Abstract
More than a dozen secreted peptides are now recognized as important hormones that coordinate and specify cellular functions in plants. Recent evidence has shown that secreted peptide hormones often undergo post-translational modification and proteolytic processing, which are critical for their function. Such ‘small post-translationally modified peptide hormones’ constitute one of the largest groups of peptide hormones in plants. This short review highlights recent progress in research on post-translationally modified peptide hormones, with particular emphasis on their structural characteristics and modification mechanisms.
Keywords: Arabinosylation, Peptide hormone, Post-translational modification, Sulfation
Introduction
Cell-to-cell signaling mediated by secreted signals and membrane-localized receptors is one of the critical mechanisms by which growth and development of multicellular organisms are cooperatively regulated. Upon binding of signals to the extracellular domain of receptors, these physicochemical inputs are converted into physiological outputs activating downstream signaling that modulates cellular functions and fates through conformational changes in the receptors triggered by signal binding. Signal molecules that specifically bind receptors are generally referred to as ligands. Because membrane-localized receptors act as master switches of complex intracellular signaling, identification of the ligand–receptor pair is one of the central issues of post-genome research.
In higher plants, a number of genes encoding small secreted peptides have been identified in Arabidopsis (Lease and Walker 2006, Silverstein et al. 2007, Ohyama et al. 2008), and a certain proportion of their products are expected to comprise candidates for ligands (i.e. hormones). One structurally characteristic group of these peptide hormones comprises small post-translationally modified peptides, including phytosulfokine (PSK) (Matsubayashi and Sakagami 1996), PSY1 (Amano et al. 2007), tracheary element differentiation inhibitory factor (TDIF) (Ito et al. 2006), CLAVATA3 (CLV3) (Fletcher et al. 1999, Ohyama et al. 2009) and root meristem growth factor (RGF) (Matsuzaki et al. 2010) (Table 1). Modifications of these peptides, such as post-translational sulfation, hydroxylation and arabinosylation, are important for their function.
Table 1.
List of structurally characterized small post-translationally modified peptide hormones in plants
| Peptide | Mature peptide structure |
|---|---|
| PSK | Tyr(SO3H)-Ile-Tyr(SO3H)-Thr-Gln |
| PSY1 | Asp-Tyr(SO3H)-Gly-Asp-Pro-Ser-Ala-Asn-Pro-Lys-His-Asp-Pro-Gly-Val-[(l-Ara)3]Hyp-Hyp-Ser |
| TDIF | His-Glu-Val-Hyp-Ser-Gly-Hyp-Asn-Pro-Ile-Ser-Asn |
| CLV3 | Arg-Thr-Val-Hyp-Ser-Gly-[(l-Ara)3]Hyp-Asp-Pro-Leu-His-His-His |
| CLE2 | Arg-Leu-Ser-Hyp-Gly-Gly-[(l-Ara)3]Hyp-Asp-Pro-Gln-His-His |
| CEP1 | Asp-Phe-Arg-Hyp-Thr-Asn-Pro-Gly-Asn-Ser-Hyp-Gly-Val-Gly-His |
| RGF1 | Asp-Tyr(SO3H)-Ser-Asn-Pro-Gly-His-His-Pro-Hyp-Arg-His-Asn |
The sulfated tyrosine residue is shown as Tyr(SO3H). The hydroxyproline residue is shown as Hyp. The hydroxyproline residue modified with three residues of l-arabinose is shown as [(l-Ara)3]Hyp.
Here, I highlight recent progress in research on post- translationally modified peptide hormones, with particular emphasis on their structural characteristics and modification mechanisms.
Structural characteristics of secreted peptide hormones
Following complete sequencing of the Arabidopsis genome, a number of genes encoding small secreted peptides have been identified by in silico database analysis. Based on our own analysis, we identified 979 putative secreted peptide genes (SignalP score >0.75) with an open reading frame (ORF) size between 50 and 150 amino acids in the Arabidopsis Information Resource (TAIR7) genome annotation release (Ohyama et al. 2008). These 979 ORFs include many functionally uncharacterized peptides, in addition to functionally characterized peptide hormones and defense-related peptides such as defensins. Although estimation of the total percentage of secreted peptides that function as peptide hormones is difficult at present, the presence of many ‘orphan receptors’ among receptor-like kinases and receptor-like proteins in Arabidopsis (Shiu and Bleecker 2001, Shiu and Bleecker 2003, Wang et al. 2008) suggests that a substantial number of ligands remain to be identified.
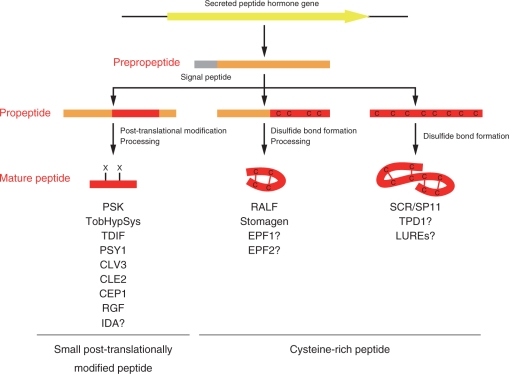
Biosynthesis of secreted peptide hormones often involves post-translational modification and proteolytic processing, hindering accurate prediction of the structure of mature peptides based on primary sequence analysis. Moreover, understanding of the process of secretion of soluble cargo to the apoplasm in plants is limited (Rojo and Denecke 2008). Nonetheless, accumulating evidence suggests that secreted peptide hormones can be divided into three groups based on their structural characteristics arising from their primary sequences and biosynthetic pathways (Fig. 1).
