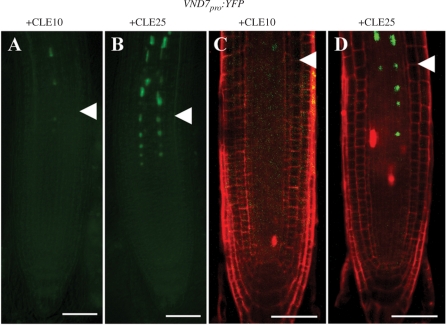
Protoxylem vessel formation is initiated by VND7, a master transcription factor (Kubo et al. 2005). To reveal whether CLE10 peptide affects the initiation step of protoxylem vessel formation or later stages of the differentiation, we examined the effect of CLE10 on the expression of VND7pro:YFP in roots. The fluorescence signal of VND7pro:YFP was detected in two cell files in the RBM and EZ in this line, as previously reported (Yamaguchi et al. 2008). Interestingly, CLE10 treatment reduced expression levels of VND7pro:YFP in the RBM when compared with CLE25 treatment (Fig. 5A–D). These results suggest that CLE10 inhibits an early stage of protoxylem development.
Fig. 5.
Inhibition of VND7pro:YFP expression by CLE10 peptide. (A–D) Seedlings were grown for 5 d in liquid medium with 1 μM CLE10 (A, C) or CLE25 (B, D). Fluorescence from VND7pro:YFP was detected in the non-stained primary root under a fluorescence microscope (A, B), and in the PI-stained primary root under a confocal microscope (C, D). White arrowheads represent the boundary between the RBM and the EZ. Scale bars are 50 μm.
Interaction between cytokinin and CLE10 signaling in regulating protoxylem differentiation
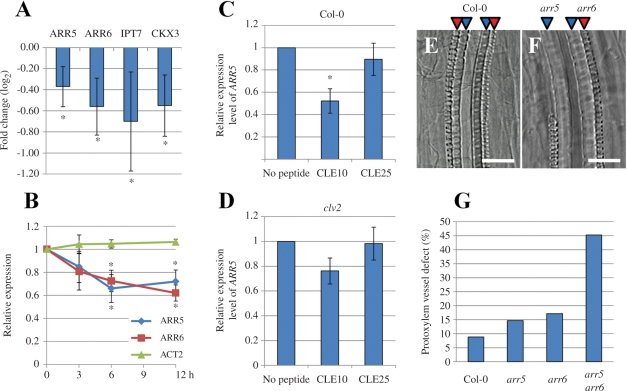
It has been reported that cytokinin also inhibits protoxylem vessel differentiation (Mähönen et al. 2006, Yokoyama et al. 2007). To examine the interaction between cytokinin and CLE10 signaling, we performed a GeneChip analysis of transcripts from CLE10 peptide-treated roots with the Arabidopsis 23K chip. CLE25 peptide treatment was used as a control, because it repressed root growth without affecting protoxylem vessel formation. A subset of the genes differentially expressed between CLE10- and CLE25-treated seedlings should be involved in the regulation of protoxylem vessel formation. The GeneChip analysis has uncovered that only four genes, ARR5, ARR6, adenylate isopentenyltransferase 7 (IPT7) and cytokinin oxidase 3 (CKX3) are significantly and reproducibly decreased upon CLE10 treatment (Fig. 6A, Supplementary Table S1). However, CLE10 treatment upregulated significantly no cytokinin-related genes. Both ARR5 and ARR6 belong to type-A ARRs, which act as negative regulators in cytokinin signaling (To et al. 2007). On the other hand, IPT7 and CKX3 act in cytokinin synthesis and degradation, respectively (Kakimoto 2001, Mok and Mok 2001, Takei et al. 2001). These results suggest that CLE10 may modulate endogenous cytokinin levels and/or cytokinin signaling by affecting the expression levels of cytokinin-related genes.
Fig. 6.
