Abstract
mTORC1 is a validated therapeutic target for renal cell carcinoma (RCC). Here, analysis of Tsc1 deficient (mTORC1 hyperactivation) mice uncovered a FoxO-dependent negative feedback circuit constraining mTORC1-mediated renal tumorigenesis. We document robust FoxO activation in Tsc1 deficient benign polycystic kidneys and FoxO extinction upon progression to murine renal tumors; murine renal tumor progression upon genetic deletion of both Tsc1 and FoxOs; and down-regulated FoxO expression in most human renal clear cell and papillary carcinomas, yet continued expression in less aggressive RCCs and benign renal tumor subtypes. Mechanistically, integrated analyses revealed that FoxO-mediated block operates via suppression of Myc through up-regulation of the Myc antagonists, Mxi1-SRα and mir-145, establishing a FoxO-Mxi1-SRα/mir-145 axis as a major progression block in renal tumor development.
Introduction
The mammalian target of rapamycin complex 1 (mTORC1) serves as the key regulator of protein synthesis and cell growth via phosphorylation of a variety of downstream targets, including S6 Kinase and 4E-BP1 (Ma and Blenis, 2009; Wullschleger et al., 2006), and plays a critical role in the regulation of cell growth, angiogenesis and metabolism in many human cancers, including RCC (Guertin and Sabatini, 2007; Hanna et al., 2008; Inoki et al., 2005; Sabatini, 2006). RCC comprises approximately 3% of all adult malignancies, ranks among the top ten cancers in the United States, and continues to show modest responses to most conventional cancer treatments (Linehan and Zbar, 2004; Rini et al., 2009). mTORC1 hyper-activation is observed in the majority of human RCC samples (Pantuck et al., 2007; Robb et al., 2007) and has emerged as a therapeutic target for RCC following several clinical trials establishing clinical benefit of mTORC1 inhibitors (Atkins et al., 2004; Hudes et al., 2007; Motzer et al., 2008). Given the clinical relevance of the mTORC1 target in RCC, an understanding of mTORC1’s complex signaling circuitry in vivo may inform its role in RCC pathogenesis and guide further drug development efforts.
Increasing knowledge of mTORC1 signaling has demonstrated that mTORC1 acts both downstream and upstream of PI3K-AKT signaling. While activated PI3K-AKT signaling promotes mTORC1 signaling through AKT-mediated phosphorylation of both TSC2 and PRAS40, mTORC1 hyper-activation also leads to feedback shutoff of PI3K/AKT signaling via a S6 Kinase-dependent down-regulation of upstream activators of PI3K including the PDGF receptor and IRS-1 (Bhaskar and Hay, 2007; Guertin and Sabatini, 2007; Hay and Sonenberg, 2004; Manning, 2004; Um et al., 2006). A major upstream regulator of mTORC1 is the TSC1-TSC2 complex, which functions to inhibit the mTORC1 activity via stimulation of GTP hydrolysis and inactivation of small GTPase Rheb, an activator of mTORC1 (Huang and Manning, 2008; Kwiatkowski and Manning, 2005; Li et al., 2004). These observations hold important therapeutic implications in that, in specific genotypic contexts, mTORC1 inhibitor treatment alone might enhance tumorigenesis in mTORC1 hyper-activation-driven tumors by stimulating PI3K-AKT-dependent survival and cell cycle entry, thereby prompting calls for combination therapeutic regimens in the clinic (Shaw and Cantley, 2006). Along these lines, the rational design and effective implementation of such combinations requires a more definitive understanding of the key downstream effector(s) of AKT that mediate this mTORC1-directed negative feedback circuit.
The PI3K-AKT axis is activated in virtually all human cancers (Cully et al., 2006; Luo et al., 2003; Salmena et al., 2008; Samuels et al., 2004). The wide range of tumorigenic phenotypes mediated by PI3K-AKT signaling is consistent with the existence of diverse downstream effectors including TSC1-TSC2 complex, FoxOs, GSK3 and MDM2. These effectors operate in a highly context-specific manner, i.e., effectors are coordinately or differentially utilized in conferring neoplastic phenotypes in distinct cell lineages and genotypes (Manning and Cantley, 2007). The mammalian FoxO transcription factors – FoxO1, FoxO3, FoxO4 – function in the nucleus to direct transcription of specific gene targets governing cellular survival, proliferation, metabolism, differentiation and oxidative defense. Activation of PI3K by extracellular growth factors leads to AKT-mediated phosphorylation of FoxO1, FoxO3 and FoxO4, resulting in their sequestration in the cytoplasm such that they are unable to regulate their gene targets (Accili and Arden, 2004; Greer and Brunet, 2005). The role and essentiality of the FoxOs in tumor suppression in vivo has received formal proof from murine genetic studies wherein broad somatic deletion of all three FoxOs was shown to engender a cancer-prone condition dominated by hemangiomas and lymphomas (Paik et al., 2007). However, the highly context and cell-lineage specific functions of FoxO as revealed from this study also highlights the necessity to fully characterize its tumor suppression function in other cell types and tissue contexts. Here we studied FoxO tumor suppression function in the context of mTORC1-mediated renal tumorigenesis.
Results
FoxOs are activated in Tsc1 deficient polycystic kidneys, but lost in Tsc1 deficient renal adenomas and carcinomas
To better understand the molecular and biological role of mTORC1 hyper-activation in renal cancer development, we assessed the impact of homozygous deletion of Tsc1 conditional knockout allele (Tsc1L) (Kwiatkowski et al., 2002) using the Rosa26-CreERT2 knock-in deletor allele which enables tamoxifen-inducible Cre-mediated excision of conditional knockout alleles in most tissues, including kidneys (Vooijs et al., 2001). Tamoxifen-treatment of adult Tsc1L/L, Rosa26-CreERT2 mice resulted in efficient deletion of Tsc1 in the kidney (Fig. S1A and 1B) as well as other organs (data not shown). As recently reported (Gan et al., 2008), somatic deletion of Tsc1 in adult mice (Tsc1L/L, Rosa26-CreERT2, hereafter referred to as “Tsc1 KO”) led to the development of polycystic kidney disease (Fig. 1A a–c), as well as severe hematopoietic defects (data not shown) compared with littermate “Tsc1 WT” control mice (Tsc1L/L, or Tsc1+/+, Rosa26-CreERT2 mice treated with tamoxifen). All Tsc1 KO mice developed severe renal and bone marrow failure and died within 7 weeks (Gan et al., 2008). Notably, all Tsc1 KO mice developed bilateral polycystic kidneys with >10-fold greater weights relative to littermate controls (Fig. 1Ac) and significantly dilated renal tubules on histopathological analysis (Fig. 1Aa and b), which is consistent with the documented connection between tuberous sclerosis and polycystic kidney disease (Cai and Walker, 2006).