Fig. 1.
Three distinct biosynthetic pathways of secreted peptide hormones categorized by their structural characteristics. Secreted peptide hormones can be categorized into the following three groups: peptides involving complex post-translational modifications followed by extensive proteolytic processing; peptides involving intramolecular disulfide bond formation followed by proteolytic processing; and peptides involving multiple intramolecular disulfide bonds without proteolytic processing. The first group of peptides are called small post-translationally modified peptides; the latter two groups are defined as cysteine-rich peptides.
In general, secreted peptide hormone genes are initially translated as pre-propeptides, followed by removal of the N-terminal signal peptide by signal peptidase to afford propeptides. The sites of cleavage in pre-propeptides can be predicted with a high degree of accuracy by SignalP software (Bendtsen et al. 2004). Propeptides are further structurally modified by several modification enzymes to give biologically functional mature peptides.
From the structural point of view, one major group of secreted peptide hormones are those characterized by the presence of post-translational modifications mediated by specific transferases and by their small size (<20 amino acids) resulting from proteolytic processing (Fig. 1). This peptide hormone group includes PSK (Matsubayashi and Sakagami 1996), tobacco hydroxyproline-rich systemin (TobHypSys) (Pearce et al. 2001a), PSY1 (Amano et al. 2007), TDIF (Ito et al. 2006), CLV3 (Fletcher et al. 1999, Ohyama et al. 2009), CLAVATA3/ESR-Related 2 (CLE2) (Ohyama et al. 2009), C-terminally encoded peptide 1 (CEP1) (Ohyama et al. 2008) and RGF1 (Matsuzaki et al. 2010) (their functions are briefly described below). Hereinafter, these peptides are designated as ‘small post-translationally modified peptides’.
Interestingly, primary sequences of these peptides have common structural features. Their multiple paralogous genes primarily encode approximately 70–110 cysteine-poor secreted peptides that exhibit significant sequence diversity, with the exception of the conserved C-terminal domains that correspond to the mature peptide sequences (Fig. 2). I speculate that a number of amino acid substitutions were accumulated within the sequences digested by proteolytic processing during molecular evolution. In this context, genes encoding paralogous secreted peptides with similar features may encode small post-translationally modified peptides. Indeed, CEP1 and RGF1 were identified by in silico screening of the peptide families with these characteristics (Ohyama et al. 2008, Matsuzaki et al. 2010). Another such candidate is INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) that positively regulates floral organ abscission (Butenko et al. 2003, Stenvik et al. 2008). IDA and its five homologs, IDA-LIKE (IDLs), share conserved domains near the C-terminus, and short synthetic peptides of this conserved domain mimic IDA functions.
Fig. 2.
Structural characteristics of primary amino acid sequences of precursor polypeptides that represent small post-translationally modified peptides. Deduced amino acid sequences of (A) PSK, (B) PSY1, (C) CLV3 and CLE2, (D) CEP1 (E) RGF1, and their representative homologs are shown. Domains encoding mature peptides are underlined. Identical amino acid residues are highlighted in black, and similar amino acid residues are highlighted in gray.
In contrast, another group of secreted peptide hormones is characterized by the presence of an even number of cysteine residues (typically six or eight) that participate in the formation of intramolecular disulfide bonds (Fig. 1). Many secreted peptides including antibiotic peptide defensins belong to this group and are referred to as ‘cysteine-rich peptides’. A representative peptide hormone in this group is S-locus cysteine-rich protein/S-locus protein 11 (SCR/SP11), a male determinant of self-incompatibility in Brassica species (Schopfer et al. 1999, Takayama et al. 2000). SCR/SP11 has been chemically confirmed to be secreted after disulfide bond formation without proteolytic processing (Mishima et al. 2003; Takayama et al. 2001). I speculate that TAPETUM DETERMINANT1 (TPD1), a peptide required for the specialization of tapetal cells in the anther (Yang et al. 2003), and LUREs, polypeptides involved in pollen tube guidance (Okuda et al. 2009), also belong to this group due to their total number and location of cysteine residues, although their in vivo structures have not been chemically characterized.