Cross-talk between type-A ARRs and CLE10 signaling in protoxylem vessel formation. (A, B) Reduction of transcript levels of cytokinin-related genes including type-A ARRs by CLE10 peptide. Arabidopsis seedlings were grown in liquid medium containing 1 μM CLE10 or 1 μM CLE25 for 3–12 h, and transcript levels of cytokinin-related genes were measured by GeneChip (A) or with quantitative PCR (B). (A) Expression levels of four genes (ARR5, ARR6, IPT7 and CKX3) were significantly decreased by 12 h treatment of CLE10. The fold change was calculated as the ratio of transcript levels of plants treated with CLE10 compared with CLE25 (in log2 scale). Asterisks indicate statistically significant differences (P < 0.1). (B) Changes in two type-A ARR transcripts by 3, 6 and 12 h treatment of 1 μM CLE10 or CLE25. ACT2 was used as a control. The relative expression was calculated as the ratio of transcript levels of plants treated with CLE10 when compared with CLE25. Asterisks indicate statistically significant differences (P < 0.01). (C, D) ARR5 expression levels of plants treated with no peptide, 1 μM CLE10 or 1 μM CLE25 for 6 h in the WT (C) or clv2 (D). The relative expression was calculated as the ratio of transcript levels of plants treated with CLE peptides when compared with no peptide. Asterisks indicate statistically significant differences when compared with no peptide treatment (P < 0.01). (A–D) Error bars represent SD (N = 3). (E–G) Defects in protoxylem vessel formation in lateral roots. (E, F) Protoxylem vessel formation in lateral roots of 8-day-old WT and arr5 arr6 seedlings. (G) The frequency of suppressed protoxylem vessel formation in lateral roots of WT, arr5, arr6, arr5 arr6 (N = 32–53). One to four lateral roots located next to the hypocotyl, per seedling, were used for measurement. Scale bars are 25 μm.
The ahk2 ahk4 mutant is sensitive to CLE10 in protoxylem vessel formation
Cytokinins are perceived by three Arabidopsis histidine kinase (AHK) receptors, and upon activation the signal is transduced by a phosphorelay to downstream components (Inoue et al. 2001, Suzuki et al. 2001, Ueguchi et al. 2001, Yamada et al. 2001). To investigate whether an increase in cytokinin levels by CLE10 contributes to the protoxylem vessel defects, we examined the effect of CLE10 peptide on protoxylem vessels in a cytokinin receptor mutant. The ahk2 ahk4 mutant was sensitive to the effects of CLE10 on protoxylem vessel formation when compared with the effects of benzyladenine (BA), while the wild type (WT) showed high sensitivity to both CLE10 and BA (Fig. 7A, B). In addition, this inhibition by CLE10 in the double mutant occurred in a dose-dependent manner, although the inhibition was not as severe as that in the WT (Fig. 7A, B). These results suggest that CLE10 may modulate not endogenous cytokinin levels but intracellular cytokinin signaling.
Fig. 7.
Effects of BA and CLE10 peptide on protoxylem vessel formation in ahk2 ahk4, clv2 and crn. (A, B) Seedlings of WT (A) and ahk2 ahk4 (B) were grown in liquid medium containing BA and CLE10 at 10, 100 or 1000 nM for 7 d. (C–F) Seedlings of WT (Col-0) (C), clv2 (D), WT (Utr) (E) and crn (F) were treated with no peptide, BA or CLE10 at 1000 nM for 7 d. Black, gray and white bars are explained in Fig. 3 (N = 12–18).
Expression levels of type-A ARRs are rapidly decreased by CLE10 treatment
Next, we examined the role of the type-A ARRs. Because type-A ARRs act as negative regulators of cytokinin signaling (Kiba et al. 2003, To et al. 2004, To et al. 2007, Ren et al. 2009), the repression of type-A ARRs by CLE10 may lead to the activation of cytokinin signaling and consequently inhibit protoxylem differentiation. We quantified the effect of CLE10 on the expression of type-A ARRs. Transcript levels of two type-A ARRs (ARR5 and ARR6) were significantly decreased by CLE10 (Fig. 6B; P < 0.01), which is consistent with the GeneChip analysis. This decrease occurred as early as 6 h after CLE10 treatment (Fig. 6B). In contrast, CLE25 treatment did not alter transcript levels of ARR5 when compared with the result from the treatment without peptide (Fig. 6C).
To investigate whether the decreased expression of these two type-A ARRs (ARR5 and ARR6) by CLE10 contributes to the protoxylem vessel defects, we observed the primary root and lateral roots of the single arr5 or arr6 null mutant and the arr5 arr6 double null mutant. Each single mutant exhibited no obvious defect in protoxylem vessel formation in the primary root and lateral roots (Fig. 6E–G). However, the arr5 arr6 double mutant occasionally displayed the loss of protoxylem vessels in lateral roots but not in the primary root (Fig. 6E–G, Supplementary Fig. S1). In addition, arr5 arr6 was more sensitive to CLE10 than the WT in protoxylem vessel formation in both the primary root and lateral roots (Supplementary Fig. S1). These results suggest that ARR5 and ARR6 may be downstream components of CLE10 signaling to cause protoxylem vessel defects, although another factor (or factors), probably another type-A ARR(s) is also involved in its signaling.