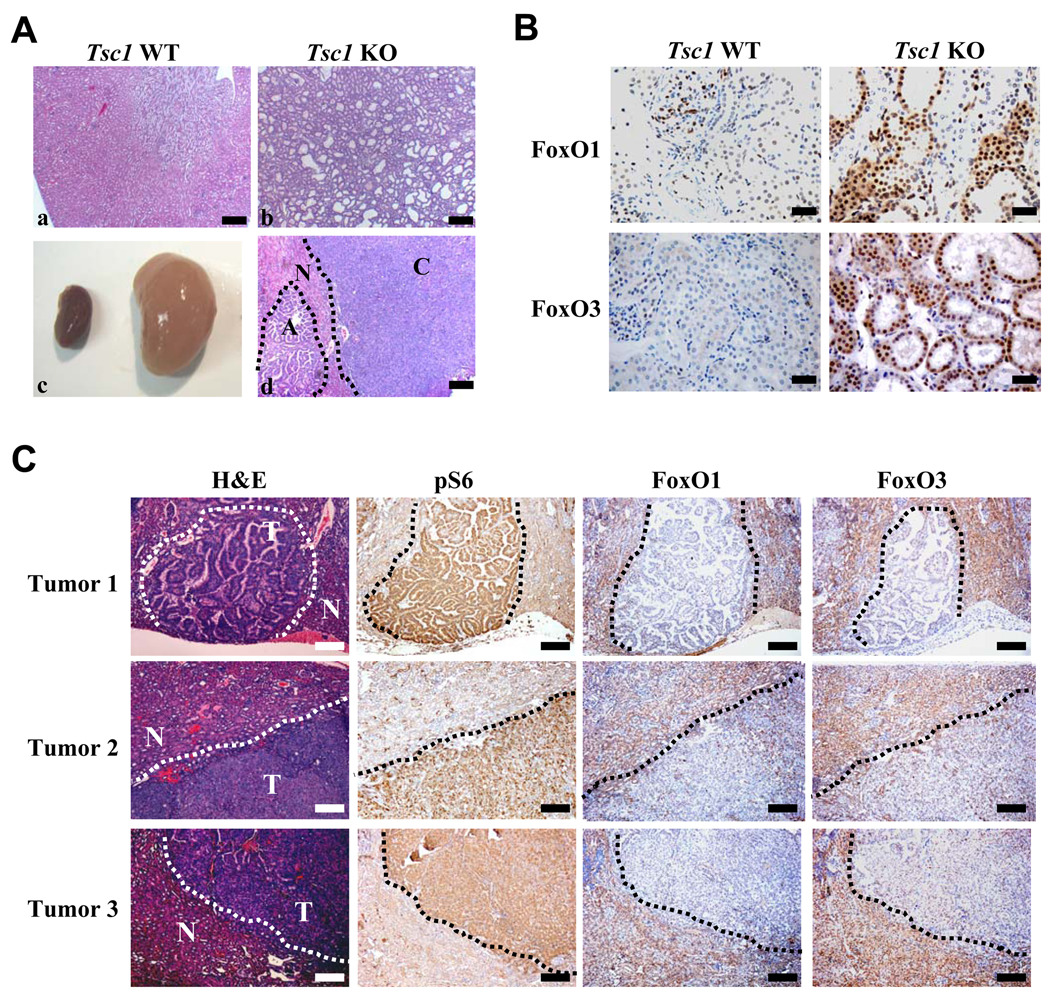
Figure 1. FoxOs are activated in Tsc1 deficient polycystic kidneys, but lost in Tsc1 deficient renal adenomas and carcinomas.
(A) Gross view of Tsc1 WT (left) and KO kidneys (right) at 30 days post tamoxifen injection (DPI) at 9 weeks of age. (c). Haematoxylin and eosin (H&E) staining of Tsc1 WT and KO kidney sections (a and b), and Tsc1 deficient kidney tumors (d). “C” denotes carcinoma, “A” denotes adenoma, and “N” denotes surrounding normal kidney cells. Scale bars: 200 um. (B) Immunohistochemical staining of Tsc1 WT and KO kidney sections with antibodies against FoxO1 and FoxO3, showing predominant nuclear localization of FoxO1 and FoxO3 in Tsc1 KO kidneys compared with WT kidneys. Scale bars: 20 um. (C) H&E and immunohistochemical staining of Tsc1 KO renal tumors with antibodies against phospho S6, FoxO1 and FoxO3, showing increased phospho S6 staining and marked reduction or absence of FoxO1 and FoxO3 staining in Tsc1 deficient renal tumors compared with adjacent normal kidney cells. In H&E staining sections, “T” denotes tumor region, “N” denotes adjacent normal kidney region. Scale bars: 100 um. See also Figure S1.
The Tsc1 KO polycystic kidneys exhibited increased phospho-S6 staining compared with WT kidneys (Fig. S1C), and rapamycin treatment of Tsc1 KO mice resulted in full rescue of the polycystic kidney defect (data not shown), strongly suggesting that mTORC1 hyper-activation upon loss of Tsc1 drives the polycystic kidney phenotype in the Tsc1 KO model. Given the lethality of the Tsc1 KO mice, we also established a large cohort of Tsc1L/+, Rosa26-CreERT2 mice (hereafter referred to as “Tsc1+/−”) for long-term tumor studies. Consistent with previous reports from mice heterozygous for germ line Tsc1 null allele (Kobayashi et al., 2001; Kwiatkowski et al., 2002), our conditional Tsc1+/− model showed renal adenomas and/or carcinomas following long latency (>14 months) (Fig. 1Ad, and Fig. 2B). These renal adenomas and carcinomas demonstrated loss of heterozygosity (LOH) for the remaining Tsc1 allele (6 of 6 samples examined; data not shown) as well as increased phospho-S6 staining indicative of mTORC1 hyper-activation (Fig. 1C).