As a third group, some cysteine-rich peptide, such as rapid alkalinization factor (RALF) (four cysteine residues), a 49 amino acid peptide suggested to be involved in various aspects of plant development (Pearce et al. 2001b, Wu et al. 2007, Covey et al. 2010), and stomagen (six cysteine residues), a 45 amino acid peptide that positively regulates stomatal density (Kondo et al. 2010, Sugano et al. 2010), involve proteolytic processing even though they have intramolecular disulfide bonds (Fig. 1). In both of these peptides, cysteine residues are clustered within the C-terminal region of the propeptides and the N-terminal cysteine-poor region is digested by processing, suggesting that the cysteine-rich region stabilized by intramolecular disulfide bonds is resistant to proteolytic digestion by the processing enzymes. Stomagen belongs to the EPIDERMAL PATTERNING FACTOR (EPF) peptide family, and EPF1 and EPF2, cysteine-rich peptides that act as negative regulators of stomatal clustering and density (Hara et al. 2007, Hara et al. 2009, Hunt and Gray 2009), may also involve proteolytic processing.
Post-translational modifications found in secreted peptide hormones
Accumulating evidence indicates that all small secreted peptides lacking cysteine residues in the mature peptide sequence always involve some post-translational modifications. Post-translational modifications are known to affect peptide conformation through steric interactions with the peptide backbone, thereby modulating the binding ability and specificity of peptides for target proteins. To date, three types of post-translational modifications have been identified in secreted peptide hormones from plants: tyrosine sulfation, proline hydroxylation and hydroxyproline arabinosylation.
Tyrosine sulfation
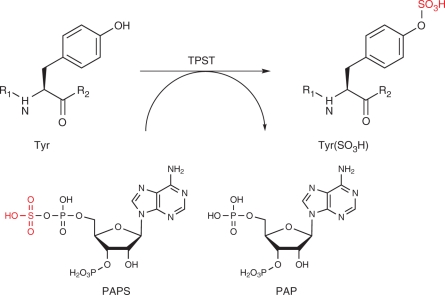
Tyrosine sulfation is a post-translational modification found in peptides and proteins synthesized through the secretory pathway of most eukaryotes, including higher plants. This modification is mediated by a specific enzyme, tyrosylprotein sulfotransferase (TPST), which catalyzes the transfer of sulfate from 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to the phenolic group of tyrosine (Moore 2003) (Fig. 3). Although the tyrosine sulfation motif in peptides is not clear-cut, the minimum requirement for tyrosine sulfation in plants is the presence of an aspartic acid residue N-terminally adjacent to a tyrosine residue, namely the Asp–Tyr sequence. Multiple acidic amino acids near this tyrosine residue also significantly enhance sulfation (Hanai et al. 2000).
Fig. 3.
Tyrosine sulfation by tyrosylprotein sulfotransferase (TPST). TPST catalyzes the transfer of sulfate from the sulfate donor 3′-phosphoadenosine-5′-phosphosulfate (PAPS) to the hydroxyl group of a tyrosine residue to form a tyrosine sulfate ester and adenosine 3′, 5′-diphosphate (PAP). R1 and R2 represent peptide chains.
In mice and humans, tyrosine sulfation is mediated by two structurally related proteins, TPST-1 and TPST-2 (Beisswanger et al. 1998, Ouyang et al. 1998). TPSTs are approximately 50 kDa type II transmembrane proteins localized in the trans-Golgi network. Their orthologs have been found in invertebrates such as Caenorhabditis elegans and Drosophila melanogaster (Moore 2003). However, no orthologs of TPST-1 and TPST-2 have been identified in Arabidopsis or other plant species. Nevertheless, significant TPST activity has been detected in microsomal fractions of various plant species (Hanai et al. 2000), suggesting that plant TPST has evolved in a manner distinct from its animal counterpart.
Recent affinity purification studies revealed that Arabidopsis TPST (AtTPST) is a Golgi-localized 62 kDa transmembrane protein (Komori et al. 2009). AtTPST is expressed throughout the plant body, and the highest levels of expression are in the root apical meristem. AtTPST orthologs were found in other higher plants, such as rice and maize, and in the moss Physcomitrella patens, but not in yeast or animals. Indeed, AtTPST shows no sequence similarity with animal TPSTs, even though both enzymes catalyze identical sulfate transfer reactions using the same co-substrate, PAPS. Moreover, AtTPST is a type I transmembrane protein with the transmembrane domain located near its C-terminus, whereas animal TPSTs are type II transmembrane proteins with the transmembrane domain located near its N-terminus. This structural diversity strongly suggests that the AtTPST gene has evolved from an ancestral gene distinct from that of animal TPSTs. In other words, plants and animals independently acquired enzymes for tyrosine sulfation through convergent evolution. Moreover, a loss-of-function mutant of AtTPST (tpst-1) displayed a marked dwarf phenotype accompanied by stunted roots, disorganized root meristem, pale green leaves, reduction in higher order veins and early senescence, suggesting diverse roles for sulfated peptides in plant growth and development (Komori et al. 2009).