CLE10 causing enhancement of cytokinin signaling suppresses protoxylem vessel formation
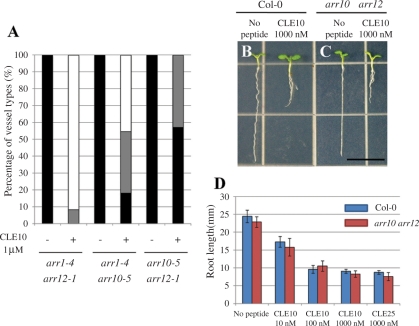
Type-B ARRs are transcription factors that act as positive elements in cytokinin signaling to mediate cytokinin-regulated gene expression, including the transcriptional induction of type-A ARRs in response to cytokinin (Yokoyama et al. 2007). Because type-A ARRs and type-B ARRs probably compete with each other for phosphotransfer (To et al. 2004), the repression of type-A ARR expression by the CLE10 peptide may activate type-B ARR activities, which will in turn cause cytokinin- dependent suppression of protoxylem vessel formation. Two type-B ARRs, ARR10 and ARR12, are key transcription factors mediating the effect of cytokinin on root vascular development (Argyros et al. 2008). Cytokinin does not inhibit protoxylem vessel differentiation in an arr10-5 arr12-1 double null mutant (Yokoyama et al. 2007, Argyros et al. 2008). We then examined the effect of CLE10 on protoxylem vessel formation in the arr10-5 arr12-1 double mutant. Similarly, CLE10 did not suppress protoxylem vessel formation in the arr10-5 arr12-1 mutant roots (Fig. 3E–H). Other type-B ARR double mutants (arr1-4 arr10-5 and arr1-4 arr12-1) showed weaker resistance to CLE10 in protoxylem vessel formation (Fig. 8A). In contrast, CLE10 peptide still inhibited root growth in arr10-5 arr12-1 (Fig. 8B–D), suggesting that root growth suppression by CLE10 is independent of these type-B ARRs. These results indicate that ARR10 and ARR12 are necessary for the CLE10-mediated suppression of protoxylem formation. Furthermore, these results are consistent with the notion that CLE10 peptide inhibits protoxylem vessel formation by activating cytokinin signaling through repression of the expression of type-A ARRs.
Fig. 8.
Different sensitivities to CLE10 among three type-B ARR double mutants. (A) Seedlings of arr1 arr12, arr1 arr10 and arr10 arr12 were grown in liquid medium with or without 1 μM CLE10 for 7 d, and the numbers of primary roots with defects in protoxylem vessels was counted. Black, gray and white bars are explained in Fig. 3 (N = 7–12). (B–D) CLE10-induced root growth inhibition in arr10 arr12. The WT and arr10 arr12 seedlings were grown for 7 d on agar plates (with 1.5% agar) containing no peptide and 10–1000 nM CLE10. (A, B) Seedlings of the WT (A) and arr10 arr12 (B) grown with no peptide (left) and with 1 μM CLE10 (right). (C) Root length in WT and arr10 arr12 seedlings. Error bars represent SD (N = 7–11).
CLE10 activates cytokinin signaling via an AHP6-independent pathway
Mähönen et al. (2006) reported that the ahp6-1 mutation inhibits protoxylem vessel formation, and the pseudophosphotransfer protein AHP6, which interferes with phosphotransfer from AHKs to AHPs in cytokinin signaling, promotes the formation of protoxylem vessel. To more closely examine the relationship between cytokinin and CLE10 signaling, we tested the effect of CLE10 on protoxylem vessels in the ahp6-1 mutant. CLE10 peptide enhanced suppression of protoxylem vessel formation in ahp6-1 (Fig. 3I–L), suggesting that CLE10 signaling acts at least partially independently of AHP6- mediated cytokinin signaling pathways.