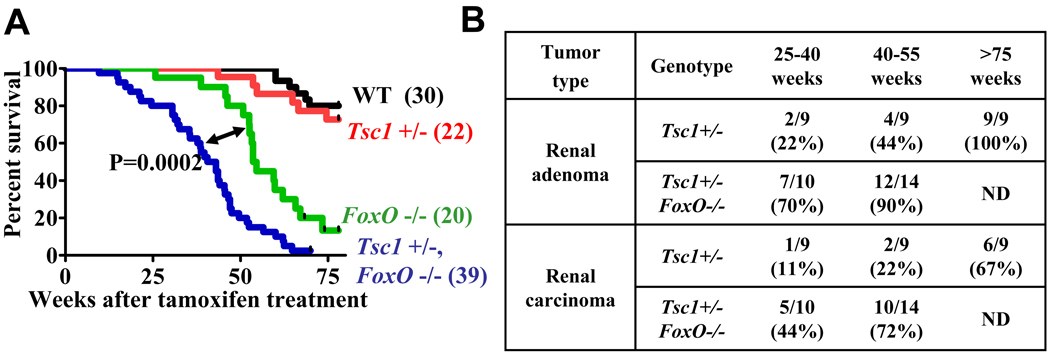
Figure 2. Dual inactivation of FoxO and Tsc1 dramatically drives renal tumor progression.
(A) Kaplan-Meier overall survival analysis for mice of indicated genotypes as a function of weeks after tamoxifen treatment. Cohort size for each mice colony is also indicated. (B) Table showing the incidence of renal adenoma and carcinoma in Tsc1+/− and Tsc1+/− FoxO−/− mice at various stages. ND: not determined.
The long latency required for the development of Tsc1 deficient renal carcinomas prompted us to consider that this model could prove useful in the identification of physiologically relevant checkpoint pathways activated in Tsc1 KO premalignant polycystic kidneys and enable the validation of such checkpoints through a detailed comparative analysis of activation status of TSC-mTORC1-related signaling surrogates in Tsc1 deficient polycystic lesions versus tumor samples. These comparative analyses revealed strong FoxO1 and FoxO3 staining with predominant nuclear localization (Fig. 1B), a pattern consistent with strong FoxO activation in Tsc1 KO polycystic kidneys relative to WT kidney samples.
In line with these tissue findings, Tsc1 and Tsc2 KO MEFs exhibited decreased FoxO1/3 phosphorylation compared with WT MEFs (Fig. S1D) and, accordingly, showed predominant FoxO1/3 nuclear localization under both serum starvation and stimulation conditions (Fig. S1E). This profile corresponds well with the anticipated constitutively high mTORC1 activity and dramatically reduced AKT phosphorylation in Tsc deficient MEFs (Fig. S1D). On this basis, we considered a model in which the regulation of FoxO activity might play a critical effector role in the mTORC1-AKT negative feedback circuit and that FoxO extinction would promote tumorigenesis. In line with this hypothesis, we observed marked reduction/loss of both FoxO1 and FoxO3 staining in all Tsc1 KO renal adenoma/carcinoma samples examined (10 of 10 bilateral kidneys, each with multiple renal adenomas/carcinomas) compared with surrounding non-malignant cells (Fig. 1C). Furthermore, western blotting analysis revealed significant reduction of FoxO4 protein levels in Tsc1 deficient renal tumors compared with adjacent normal kidney samples (Fig. S1F). Together, these data are consistent with the possibility that FoxO activation participates in a feedback checkpoint functioning to restrain the renal tumorigenesis initiated by loss of Tsc1, and that subsequent loss of FoxO may be required for renal cancer development in Tsc1 deficient mice.
Dual inactivation of FoxO and Tsc1 dramatically drives renal tumor progression
To secure genetic evidence for the hypothetical FoxO-dependent block of Tsc1 KO renal carcinoma development, we engineered mice harboring conditional knockout alleles of Tsc1 and/or FoxO1/3/4, and Rosa26-CreERT2 (For simplicity, after tamoxifen treatment to induce gene deletion, “FoxO1/3/4L/L, Rosa26-CreERT2” mice will be referred to as “FoxO−/−” hereafter.) We established Tsc1+/+ FoxO+/+ (WT), Tsc1+/− FoxO−/−, Tsc1+/− FoxO+/+ and Tsc1+/+ FoxO−/− cohorts and monitored survival and cancer predisposition over a period of 80 weeks. Although there was no significant survival difference between Tsc1+/− and WT mice, there was a marked Tsc1-dependent reduction in lifespan of FoxO−/− mice (FoxO−/− mean survival 54.1 weeks vs Tsc1+/− FoxO−/− mean survival 41.8 weeks, P=0.0002) (Fig. 2A). The decrease in survival in FoxO−/− mice relates to hemagiosarcomas and thymic lymphomas as previously reported (Paik et al., 2007) and careful inspection of the kidneys showed no evidence of renal adenoma or carcinoma (data not shown). In contrast, Tsc1+/− FoxO−/− mice showed dramatic reduction in latency and increased penetrance of renal adenomas and carcinomas (Fig. 2B). Notably, the penetrance and onset of FoxO−/− cancer phenotypes (hemagiosarcomas and thymic lymphomas) were not affected in Tsc1+/− FoxO−/− mice. The accelerated renal tumor development in Tsc1+/−FoxO−/− mice significantly contributed to the shorter life span of Tsc1+/− FoxO−/− mice compared with FoxO−/− mice. These genetic studies establish that FoxO activation is a potent block in Tsc1 null renal tumor development in the in vivo setting.
FoxOs are extinguished in the majority of human renal tumor samples
The above data from murine model systems prompted detailed examination of FoxO expression/activation status in human kidney tumor samples. RCC is the most common malignancy of the adult kidney and presents as a heterogeneous group of tumors with distinct histological features, including clear cell, papillary (including both type 1 and type 2), and chromophobe RCC as well as rarer subtypes (Lopez-Beltran et al., 2006). Clear cell RCC (ccRCC) is the predominant subtype and accounts for up to 75% of total RCC cases (Linehan and Zbar, 2004; Rini et al., 2009). It is generally considered that all the clear-cell tumors are carcinomas, with greater or lesser aggressiveness, and an adenoma state of clear cells is not accepted (Algaba, 2008). Compared with more aggressive clear cell and papillary subtypes, chromophobe RCCs tend to have a benign course after surgery, provided that the tumor stage and grade are favorable at the time of surgery (Cohen and McGovern, 2005). Benign epithelial neoplasms also occur in the kidney, including oncocytoma and metanephric adenoma, among others.