To date, three tyrosine-sulfated peptide hormones, PSK, PSY1 and RGF1, have been found in plants. PSK is a five amino acid secreted peptide containing two sulfated tyrosines (Table 1). It was initially identified as a growth-promoting signal involved in the ‘density effect’ in plant cell cultures (Matsubayashi and Sakagami 1996). In general, proliferation of plant cells is suppressed under low-density culture conditions but promoted by the external addition of ‘conditioned’ medium derived from the rapidly growing high-density culture, suggesting that individual cells in culture secrete the growth-promoting signal into the medium. This growth-promoting signal was purified and named PSK. PSK is produced from ∼80 amino acid precursor peptides via post-translational sulfation by TPST and proteolytic processing (Fig. 2A) (Yang et al. 1999, Matsubayashi et al. 2006). In vitro studies suggest that this proteolytic processing is mediated, at least in part, by a subtilisin-like serine protease, AtSBT1.1 (Srivastava et al. 2008). Genes encoding PSK precursors are widely expressed in a variety of tissues and up-regulated by wounding (Matsubayashi et al. 2006). Further biochemical analysis reveals that PSK is recognized by a membrane-localized leucine-rich repeat receptor kinase (LRR-RK), PSKR1 (Matsubayashi et al. 2002). Disruption of PSKR1 and its two homologs in Arabidopsis causes pleiotropic growth defects such as short roots, smaller leaves and early senescence (Matsubayashi et al. 2006, Amano et al. 2007).
The second sulfated peptide, PSY1, is an 18 amino acid secreted glycopeptide containing one residue of sulfated tyrosine (Table 1) (Fig. 2B); this glycopeptide was identified by exhaustive analysis of tyrosine-sulfated peptides in plant cell culture media (Amano et al. 2007). PSY1 is expressed in various Arabidopsis tissues and promotes cellular proliferation and expansion at nanomolar concentrations.
The third sulfated peptide, RGF1, is a 13 amino acid secreted peptide involved in maintenance of the root stem cell niche in Arabidopsis (Table 1) (Matsuzaki et al. 2010). RGFs are produced from ∼100 amino acid precursor peptides via post- translational sulfation and proteolytic processing (Fig. 2E). RGF1 was identified by a search of the sulfated peptide(s) that recovers root meristem defects of the tpst-1 mutant in combination with in silico screening of genes encoding sulfated peptides and practical bioassays using synthetic sulfated peptides. This approach is based on the assumption that phenotypes of the tpst-1 mutant reflect the deficiency in the biosynthesis of all functional tyrosine-sulfated peptides. As mentioned above, tpst-1 shows a stunted root phenotype accompanied by loss of maintenance of stem cells and a considerable decrease in meristematic activity. Interestingly, two sulfated peptide hormones, PSK and PSY1, promoted cell elongation activity of tpst-1 roots but did not restore meristematic activity, indicating that tyrosine-sulfated peptide(s) other than PSK and PSY1 is involved in maintenance of root stem cells and regulation of meristematic activity.
RGF family peptides are expressed mainly in the stem cell area and the innermost layer of central columella cells, and diffuse into the meristematic region through the apoplast. RGF peptides regulate root development by stabilizing PLETHORA transcription factor proteins which are specifically expressed in root meristem and mediate patterning of the root stem cell niche.
Proline hydroxylation
Another important class of post-translational modifications in plant peptide hormones is proline hydroxylation. This modification is mediated by prolyl 4-hydroxylase (P4H), which catalyzes oxidation of proline residues exclusively at the fourth position carbon. P4H belongs to a family of 2-oxoglutarate-dependent dioxygenases that requires 2-oxoglutarate and O2 as co-substrates (Myllyharju 2003). P4H is a type II membrane protein with the transmembrane domain located near its N-terminus. It localizes in both the endoplasmic reticulum (ER) and Golgi. To date, two P4H genes and nine homologs have been identified in Arabidopsis (Hieta and Myllyharju 2002, Tiainen et al. 2005, Yuasa et al. 2005). Arabidopsis P4H proteins show an approximately 25% identity to the catalytically important C-terminal regions of the human P4H (I) and (II) subunits. Although some sequence motifs have been reported for efficient proline hydroxylation (Shimizu et al. 2005), no consensus sequence has been determined for proline hydroxylation of secreted peptide hormones in plants. Hydroxyproline (Hyp) residues have been found in TobHypSys (Pearce et al. 2001a), TomHypSys (Pearce and Ryan 2003), PSY1 (Amano et al. 2007), TDIF (Ito et al. 2006), CEP1 (Ohyama et al. 2008), CLV3 (Kondo et al. 2006, Ohyama et al. 2009), CLE2 (Ohyama et al. 2009) and RGF1 (Matsuzaki et al. 2010). Among them, Hyp residues of TobHypSys, TomHypSys, PSY1, CLV3 and CLE2 are further modified with a pentose sugar such as l-arabinose, as described in the next section.
TDIF is a 12 amino acid peptide in which two proline residues are hydroxylated. This peptide was initially identified as an inhibitory factor of transdifferentiation of dispersed Zinnia (Zinnia elegans L.) mesophyll cells into tracheary elements (the main conductive cells of the xylem) (Ito et al. 2006). The TDIF peptide suppresses xylem cell development at subnanomolar concentrations and promotes cell division in vitro. The TDIF sequence was identical to the C-terminal 12 amino acids of Arabidopsis CLE41 and CLE44. Further in vivo studies have revealed that the TDIF/CLE41/CLE44 peptides expressed in phloem and neighboring cells are recognized by TDR/PXY, an LRR-RK located in the plasma membrane of procambial cells, and control the procambial cell fate (Hirakawa et al. 2008). This signal suppresses xylem cell differentiation of procambial cells and promotes their proliferation. Recent genetic analyses have shown that WOX4 transcription factor is required for promoting the proliferation of procambial/cambial stem cells in response to the TDIF/CLE41/CLE44 signal (Hirakawa et al. 2010).