CLE10 inhibits protoxylem vessel formation via CLV2
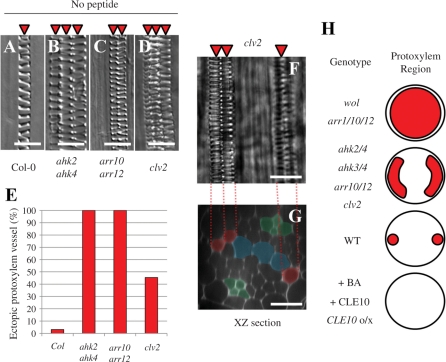
Plants defective in the receptor for the CLE10 peptide should be insensitive to exogenous CLE10. Therefore, we analyzed the effect of CLE10 on protoxylem vessel formation in a series of T-DNA insertion lines of the CLE receptor, CLV2, and many members of subclass XI of the leucine-rich repeat receptor-like kinase (LRR-RLK) family, including CLV1 and TDIF receptor (TDR/PXY) (Fisher and Turner 2007, Hirakawa et al. 2008, Butenko et al. 2009). Of these, only the clv2 null mutant exhibited insensitivity to CLE10 in the suppression of protoxylem vessel formation (Fig. 7C, D). Indeed, ARR5 expression level in the clv2 mutant was also resistant to CLE10 (Fig. 6C, D). Interestingly, in clv2 roots, the ectopic protoxylem vessels were formed in the DZ where metaxylem vessels had not yet differentiated (Fig. 9D–G). The cytokinin receptor mutant ahk2 ahk4 and type-B ARR mutant arr10 arr12 also produced ectopic protoxylem vessels, although these mutants produced them more frequently than the clv2 mutant (Fig. 9A–E). These results indicate that endogenous CLV2 functions in the suppression of protoxylem vessel formation and mediates CLE10 signaling.
Fig. 9.
Ectopic protoxylem vessel formation in the primary root of mutants. (A–D) Protoxylem vessel formation in the WT (A), ahk2 ahk4 (B), arr10 arr12 (C) and clv2 (D). Arabidopsis roots exhibit diarch symmetry of the vascular bundle in which a protoxylem vessel file per arch is formed. However, ahk2 ahk4, arr10 arr12 and clv2 roots often have two or three protoxylem vessel files per arch. (E) The frequency of the primary root with ectopic protoxylem vessel files in the WT, ahk2 ahk4, arr10 arr12 and clv2 (N = 32–33). (F, G) Confocal images of the primary root of clv2. (F) A bright field image of the mPS–PI-stained stele. (G) An optical section of (F). Red and green indicate protoxylem vessels and phloem cells, respectively. Blue indicates undifferentiated metaxylem vessel cells. (H) Protoxylem vessel phenotypes in the primary root caused by loss-of-function and gain-of-function of CLE and cytokinin signaling. Circles show the root stele. Red indicates the area that protoxylem vessel cells occupy. CLE10o/x shows plants in which CLE10 is overexpressed. Scale bars are 25 μm.
CLV2 cannot function alone as a receptor kinase, because of lack of an intracellular kinase domain. It has been proposed that in the CLV3 signaling pathway, a protein kinase CORYNE (CRN)/suppressor of LLP2 (SOL2), which carries a transmembrane domain and a short extracellular domain, possibly functions as a heterodimer with CLV2 in the shoot apical meristem (SAM) (Müller et al. 2008, Miwa et al. 2008, Bleckmann et al. 2010, Zhu et al. 2010). Therefore we examined the CLE10 sensitivity of the sol2-1 mutant. This mutant showed insensitivity to exogenous CLE10 in protoxylem vessel formation (Fig. 7E, F). Because CRN/SOL2 is expressed in the root stele (Müller et al. 2008), CLV2 may function together with CRN in the CLE10 signaling pathway regulating protoxylem development, as in the CLV3 signaling pathway in SAM.
Discussion
Novel function of CLE peptides in vascular development
In this study, we found that CLE peptides including CLE10 inhibited protoxylem vessel formation in roots as well as root growth, while CLE25 and CLE26 repressed root growth without inhibiting protoxylem vessel formation. However, TDIF, which inhibits vessel differentiation in leaves and hypocotyls (Ito et al. 2006), did not affect protoxylem vessel formation in roots or root growth. These results suggest that the machinery underlining vessel development may differ between roots and leaves/hypocotyls. Further comprehensive analyses indicated that the CLE10-dependent suppression of protoxylem vessel formation in roots is an important regulatory machinery, which activates cytokinin signaling, and is not the secondary effect of the repression of root growth.
Type-A ARRs including ARR5 and ARR6, positively regulate protoxylem vessel formation in the CLE10 signaling pathway
The analysis with GeneChip and quantitative real-time reverse transcription (RT)–PCR revealed that CLE10 treatment significantly reduced transcripts of ARR5 and ARR6, which share high similarity and function as negative regulators in cytokinin signaling (Kiba et al. 2003, To et al. 2004, To et al. 2007, Ren et al. 2009). In lateral roots of a double mutant but not of single mutants of arr5 and arr6, protoxylem vessel formation was strongly inhibited, indicating that ARR5 and ARR6 function in the promotion of protoxylem vessel formation redundantly. This result is consistent with the idea that the downregulation of ARR5 and ARR6 by CLE10 peptide causes the suppression of protoxylem vessel formation. However, the facts that the suppression of protoxylem vessel formation in arr5 arr6 is weaker than that in CLE10-treated plants and that protoxylem vessel defects in arr5 arr6 are not observed in the primary root suggest the contribution of other type-A ARRs to protoxylem vessel formation.