To assess FoxO expression in this spectrum of benign and aggressive kidney tumor types, FoxO1 and FoxO3 protein abundance was determined by immunohistochemistry (IHC) in a tissue microarray (TMA) which contained 21 normal kidney samples, 68 clear cell RCC, 10 papillary RCC, 8 chromophobe RCC, 5 oncocytomas as well as several rare renal tumor subtypes. TMA-IHC analysis revealed FoxO1 and FoxO3 cytoplasmic and nuclear signal in both the proximal and distal tubular epithelium in normal kidney samples (Fig. 3Aa and 3Ca). In contrast, FoxO1 and FoxO3 signal was not detectable in the majority of clear cell and papillary RCC samples (FoxO1: 64 of 68 clear cell and 8 of 10 papillary; FoxO3: 63 of 68 clear cell and 7 of 10 papillary) (Fig. 3A–3D). FoxO1 and FoxO3 signal was readily detected in the tumor endothelial cells, providing a positive internal staining control (Fig. S2A; data not shown). Notably, all of the less aggressive RCCs and benign renal tumors retained FoxO1 and/or FoxO3 expression (Fig. 3A–3D). FoxO1 nuclear signal was especially strong in oncocytoma samples (Fig. S2B).
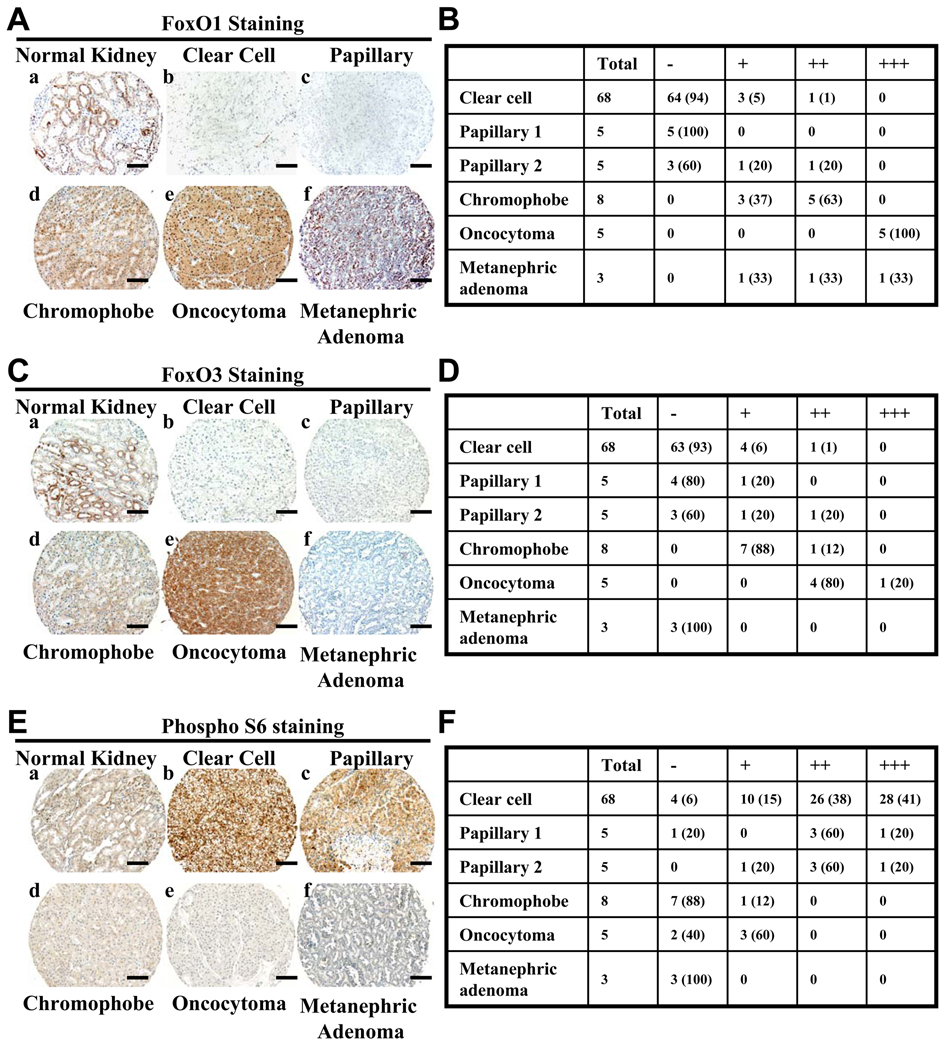
Figure 3. FoxOs are extinguished in the majority of human renal tumor samples.
(A, C and E) Representative immunohistochemical images showing FoxO1 (A), FoxO3 (C) and phospho-S6 (E) staining in normal kidney and different subtypes of RCC samples from the TMA-IHC analysis. Scale bars: 50 um. (B, D and F) Tables showing the number of tumor samples with corresponding staining scores of FoxO1 (B), FoxO3 (D) and phospho-S6 (F) in different subtypes of RCC samples. Percentage is shown in parenthesis. −: negative staining; +: low staining; ++: moderate staining; +++: strong staining. See also Figure S2.
We also assessed phospho-S6 staining on the same TMA sample set. This analysis revealed negative to weak phospho-S6 staining in most normal kidney samples and moderate to strong phospho-S6 staining in the majority of human renal clear cell and papillary carcinomas samples (Fig. 3E–3F), consistent with previous reports that mTORC1 hyper-activation is observed in the majority of human RCC samples (Pantuck et al., 2007; Robb et al., 2007). Importantly, our analysis also showed negative to weak phospho-S6 staining in chromophobe RCC, oncocytoma and metanephric adenoma samples (Fig. 3E–3F). The inverse staining pattern between FoxO1/3 and phospho-S6 from different human renal tumor types provides further support to our conclusion that FoxOs serve as a checkpoint in mTORC1-activated renal tumorigenesis.