Another Hyp-containing peptide is CEP1, a 15 amino acid peptide with two Hyp residues. CEP1 was initially identified by in silico gene screening for structural features of pre- propeptides of known small post-translationally modified peptides (Ohyama et al. 2008). The common feature of known small post-translationally modified peptide hormones is that they are encoded by multiple paralogous genes. In addition, the primary products of these genes are approximately 70–110 amino acid cysteine-poor secreted peptides that exhibit significant sequence diversity, with the exception of short conserved domains that correspond to the mature peptide sequences. CEP1 family peptides fulfill these criteria. CEP1 is mainly expressed in the lateral root primordia and, when overexpressed or externally applied, significantly arrests root growth. Therefore, it is a strong candidate as a novel peptide hormone.
Hydroxyproline arabinosylation
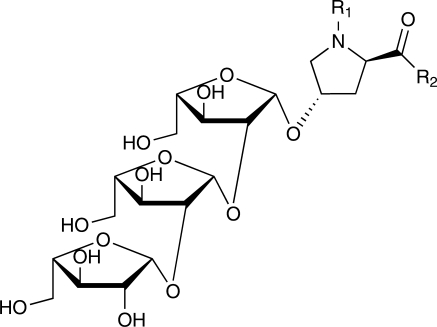
Recent biochemical analysis revealed that Hyp residues of several secreted peptide hormones such as PSY1, CLV3 and CLE2 are further modified with an O-linked l-arabinose chain (Amano et al. 2007, Ohyama et al. 2009). Linkage analysis of the sugar moiety of CLV3 by gas chromatography–mass spectometry (GC-MS) and α-l-arabinofuranosidase treatment suggests that arabinose residues are linked to each other via β-1,2-bonds (Ohyama et al. 2009) (Fig. 4). Nuclear magnetic resonance (NMR) analysis of Hyp-bound linear triarabinoside isolated from cell wall preparations of Arabidopsis cell suspension cultures also indicates the presence of β-1,2-linked sugars (Bollig et al. 2007). Linear β-1,2-linked triarabinoside has been found in lectins derived from potato tubers (Ashford et al. 1982), and from Hyp-rich glycoproteins derived from cell wall preparations of suspension-cultured tobacco cells (Akiyama and Kato 1976, Akiyama et al. 1980). In contrast, N-linked glycans that are often found in many extracellular proteins have never been detected in secreted peptide hormones.
Fig. 4.
Proposed structure of hydroxyproline-bound triarabinoside. Linear β-1,2-linked triarabinoside is suggested to be a common structure in glycopeptide hormones. R1 and R2 represent peptide chains.
PSY1 was the first structurally characterized arabinosylated peptide hormone. This 18 amino acid secreted glycopeptide contains one sulfated tyrosine residue (described above) and two Hyp residues, one of which is further modified with three l-arabinose residues (Amano et al. 2007).
CLV3 is a peptide signal that regulates stem cell fate in Arabidopsis shoot apical meristem (Fletcher et al. 1999). It was also determined to be post-translationally modified with three l-arabinose residues (Ohyama et al. 2009). Arabinosylated CLV3 peptide was identified in the culture medium of whole-plant submerged cultures of CLV3-overexpressing Arabidopsis plants. In submerged cultures, Arabidopsis plants develop cuticle-less hyperhydric leaves with large intercellular spaces filled with water, through which secreted peptides in the apoplast diffuse into the culture medium (Ohyama et al. 2008). The mature CLV3 peptide was determined to be a 13 amino acid CLE domain peptide with the seventh Hyp residue post- translationally modified with three l-arabinose residues (Ohyama et al. 2009). This arabinosylated CLV3 peptide interacts more strongly with the ectodomain of CLV1, one of the receptor components for CLV3, than non-arabinosylated forms.
By the same approach, the mature CLE2 peptide was investigated in whole-plant submerged cultures of CLE2- overexpressing Arabidopsis plants. As is the case with CLV3, mature CLE2 was determined to be a 12 amino acid peptide with the seventh Hyp residue post-translationally modified with three l-arabinose residues (Ohyama et al. 2009). Interestingly, the CLE2 glycopeptide binds CLV1 with nanomolar binding affinity. Although the expression pattern and physiological function of CLE2 in Arabidopsis plants is yet to be characterized, these results suggest that CLE2 functions as a ligand for CLV1. In legume plants, CLE2 orthologs, LjCLE-RS1 and LjCLE-RS2, are thought to act as root-derived mobile signals that systemically regulate nodule numbers through the CLV1 ortholog, HAR1 receptor kinase (Okamoto et al. 2009). Binding of the CLE2 glycopeptide to CLV1 in Arabidopsis strongly supports the molecular basis of this autoregulation model.