CLE10 has little effect on the suppression of protoxylem vessel formation in the double type-B ARR mutant, arr10 arr12, which supports the idea that CLE10 regulates protoxylem vessel formation through cytokinin signaling. ARR1, ARR10 and ARR12 are major members of the type-B ARR family, which function as positive transcription factors in cytokinin signaling (Mason et al. 2005). Indeed, the triple mutant arr1 arr10 arr12 shows high resistance to exogenous cytokinin (Argyros et al. 2008, Ishida et al. 2008). Our results indicated that arr10 arr12 was the most resistant of all type-B ARR double mutants (arr1 arr10, arr1 arr12 and arr10 arr12) against CLE10 in terms of the inhibition of protoxylem development. This is consistent with the observation that the cytokinin-causing vascular defect is overcome more effectively in arr10 arr12 than in arr1 arr12 and arr1 arr10 (Argyros et al. 2008). Taken together with the fact that ARR10 and ARR12 are expressed in the root stele (Mason et al. 2004, Yokoyama et al. 2007), we present a model that cross-talk between CLE10 peptide and cytokinin signaling via type-B ARRs (ARR10 and ARR12) and type-A ARRs (ARR5 and ARR6) regulates protoxylem vessel development in roots (Fig. 10).
Fig. 10.
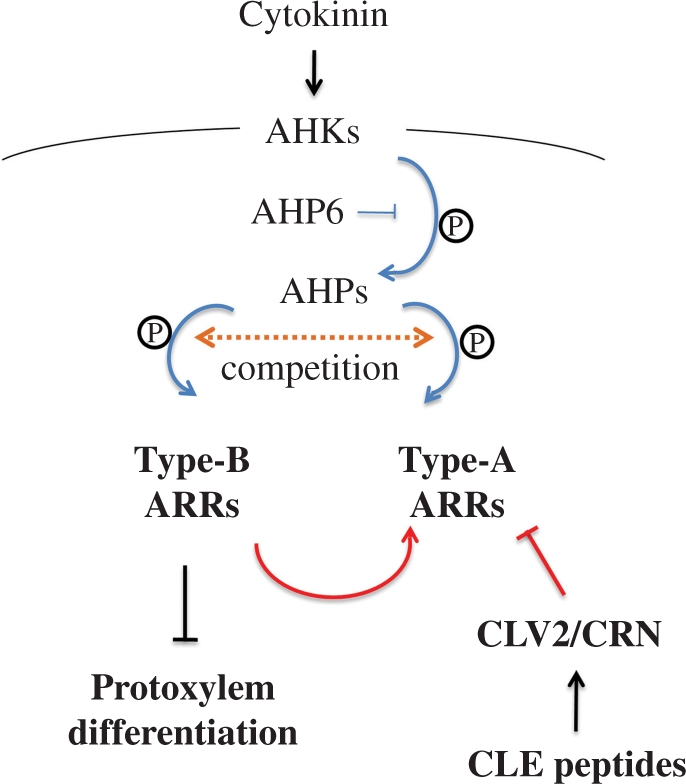
A model illustrating the cross-talk between cytokinin and CLE peptide signaling pathways to suppress protoxylem vessel formation. CLE peptides positively regulate cytokinin signaling via type-A ARRs in protoxylem vessel formation. Blue and red lines indicate phosphorylational and transcriptional regulation, respectively.
CLE peptides and cytokinins may suppress the initiation of protoxylem vessel formation in the root meristem
Generally, the initiation of protoxylem vessel formation occurs in the root meristem (Carlsbecker et al. 2010). Most of the genes encoding signaling components that we found as key regulators of protoxylem vessel formation (CLV2, CRN, ARR5, ARR6, ARR10 and ARR12) were expressed in the RBM (Supplementary Fig. S2C–I). Furthermore, the CLE10 peptide could suppress the expression of VND7, which directs protoxylem differentiation, in the RBM. Considering that CLE9 and CLE10 genes are expressed in vascular tissues of the EZ near the RBM (Supplementary Fig. S2A, B), CLE10 peptide might move down to the RBM and inhibit protoxylem vessel formation in the procambium, together with other factors such as ARRs and CLV2.