FoxO activation promotes cell cycle arrest and apoptosis in human RCC cells
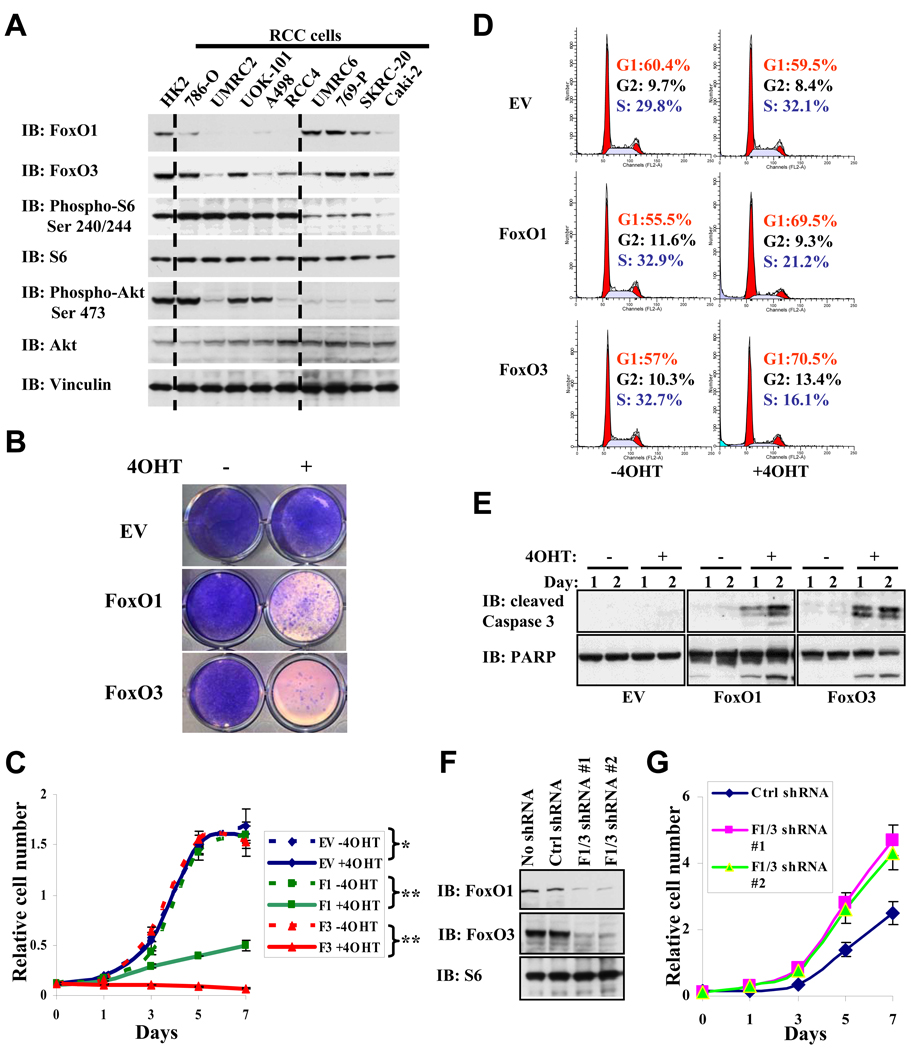
The above data prompted further examination of FoxO function in human RCC cell lines. We first examined the expression levels of FoxO1/3 and mTORC1 activation status in a panel of human RCC cell lines and the immortalized WT human kidney cells (HK2). Consistent with the data from murine renal tumors (Fig. 1C), these analyses revealed that high phosphorylation levels of S6 (surrogate of mTORC1 activation) in general correlated with low expression levels of FoxO1 and FoxO3 in these human RCC cell lines (Fig. 4A). We then assessed the impact of FoxO reactivation in RCC4 and UMRC2 cells which harbor low FoxO1/3 expression and high mTORC1 activation. To this end, we generated RCC4 and UMRC2 cell lines with stable expression of FoxO1(TA)ERT2 or FoxO3(TA)ERT2 construct, which expressed a fusion protein consisting of FoxO(TA) (containing three Ser/Thr AKT phosphorylation sites mutated to alanine) fused to the T2-modified estrogen receptor (ERT2) moiety (Fig. S3A). We documented that FoxO(TA)ERT2 fusion protein sequestered FoxO(TA) in the cytoplasm and that 4OHT treatment resulted in rapid translocation of FoxO(TA)ERT2 into nucleus (Fig. S3B). We also established stable cell lines with ERT2 expression as control cell lines (For simplicity, ERT2, FoxO1(TA)ERT2 and FoxO3(TA)ERT2 cell lines will be referred to as EV (empty vector), FoxO1 and FoxO3 thereafter). 4OHT treatment of FoxO1 or FoxO3 RCC4 cells resulted in marked cell growth suppression compared with vehicle-treated cells or 4OHT-treated EV RCC4 cells (Fig. 4B and 4C). Further analysis demonstrated that FoxO reactivation in RCC4 cells led to potent G1/S cell cycle arrest (Fig. 4D) and enhanced apoptosis (Fig. 4E). Reactivation of FoxO1/3 in UMRC2 cells resulted in similar phenotypes (Fig. S3C–3E). Finally, in UOK101 cells which maintain FoxO3 and FoxO1 expression, knockdown of both FoxO1 and FoxO3 led to increased cell proliferation (Fig. 4F–4G). These human RCC cell line data align with our murine data, strongly supporting the view that FoxOs function as tumor suppressors in the progression to human RCC.
Figure 4. FoxO activation promotes cell cycle arrest and apoptosis in human RCC cells.
(A) FoxO1/3 expression and mTORC1 activation status in human RCC cells. Western blotting by various antibodies was performed on cell lysates isolated from HK2 normal kidney cells and a panel of human RCC cell lines as indicated. (B) Image showing the crystal violet staining of RCC4 EV, RCC4 FoxO1, RCC4 FoxO3 cells with or without 4OHT treatment for 7 days. EV: empty vector. (C) Cell proliferation assay of RCC4 EV, RCC4 FoxO1, RCC4 FoxO3 cells with or without 4OHT treatment. *: P > 0.5; **: P = 6.6 × 10−5 (all refer to P values at 7th day). (D) FACS analysis showing the cell cycle profiling of RCC4 EV, RCC4 FoxO1 (F1), RCC4 FoxO3 (F3) cells with or without 4OHT treatment for 2 days. (E) RCC4 EV, RCC4 FoxO1, RCC4 FoxO3 cells were treated with or without 4OHT for 1 or 2 days as indicated. Cell lysates were then analyzed by Western blotting with cleaved Caspase-3 and PARP as indicated. (F) UOK101 cells were infected with ctrl (scramble) shRNA, FoxO1 and FoxO3 shRNA #1, or FoxO1 and FoxO3 shRNA #2-containing lentivirus. Cell lysates were extracted from stable cell lines and analyzed by Western blotting with FoxO1, FoxO3 and S6 as indicated. (G) Cell proliferation assay of UOK101 ctrl shRNA cells, UOK101 FoxO1/3 shRNA #1 cells, and UOK101 FoxO1/3 shRNA #2 cells. P < 10−4 for comparison between ctrl shRNA and F1/3 shRNA cells (all refer to P values at 7th day). See also Figure S3.