Although sugar composition and linkage have not been determined, defense-related peptides TobHypSys and TomHypSys also contain multiple Hyp residues modified with multiple pentose residues (Pearce et al. 2001a, Pearce and Ryan 2003). TobHypSys and TomHypSys were isolated from tobacco or tomato leaf extracts due to their ability to cause a pH increase when added to the medium of suspension-cultured tobacco cells. A link has been suggested between proton flux across the plasma membrane and the induction of defense genes (Schaller and Oecking 1999). Both TobHypSys and TomHypSys show proteinase inhibitor-inducing activities at nanomolar concentrations. Chemically synthesized TobHypSys peptides lacking sugar chains were found to be 10,000 times less active than native polypeptides, suggesting that the sugar chains are important for their activity.
In general, O-glycosylation occurs from the successive addition of nucleotide-activated sugars catalyzed by glycosyltransferases in the Golgi. Biosynthesis of Hyp-bound β-1,2-linked triarabinoside is thought to involve two distinct arabinosyltransferases. The first one is responsible for the formation of a β-linkage with a 4-position Hyp (hydroxyproline arabinosyltransferase), and the second one mediates a β-1,2-linkage between arabinofuranose residues (arabinosyltransferase). Recent chemical genetic screening suggests that XEG113 (At2g35610) may encode β-1,2-arabinosyltransferase (Gille et al. 2009). In contrast, there are no reports yet on hydroxyproline arabinosyltransferase.
Future perspective
In a classical research approach, candidates of hormones have been identified by repeated purifications based on their specific biological activities. The feasibility of using this approach, however, largely depends on the quality and sensitivity of the bioassay system. Another major limitation of this approach is the lack of an established bioassay system to detect peptide hormones with an unexpected class of functions. In this context, an alternative strategy for identification of novel hormones is needed to overcome limitations of bioassay-based approaches.
From a structural point of view, small post-translationally modified peptides are produced through a secretory pathway after post-translational modification and then proteolytic processing. These processes involve at least two enzymes, namely a post-translational modification enzyme and a proteolytic processing enzyme. Post-translational modification also requires co-substrates such as the sulfation donor PAPS and the arabinosylation donor UDP-l-arabinose synthesized using ATP. Thus, biosynthesis of small post-translationally modified peptides requires considerably higher energy compared with normal proteins and peptides. Nevertheless, a number of post-translationally modified peptides have been evolutionarily conserved, suggesting that these ‘expensive’ modified peptides afford physiological merits to the plants greater than their energy costs. In this context, post-translational modifications (as well as proteolytic processing) can be indicative of biologically active peptides. Indeed, the peptidomics approach targeting sulfated peptides has successfully identified a novel peptide hormone, PSY1 (Amano et al. 2007), and an in silico gene screening approach has uncovered a Hyp-containing peptide hormone candidate, CEP1 (Ohyama et al. 2008). Thus, post-translationally modified peptides appear to be good candidates for peptide hormones.
Another important aspect of post-translationally modified peptide research involves phenotypic analysis of loss-of- function mutants of post-translational modification enzymes. Because post-translational modification enzymes recognize particular sequences within multiple target peptides, their loss of function should be reflected as deficiencies in the biosynthesis of modified peptides. Thus, the presence of novel post-translationally modified peptide hormones should be revealed through phenotypic analysis of such mutants. Indeed, a novel peptide hormone, RGF1, was successfully identified by a search of the sulfated peptide(s) that recovers root meristem defects of the tpst-1 mutant (Matsuzaki et al. 2010). In this context, identification of hydroxyproline arabinosyltransferase, followed by phenotypic analysis of loss-of-function mutants of this enzyme, could provide a clearer picture of the functions of arabinosylated peptide hormones in plants.
Funding
This research was supported by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) [Grant-in-Aid for Scientific Research for Priority Areas (No. 19060010)]; Japan Society for the Promotion of Science [Grant-in-Aid for Creative Scientific Research (No. 19GS0315)].
Glossary
Abbreviations
- CLV3
CLAVATA3
- Hyp
hydroxyproline
- IDA
INFLORESCENCE DEFICIENT IN ABSCISSION
- LRR-RK
leucine-rich repeat receptor kinase
- ORF
open reading frame
- PAPS
3′-phosphoadenosine-5′-phosphosulfate
- P4H
prolyl 4-hydroxylase
- PSK
phytosulfokine
- RGF
root meristem growth factor
- TDIF
tracheary element differentiation inhibitory factor
- TPST
tyrosylprotein sulfotransferase.