Ectopic protoxylem vessel formation in clv2 and some mutants of cytokinin-related genes occurred in procambial cells adjacent to original protoxylem vessels, but not in metaxylem vessel cell files (Fig. 9A–G). This ectopic differentiation from procambial cells is also observed in ahk3 ahk4 (Mähönen et al. 2006). In more severe cytokinin mutants such as woodenleg (wol) and a type-B ARR triple mutant, not only procambial cells adjacent to original protoxylem vessels but also most procambial and metaxylem precursor cells differentiate into protoxylem vessels (Fig. 9H) (Yokoyama et al. 2007, Ishida et al. 2008). In contrast, the supply of cytokinin and CLE10 and CLE10 overexpression reduced the number of protoxylem vessels. Therefore, it is probable that basically cytokinin signaling inhibits protoxylem vessel formation in the whole root stele and CLE peptide signaling modulates cytokinin signaling locally (Fig. 9H).
Relationships between CLE peptides and type-A ARRs
Similar cross-talk between CLE signaling and type-A ARRs has been reported in SAM maintenance, in which the transcription factor WUSCHEL (WUS) acts downstream of CLV3 signaling (Schoof et al. 2000). WUS directly represses the expression of several type-A ARRs (ARR5, ARR6, ARR7 and ARR15) in SAM (Leibfried et al. 2005). These suggest that CLE peptides may commonly regulate cytokinin signaling through type-A ARRs during plant development, although signaling pathways of many other CLE peptides remain to be elucidated. WUS also upregulates CLV3 expression in SAM maintenance through a negative feedback (Brand et al. 2002). In crown root development, OsWOX11 (WUS-related HOMEOBOX), which is a rice homolog of Arabidopsis WOX11, directly represses RR2, a type-A response regulator gene (Zhao et al. 2009). However, our GeneChip analysis revealed that no CLE or WOX gene was induced by CLE10 peptide application. Therefore, although some type-A ARRs may play a role as downstream factors of CLE signaling pathways, the mechanisms by which type-A ARRs are induced may vary.
Materials and Methods
Plant materials and growth conditions
All Arabidopsis thaliana plants other than sol2-1 (Utr background) were of Col-0 background. Arabidopsis seeds were grown on 1/2 Murashige and Skoog (MS) agar plates or liquid medium with or without CLE peptides at 22°C. CLE peptides were obtained and stored as described previously (Ito et al. 2006). ahp6-1 seeds were a gift from Dr Ykä Helariutta (Helsinki University) (Mähönen et al. 2006), arr1-4 arr10-5, arr1-4 arr12-1 and 10-5 arr12-1 seeds from Dr Takafumi Yamashino (Nagoya University) (Yokoyama et al. 2007), clv2 seeds (GABI_686A09) from Dr Sumie Ishiguro (Nagoya University) (Rosso et al. 2003), ahk2 ahk4 (ahk2-2tk cre1-12) seeds from Dr Tatsuo Kakimoto (Osaka University) (Mähönen et al. 2006), sol2-1 seeds from Dr Shinichiro Sawa (Miwa et al. 2008) and VND7pro:YFP seeds from Dr Kyoko Ito-Ohashi. Type-A ARR mutants were described previously (To et al. 2004).
To generate the CLE promoter:GUS reporter constructs, we used an approximately 1–2 kb DNA fragment upstream of the predicted start codon of each gene. Primer sets used are listed in Supplementary Table S2. Expression of the CLE promoter:GUS was observed in T2 or T3 plants. GUS staining was performed according to a previous protocol (Nagawa et al. 2006).
The CLE10 coding sequence was cloned into the pMDC7 vector (Curtis and Grossniklaus 2003) to generate XVE:CLE10, and introduced into Arabidopsis plants. The phenotype was observed with T3 plants. Primer sets used are listed in Supplementary Table S2.
Observation of vasculature
Roots were fixed in a 1:3 mixture of acetic acid/ethanol and mounted in a mixture of chloral hydrate/glycerol/water (8:1:2) and observed under a light microscope (BX51; Olympus, Tokyo, Japan). Sectioning was performed as previously described (Hirakawa et al. 2008).
mPS–PI staining
Modified pseudo-Schiff–propidium iodide (mPS–PI) staining was performed as previously described (Truenit et al. 2008). Samples were observed under an inverted fluorescent microscope (IX70; Olympus) equipped with a confocal unit (CSU10; Yokogawa, Tokyo, Japan).