Myc signaling is the key downstream effector of FoxOs in the regulation of renal tumorigenesis
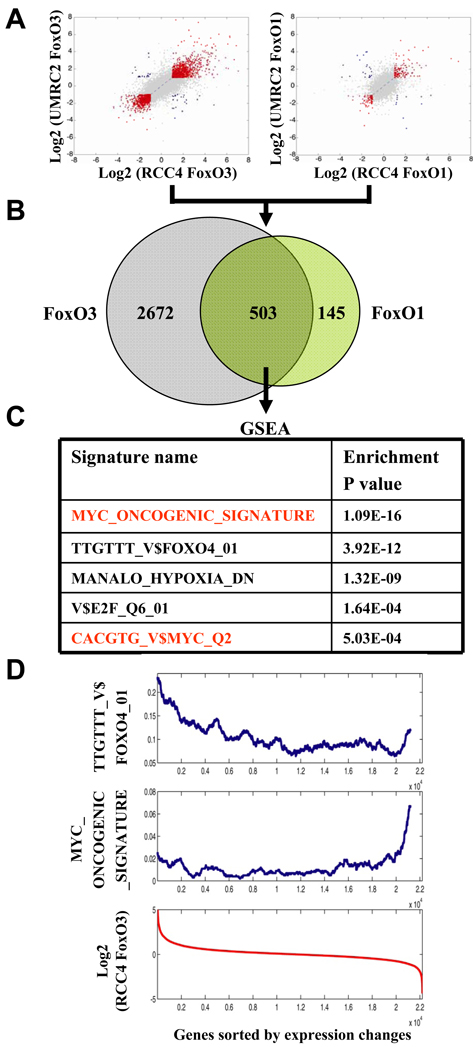
We next sought to determine the mechanisms by which FoxO might govern the biology of RCC cells. To this end, we conducted comparative transcriptome analysis of EV, FoxO1, or FoxO3-expressing RCC4 and UMRC2 cells at 12 hours with or without 4OHT treatment. To enrich for more proximal actions of FoxO, we selected the 12 hour time point as time course studies revealed dramatic transcriptional changes of known FoxO targets (such as Cyclin D1), yet no discernable cellular phenotypes (apoptosis and cell cycle arrest) (data not shown). We generated 4 transcriptome datasets: FoxO1 RCC4, FoxO3 RCC4, FoxO1 UMRC2, and FoxO3 UMRC2, and normalized these transcriptome data against 4OHT-treated EV cells which show modest 4OHT-induced transcriptional changes (Fig. 5A). As FoxO1 or FoxO3 reactivation generated comparable phenotypes, we further intersected FoxO1 and FoxO3 transcriptome datasets and focused on 503 genes exhibiting similar changes (designated as “FoxO RCC transcriptome”, Fig. 5B, Table S1).
Figure 5. Myc signaling is the key downstream effector of FoxOs in the regulation of renal tumorigenesis.
(A) Comparison of gene expression changes between RCC4 FoxO3 and UMRC2 FoxO3, RCC4 FoxO1 and UMRC2 FoxO1 transcriptome datasets. x and y axes show Log2 transformed fold changes in gene expression. Genes with fold changes greater than 1.5 or less than 0.67 for both x and y axes are highlighted (with changes in the same direction in red while the opposite direction in blue, respectively). (B) Venn diagram displaying common transcriptome changes of FoxO1 and FoxO3 transcriptome datasets. (C) The table showing the top lists of gene signatures enriched in FoxO RCC transcriptome dataset with enrichment P value indicated. (D) The distribution of putative FoxO targets (TTGTTT_V$FOXO4_01) and Myc-regulated genes (MYC_ONCOGENIC_SIGNATURE) in RCC4 FoxO3 transcriptome dataset. All genes were sorted according to expression changes in RCC4 FoxO3 transcriptome dataset (the red curve). The frequency of either putative FoxO targets (TTGTTT_V$FOXO4_01) or Myc-regulated genes (MYC_ONCOGENIC_SIGNATURE) in a sliding window of 1,000 sorted genes was calculated. See also Figure S4 and Table S1.
Next we conducted GSEA to identify gene sets that are enriched in FoxO RCC transcriptome. This analysis readily identified FoxO (TTGTTT_V$FOXO4_01), hypoxia (MANALO_HYPOXIA_DN) (Manalo et al., 2005) and E2F (V$E2F_Q6_01) gene sets in FoxO RCC transcriptome (Fig. 5C, see Fig. S4A for gene set description). Most notably, the analysis revealed a previously described Myc oncogenic signature (Bild et al., 2006) as the most significantly enriched gene set (P = 4.51 × 10−16, Fig. 5C and Fig. S4A). Accordingly, Myc binding motif-containing gene set (CACGTG_V$MYC_Q2) was also significantly enriched in FoxO RCC transcriptome (Fig. 5C and Fig. S4A). The connection of FoxO to Myc signaling in the context of RCC development is intriguing given recurrent Myc gene amplification in human RCC (Beroukhim et al., 2009), the involvement of Myc signaling in renal neoplasia as evidenced in genetically engineered mouse models (Schreiber-Agus et al., 1998; Trudel et al., 1991) and human cell culture studies (Eilers and Eisenman, 2008; Gordan et al., 2007a; Zhang et al., 2007) (see discussion). To understand how FoxO might intersect with Myc signaling, we first considered a model wherein FoxO and Myc might co-regulate a common set of transcription targets through co-binding on the same promoters. In silico promoter analysis of FoxO RCC transcriptome identified 90 putative FoxO direct targets and 81 putative Myc direct targets. However, integration of these two datasets only revealed 12 common targets, which is statistically insignificant (P > 0.5. Fig. S4B). Furthermore, the computational analysis showed that the FoxO putative targets (TTGTTT_V$FOXO4_01) are enriched in up-regulated genes of the FoxO RCC transcriptome, while Myc signature genes (MYC_ONCOGENIC_SIGNATURE) are mostly enriched in down-regulated genes of the FoxO RCC transcriptome (Fig. 5D), suggesting FoxO might function to inhibit Myc activity and/or expression.