References
- Akiyama Y., Kato K. Hydroxyproline arabinosides from suspension-cultured tobacco cell wall. Agric. Biol. Chem. 1976;40:2343–2348. [Google Scholar]
- Akiyama Y., Mori M., Kato K. 13C-NMR analysis of hydroxyproline arabinosides from Nicotiana tabacum. Agric. Biol. Chem. 1980;44:2487–2489. [Google Scholar]
- Amano Y., Tsubouchi H., Shinohara H., Ogawa M., Matsubayashi Y. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl Acad. Sci. USA. 2007;104:18333–18338. doi: 10.1073/pnas.0706403104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashford D., Desai N.N., Allen A.K., Neuberger A., O'Neill M.A., Selvendran R.R. Structural studies of the carbohydrate moieties of lectins from potato (Solanum tuberosum) tubers and thorn-apple (Datura stramonium) seeds. Biochem. J. 1982;201:199–208. doi: 10.1042/bj2010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisswanger R., Corbeil D., Vannier C., Thiele C., Dohrmann U., Kellner R., et al. Existence of distinct tyrosylprotein sulfotransferase genes: molecular characterization of tyrosylprotein sulfotransferase-2. Proc. Natl Acad. Sci. USA. 1998;95:11134–11139. doi: 10.1073/pnas.95.19.11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bollig K., Lamshoft M., Schweimer K., Marner F.J., Budzikiewicz H., Waffenschmidt S. Structural analysis of linear hydroxyproline-bound O-glycans of Chlamydomonas reinhardtii—conservation of the inner core in Chlamydomonas and land plants. Carbohydr. Res. 2007;342:2557–2566. doi: 10.1016/j.carres.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Butenko M.A., Patterson S.E., Grini P.E., Stenvik G.E., Amundsen S.S., Mandal A., et al. INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell. 2003;15:2296–2307. doi: 10.1105/tpc.014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey P.A., Subbaiah C.C., Parsons R.L., Pearce G., Lay F.T., Anderson M.A., et al. A pollen-specific RALF from tomato that regulates pollen tube elongation. Plant Physiol. 2010;153:703–715. doi: 10.1104/pp.110.155457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Gille S., Hansel U., Ziemann M., Pauly M. Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc. Natl Acad. Sci. USA. 2009;106:14699–14704. doi: 10.1073/pnas.0905434106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai H., Nakayama D., Yang H., Matsubayashi Y., Hirota Y., Sakagami Y. Existence of a plant tyrosylprotein sulfotransferase: novel plant enzyme catalyzing tyrosine O-sulfation of preprophytosulfokine variants in vitro. FEBS Lett. 2000;470:97–101. doi: 10.1016/s0014-5793(00)01299-0. [DOI] [PubMed] [Google Scholar]
- Hara K., Kajita R., Torii K.U., Bergmann D.C., Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21:1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Yokoo T., Kajita R., Onishi T., Yahata S., Peterson K.M., et al. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 2009;50:1019–1031. doi: 10.1093/pcp/pcp068. [DOI] [PubMed] [Google Scholar]
- Hieta R., Myllyharju J. Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. Effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor alpha-like peptides. J. Biol. Chem. 2002;277:23965–23971. doi: 10.1074/jbc.M201865200. [DOI] [PubMed] [Google Scholar]
- Hirakawa Y., Kondo Y., Fukuda H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell. 2010;22:2618–2629. doi: 10.1105/tpc.110.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., et al. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl Acad. Sci. USA. 2008;105:15208–15213. doi: 10.1073/pnas.0808444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L., Gray J.E. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 2009;19:864–869. doi: 10.1016/j.cub.2009.03.069. [DOI] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- Komori R., Amano Y., Ogawa-Ohnishi M., Matsubayashi Y. Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc. Natl Acad. Sci. USA. 2009;106:15067–15072. doi: 10.1073/pnas.0902801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Kajita R., Miyazaki A., Hokoyama M., Nakamura-Miura T., Mizuno S., et al. Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 2010;51:1–8. doi: 10.1093/pcp/pcp180. [DOI] [PubMed] [Google Scholar]
- Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- Lease K.A., Walker J.C. The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 2006;142:831–838. doi: 10.1104/pp.106.086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Ogawa M., Kihara H., Niwa M., Sakagami Y. Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 2006;142:45–53. doi: 10.1104/pp.106.081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Ogawa M., Morita A., Sakagami Y. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science. 2002;296:1470–1472. doi: 10.1126/science.1069607. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Sakagami Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl Acad. Sci. USA. 1996;93:7623–7627. doi: 10.1073/pnas.93.15.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science. 2010;329:1065–1067. doi: 10.1126/science.1191132. [DOI] [PubMed] [Google Scholar]
- Mishima M., Takayama S., Sasaki K., Jee J.G., Kojima C., Isogai A., et al. Structure of the male determinant factor for Brassica self-incompatibility. J. Biol. Chem. 2003;278:36389–36395. doi: 10.1074/jbc.M305305200. [DOI] [PubMed] [Google Scholar]
- Moore K.L. The biology and enzymology of protein tyrosine O-sulfation. J. Biol. Chem. 2003;278:24243–24246. doi: 10.1074/jbc.R300008200. [DOI] [PubMed] [Google Scholar]