Microarray analysis
We treated 7-day-old seedlings with 1 μM CLE10 or CLE25 (a control peptide) for 12 h. After treatment, total RNA was extracted from about 100 root tips (5 mm in length). RNA extraction and purification were performed as described previously (Miwa et al. 2008). GeneChip analyses were independently performed three times with the Arabidopsis ATH1 Genome Array (Affymetrix, Santa Clara, CA, USA) as described in the GeneChip Expression Analysis Technical Manual. Comparison analysis of samples treated with CLE25 and CLE10 was performed with the GeneChip Operating Software.
Real-time PCR
Total RNA was extracted from whole main roots of 7-day-old seedlings treated with mock or CLE peptides. Quantitative PCR was performed independently three times by Light Cycler (Roche Diagnostics, Indianapolis, IN, USA). As the internal control, the tubulin α4 gene was used. Gene-specific primer sets used are listed in Supplementary Table S2.
Supplementary Data
Supplementary data are available at PCP Online.
Funding
This work was supported in part by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan, Japan (1906009 to H.F.), the Japan Society for the Promotion of Science (20247003 to H.F., JSPS Research Fellowships for Young Scientists to Y.K. and Y.H.) and the Program of Basic Research Activities for Innovative Biosciences from Bio-oriented Technology Research Advancement Institution to H.F.
Acknowledgments
We thank Takafumi Yamashino and Takeshi Mizuno (Nagoya University) for critical discussion; Kuninori Iwamoto and Sayaka Isu for technical support; and Kyoko Ohashi-Ito, Shinichiro Sawa, Sumie Ishiguro, Takafumi Yamashino, Tatsuo Kakimoto and Ykä Helariutta for providing mutant seeds.
Glossary
Abbreviations
- AHK
Arabidopsis histidine kinase
- AHP
Arabidopsis phosphotransfer protein
- ARR
Arabidopsis response regulator
- BA
Benzyladenine
- CKX3
cytokinin oxidase 3
- CLE
CLAVATA3/EMBRYO SURROUNDING REGION-related
- CRN
CORYNE
- DZ
differentiation zone
- EZ
elongation zone
- GUS
β-glucuronidase
- IPT7
adenylate isopentenyltransferase 7
- LRR-RLK
leucine-rich repeat receptor-like kinase
- mPS–PI
modified pseudo-Schiff–propidium iodide
- RBM
root basal meristem
- RT–PCR
reverse transcription–PCR
- SAM
shoot apical meristem
- SOL2
suppressor of LLP2
- TDIF
tracheary element differentiation inhibitory factor
- WT
wild type
- WUS
WUSCHEL.
References
- Argyros R.D., Mathews D.E., Chiang Y.H., Palmer C.M., Thibault D.M., Etheridge N., et al. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell. 2008;20:2102–2116. doi: 10.1105/tpc.108.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann A., Weidtkamp-Peters S., Seidel C.A., Simon R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 2010;152:166–176. doi: 10.1104/pp.109.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U., Grünewald M., Hobe M., Simon R. Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 2002;129:565–575. doi: 10.1104/pp.001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko M.A., Vie A.K., Brembu T., Aalen R.B., Bones A.M. Plant peptides in signalling: looking for new partners. Trends Plant Sci. 2009;14:255–263. doi: 10.1016/j.tplants.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A., Lee J.Y., Roberts C.J., Dettmer J., Lehesranta S., Zhou J., et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M., Hause G., Boutilier K., Casamitjana-Martinez E., Weijers D., Offringa R., et al. Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene. 2004;327:37–49. doi: 10.1016/j.gene.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Fisher K., Turner S. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 2007;17:1061–1066. doi: 10.1016/j.cub.2007.05.049. [DOI] [PubMed] [Google Scholar]
- Fukuda H. Signals that control plant vascular cell differentiation. Nat. Rev. Mol. Cell Biol. 2004;5:379–391. doi: 10.1038/nrm1364. [DOI] [PubMed] [Google Scholar]
- Hejátko J., Ryu H., Kim G.T., Dobesová R., Choi S., Choi S.M., et al. The histidine kinases CYTOKININ-INDEPENDENT1 and ARABIDOPSIS HISTIDINE KINASE2 and 3 regulate vascular tissue development in Arabidopsis shoots. Plant Cell. 2009;21:2008–2021. doi: 10.1105/tpc.109.066696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., et al. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl Acad. Sci. USA. 2008;105:15208–15213. doi: 10.1073/pnas.0808444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C.E., Kieber J.J. Cytokinin signaling in Arabidopsis. Plant Cell. 2002;14:S47–S59. doi: 10.1105/tpc.010444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., et al. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Ishida K., Yamashino T., Yokoyama A., Mizuno T. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008;49:47–57. doi: 10.1093/pcp/pcm165. [DOI] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- Jun J.H., Fiume E., Fletcher J.C. The CLE family of plant polypeptide signaling molecules. Cell. Mol. Life Sci. 2008;65:743–755. doi: 10.1007/s00018-007-7411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001;42:677–685. doi: 10.1093/pcp/pce112. [DOI] [PubMed] [Google Scholar]