FoxOs regulate Myc through Mxi-1 and mir-145 in RCC cells
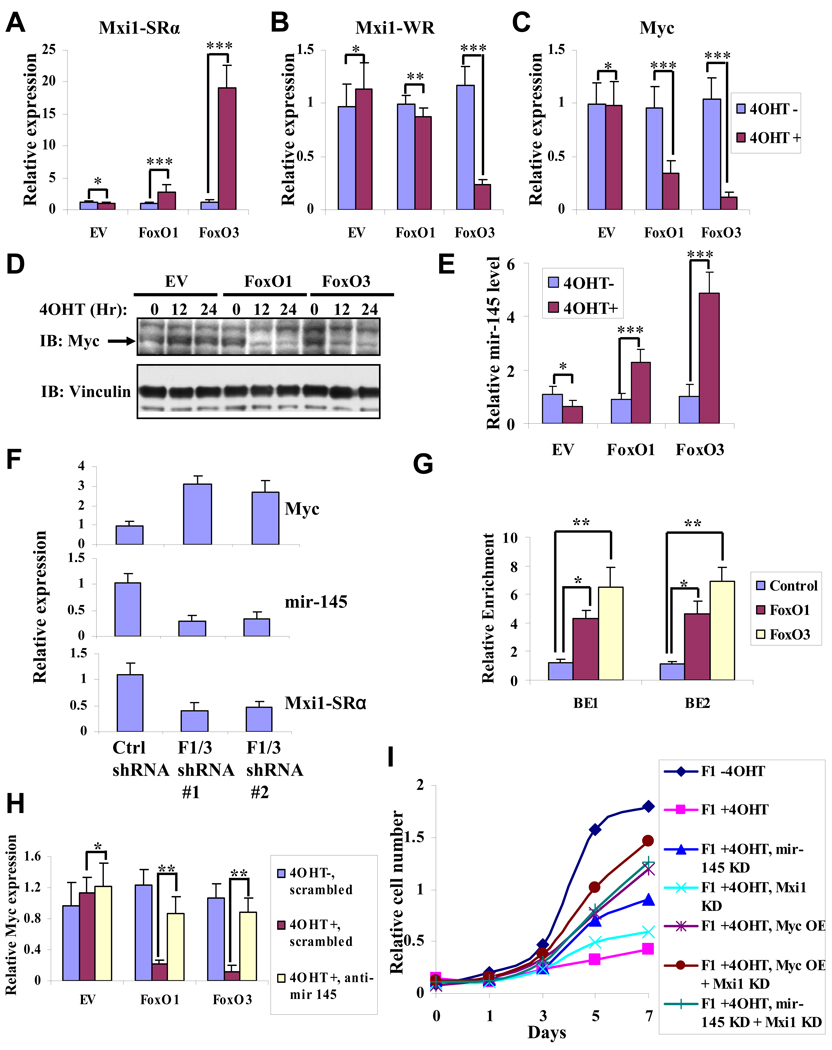
We and others previously showed that Mxi1 splicing variant strong repressor (Mxi1-SRα), but not Mxi1 weak repressor (Mxi1-WR), mediates antagonistic actions on Myc activity through recruitment of Sin3A-HDAC transcriptional repressor complex (Alland et al., 1997; Ayer et al., 1995; Heinzel et al., 1997; Schreiber-Agus et al., 1995). Mxi1-SRα was identified to be a FoxO direct target in other cellular contexts (Delpuech et al., 2007). We found FoxO activation significantly up-regulated Mxi1-SRα expression while down-regulating Mxi1-WR expression in both RCC4 and UMRC2 cells (Fig. 6A and 6B, Fig. S5A and S5B), suggesting FoxO might inhibit Myc activity via up-regulation of Mxi1-SRα expression.
Figure 6. FoxOs regulate Myc through Mxi-1 and mir-145 in RCC cells.
(A, B, C and E) Bar graph showing the relative expression changes of Mxi1-SRα (A), Mxi1-WR (B), Myc (C) and mir-145 (E) by quantitative RT-PCR in RCC4 EV, RCC4 FoxO1, and RCC4 FoxO3 cells with or without 4OHT treatment for 12 hours. *: P > 0.1; **: P = 0.04; ***: P < 0.01. (D) Western blotting showing endogenous Myc protein levels in RCC4 EV, RCC4 FoxO1, RCC4 FoxO3 cells at different time points after 4OHT treatment. (F) Bar graph showing the relative expression changes of Myc, Mxi1-SRα, and mir-145 by quantitative RT-PCR in UOK101 ctrl shRNA cells, UOK101 FoxO1/3 shRNA #1 cells, and UOK101 FoxO1/3 shRNA #2 cells. P < 0.05 for comparison between ctrl shRNA and F1/3 shRNA cells in each setting. (G) Endogenous FoxO1 and FoxO3 directly bind to two FoxO BEs located in mir-145 promoter region in UOK101 cells. Bar graph showing the relative enrichment determined by quantitative RT-PCR following ChIP analysis using IgG (control), FoxO1 and FoxO3 antibodies. *: P = 0.03; **: P = 0.01. (H) mir-145 contributes to FoxO-mediated suppression of Myc expression in RCC4 cells. Bar graph showing the relative expression levels of endogenous Myc by quantitative RT-PCR in RCC4 EV, RCC4 FoxO1 or RCC4 FoxO3 cells with anti-mir-145 or scrambled control oligo transfection. *: P > 0.5; **: P < 0.01. (I) Cell proliferation assay of RCC4 FoxO1 (F1) cells under different conditions as indicated. OE: overexpression; KD: knockdown. See also Figure S5.
In addition, we observed FoxO reactivation resulted in significant down-regulation of Myc expression in the FoxO RCC transcriptome and in confirmatory quantitative RT-PCR and western blot analysis of RCC4 cells and UMRC2 cells (Fig. 6C and 6D; Fig. S5C). The absence of FoxO binding elements in the Myc promoter region prompted us to consider that FoxO may regulate microRNA(s) governing Myc expression. To test this hypothesis, we examined the FoxO RCC transcriptome for enrichment of putative targets of microRNA genes, searched the microRNA promoter regions for putative FoxO binding elements (BEs), and determined whether any of these microRNAs could also target Myc via the targetScan algorithm and literature mining. This effort identified mir-145 which is known to target Myc (Sachdeva et al., 2009) and harbors 2 putative FoxO BEs within 2 kb upstream of mir-145 transcriptional start site (Fig. S5D). Quantitative RT-PCR confirmed that FoxO3 activation can significantly up-regulate mir-145 expression in RCC4 cells (Fig. 6E). Conversely, knockdown of FoxO1 and FoxO3 in UOK101 cells led to decrease of mir-145 and Mxi1-SRα expression, and increase of Myc expression (Fig. 6F). Furthermore, chromatin immunoprecipitation (ChIP) analysis documented that both ectopically expressed and endogenous FoxO1 and FoxO3 can directly bind to both FoxO BEs in mir-145 promoter region (Fig. 6G and Fig. S5E). Importantly, knockdown of mir-145 by anti-mir-145 (Fig. S5F) significantly normalized Myc expression upon FoxO activation (Fig. 6H), strongly suggesting that mir-145 contributes to FoxO-mediated suppression of Myc expression in RCC cells. Consistent with recent report in other cell types (Sachdeva et al., 2009), we found overexpression of mir-145 was sufficient to down-regulate Myc expression (Fig. S5G and S5H) and inhibit cell growth in RCC4 cells (Fig. S5I). Finally, knockdown of Mxi1-SRα, knockdown of mir-145, and overexpression of Myc, either individually or in combination, can significantly rescue the cell growth inhibition effected by FoxO activation in RCC4 cells (Fig. 6I). Together, these data establish that FoxO inhibits both Myc activity and expression via transcriptional regulation of Mxi1-SRα and mir-145, and that Myc signaling is at least one key downstream effector of FoxO in RCC cells.