- Myllyharju J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003;22:15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Ogawa M., Matsubayashi Y. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 2008;55:152–160. doi: 10.1111/j.1365-313X.2008.03464.x. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- Okamoto S., Ohnishi E., Sato S., Takahashi H., Nakazono M., Tabata S., et al. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 2009;50:67–77. doi: 10.1093/pcp/pcn194. [DOI] [PubMed] [Google Scholar]
- Okuda S., Tsutsui H., Shiina K., Sprunck S., Takeuchi H., Yui R., et al. Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature. 2009;458:357–361. doi: 10.1038/nature07882. [DOI] [PubMed] [Google Scholar]
- Ouyang Y., Lane W.S., Moore K.L. Tyrosylprotein sulfotransferase: purification and molecular cloning of an enzyme that catalyzes tyrosine O-sulfation, a common posttranslational modification of eukaryotic proteins. Proc. Natl Acad. Sci. USA. 1998;95:2896–2901. doi: 10.1073/pnas.95.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G., Moura D.S., Stratmann J., Ryan C.A. Production of multiple plant hormones from a single polyprotein precursor. Nature. 2001a;411:817–820. doi: 10.1038/35081107. [DOI] [PubMed] [Google Scholar]
- Pearce G., Moura D.S., Stratmann J., Ryan C.A., Jr. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl Acad. Sci. USA. 2001b;98:12843–12847. doi: 10.1073/pnas.201416998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G., Ryan C.A. Systemic signaling in tomato plants for defense against herbivores. Isolation and characterization of three novel defense-signaling glycopeptide hormones coded in a single precursor gene. J. Biol. Chem. 2003;278:30044–30050. doi: 10.1074/jbc.M304159200. [DOI] [PubMed] [Google Scholar]
- Rojo E., Denecke J. What is moving in the secretory pathway of plants? Plant Physiol. 2008;147:1493–1503. doi: 10.1104/pp.108.124552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A., Oecking C. Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell. 1999;11:263–272. doi: 10.1105/tpc.11.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer C.R., Nasrallah M.E., Nasrallah J.B. The male determinant of self-incompatibility in Brassica. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- Shimizu M., Igasaki T., Yamada M., Yuasa K., Hasegawa J., Kato T., et al. Experimental determination of proline hydroxylation and hydroxyproline arabinogalactosylation motifs in secretory proteins. Plant J. 2005;42:877–889. doi: 10.1111/j.1365-313X.2005.02419.x. [DOI] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl Acad. Sci. USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein K.A., Moskal W.A., Jr., Wu H.C., Underwood B.A., Graham M.A., Town C.D., et al. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007;51:262–280. doi: 10.1111/j.1365-313X.2007.03136.x. [DOI] [PubMed] [Google Scholar]
- Srivastava R., Liu J.X., Howell S.H. Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J. 2008;56:219–227. doi: 10.1111/j.1365-313X.2008.03598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik G.E., Tandstad N.M., Guo Y., Shi C.L., Kristiansen W., Holmgren A., et al. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell. 2008;20:1805–1817. doi: 10.1105/tpc.108.059139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S.S., Shimada T., Imai Y., Okawa K., Tamai A., Mori M., et al. Stomagen positively regulates stomatal density in Arabidopsis. Nature. 2010;463:241–244. doi: 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- Takayama S., Shiba H., Iwano M., Shimosato H., Che F.S., Kai N., et al. The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl Acad. Sci. USA. 2000;97:1920–1925. doi: 10.1073/pnas.040556397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Shimosato H., Shiba H., Funato M., Che F.S., Watanabe M., et al. Direct ligand–receptor complex interaction controls Brassica self-incompatibility. Nature. 2001;413:534–538. doi: 10.1038/35097104. [DOI] [PubMed] [Google Scholar]
- Tiainen P., Myllyharju J., Koivunen P. Characterization of a second Arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J. Biol. Chem. 2005;280:1142–1148. doi: 10.1074/jbc.M411109200. [DOI] [PubMed] [Google Scholar]
- Wang G., Ellendorff U., Kemp B., Mansfield J.W., Forsyth A., Mitcell K., et al. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 2008;147:503–517. doi: 10.1104/pp.108.119487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Kurten E.L., Monshausen G., Hummel G.M., Gilroy S., Baldwin I.T. NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. Plant J. 2007;52:877–890. doi: 10.1111/j.1365-313X.2007.03289.x. [DOI] [PubMed] [Google Scholar]
- Yang H., Matsubayashi Y., Nakamura K., Sakagami Y. Oryza sativa PSK gene encodes a precursor of phytosulfokine-α, a sulfated peptide growth factor found in plants. Proc. Natl Acad. Sci. USA. 1999;96:13560–13565. doi: 10.1073/pnas.96.23.13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.L., Xie L.F., Mao H.Z., Puah C.S., Yang W.C., Jiang L., et al. TAPETUM DETERMINANT1 is required for cell specialization in the Arabidopsis anther. Plant Cell. 2003;15:2792–2804. doi: 10.1105/tpc.016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa K., Toyooka K., Fukuda H., Matsuoka K. Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J. 2005;41:81–94. doi: 10.1111/j.1365-313X.2004.02279.x. [DOI] [PubMed] [Google Scholar]