- Kiba T., Yamada H., Sato S., Kato T., Tabata S., Yamashino T., et al. The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:868–874. doi: 10.1093/pcp/pcg108. [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Nakamura Y., Sasaki E., Kyozuka J., Fukuda H., Sawa S. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007;48:1821–1825. doi: 10.1093/pcp/pcm154. [DOI] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- Mähönen A.P., Bishopp A., Higuchi M., Nieminen K.M., Kinoshita K., Törmäkangas K., et al. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- Mähönen A.P., Bonke M., Kauppinen L., Riikonen M., Benfey P.N., Helariutta Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Li J., Mathews D.E., Kieber J.J., Schaller G.E. Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol. 2004;135:927–937. doi: 10.1104/pp.103.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., et al. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17:3007–3018. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Kitano M., Kusumoto T., Tarkowski P., Kinoshita-Tsujimura K., Václavíková K., Miyawaki K., et al. Cytokinins are central regulators of cambial activity. Proc. Natl Acad. Sci. USA. 2008;105:20027–20031. doi: 10.1073/pnas.0805619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H., Betsuyaku S., Iwamoto K., Kinoshita A., Fukuda H., Sawa S. The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol. 2008;49:1752–1757. doi: 10.1093/pcp/pcn148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok D.W., Mok M.C. Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- Müller R., Bleckmann A., Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20:934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa S., Sawa S., Sato S., Kato T., Tabata S., Fukuda H. Gene trapping in Arabidopsis reveals genes involved in vascular development. Plant Cell Physiol. 2006;47:1394–1405. doi: 10.1093/pcp/pcl009. [DOI] [PubMed] [Google Scholar]
- Nieminen K., Immanen J., Laxell M., Kauppinen L., Tarkowski P., Dolezal K., et al. Cytokinin signaling regulates cambial development in poplar. Proc. Natl Acad. Sci. USA. 2008;105:20032–20037. doi: 10.1073/pnas.0805617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- Ren B., Liang Y., Deng Y., Chen Q., Zhang J., Yang X., et al. Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res. 2009;19:1178–1190. doi: 10.1038/cr.2009.88. [DOI] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F., Jürgens G., Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Miwa K., Ishikawa K., Yamada H., Aiba H., Mizuno T. The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant Cell Physiol. 2001;42:107–113. doi: 10.1093/pcp/pce037. [DOI] [PubMed] [Google Scholar]
- Takei K., Sakakibara H., Sugiyama T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 2001;276:26405–26410. doi: 10.1074/jbc.M102130200. [DOI] [PubMed] [Google Scholar]
- To J.P., Haberer G., Ferreira F.J., Deruère J., Mason M.G., Schaller G.E., et al. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To J.P., Deruère J., Maxwell B.B., Morris V.F., Hutchison C.E., Ferreira F.J., et al. Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell. 2007;19:3901–3914. doi: 10.1105/tpc.107.052662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., et al. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell. 2008;20:1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi C., Sato S., Kato T., Tabata S. The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:751–755. doi: 10.1093/pcp/pce094. [DOI] [PubMed] [Google Scholar]
- Whitford R., Fernandez A., De Groodt R., Ortega E., Hilson P. Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl Acad. Sci. USA. 2008;105:18625–18630. doi: 10.1073/pnas.0809395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H., Suzuki T., Terada K., Takei K., Ishikawa K., Miwa K., et al. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 2001;42:1017–1023. doi: 10.1093/pcp/pce127. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Kubo M., Fukuda H., Demura T. Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008;55:652–664. doi: 10.1111/j.1365-313X.2008.03533.x. [DOI] [PubMed] [Google Scholar]
- Yokoyama A., Yamashino T., Amano Y., Tajima Y., Imamura A., Sakakibara H., et al. Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 2007;48:84–96. doi: 10.1093/pcp/pcl040. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Hu Y., Dai M., Huang L., Zhou D.X. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell. 2009;21:736–748. doi: 10.1105/tpc.108.061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Wang Y., Li R., Song X., Wang Q., Huang S., et al. Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 2010;61:223–233. doi: 10.1111/j.1365-313X.2009.04049.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.