Myc/ Mxi1-SRα/mir-145 alterations in Tsc1 renal cancer mouse models and human RCCs
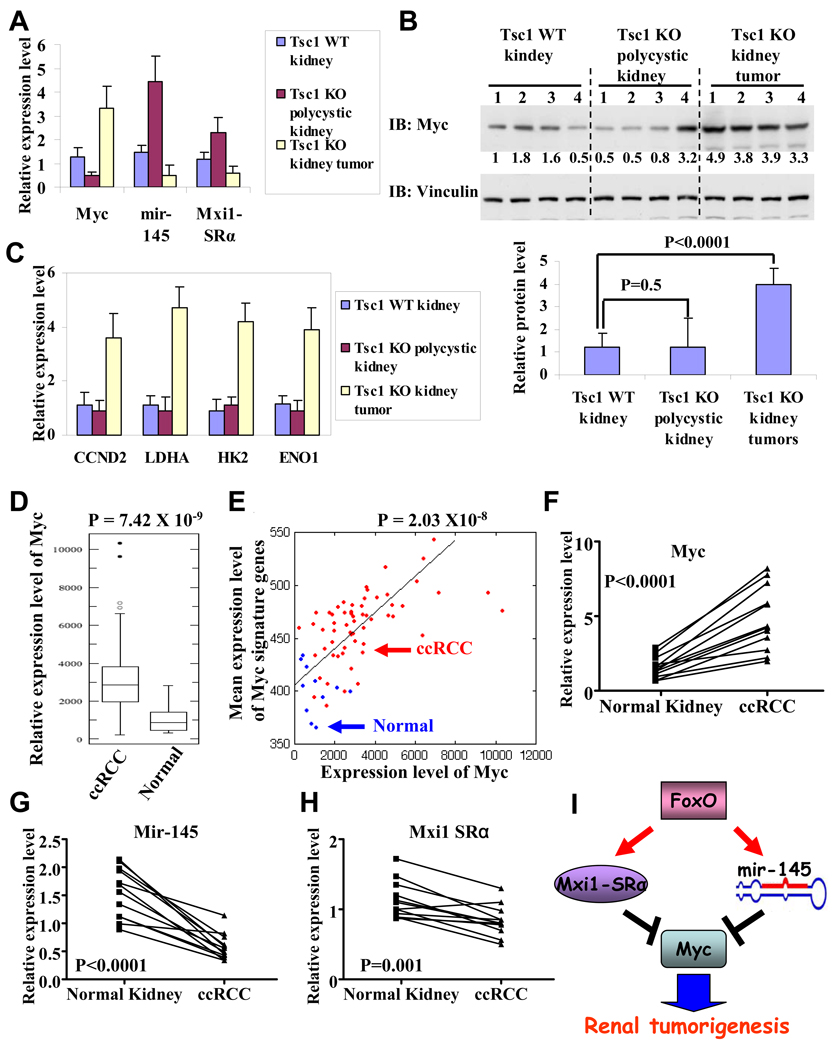
The above mechanistic studies in renal cancer cell culture systems prompted further investigation of Myc signaling in vivo in the context of RCC development in mice and humans. Examination of Myc/Mxi1-SRα/mir-145 status in Tsc1 mouse models revealed that mir-145 and Mxi1-SRα expression levels were upregulated in Tsc1 KO polycystic kidneys (with FoxO activation) and downregulated in Tsc1 KO kidney tumors (with FoxO extinction) compared with Tsc1 WT kidneys (Fig. 7A). Furthermore, while Myc expression was down-regulated in Tsc1 KO polycystic kidneys compared with Tsc1 WT kidneys (Fig. 7A), there was no obvious difference in Myc protein levels and in expression levels of Myc targets between Tsc1 KO polycystic kidneys and Tsc1 WT kidneys (Fig. 7B–7C). Myc expression and protein levels, and Myc target expression levels, are dramatically increased in Tsc1 KO kidney tumors compared with either Tsc1 WT kidneys or Tsc1 KO polycystic kidneys (Fig. 7A–7C). Since mTORC1 hyper-activation is also known to promote Myc translation (West et al., 1998), our data together suggest that, in Tsc1 KO polycystic kidneys with mTORC1 hyperactivation and FoxO activation, Myc protein level is balanced by both increased protein synthesis afforded by mTORC1 hyperactivation and decreased mRNA levels via FoxO activation. While in Tsc1 KO kidney tumors, loss of FoxO expression leads to increased Myc expression. This, together with increased protein synthesis, leads to further increased Myc protein level, which contributed to the renal tumorigenesis in Tsc1 KO mice.
Figure 7. Myc/Mxi1-SRα/mir-145 alterations in Tsc1 renal cancer mouse models and human RCCs.
(A) Bar graphs showing relative expression levels of Myc, Mxi1-SRα, mir-145 in Tsc1 WT kidneys, Tsc1 KO polycystic kidneys and Tsc1 KO kidney tumors. 4 samples were analyzed in each setting. P < 0.01 for comparison between Tsc1 WT kidneys and Tsc1 KO polycystic kidneys (or Tsc1 KO kidney tumors) for each gene. (B) Western blotting showing Myc and Vinculin protein levels in Tsc1 WT kidneys, Tsc1 KO polycystic kidneys and Tsc1 KO kidney tumors. Quantified Myc protein levels are also shown in the bar graph. (C) Bar graphs showing relative expression levels of Myc targets in Tsc1 WT kidneys, Tsc1 KO polycystic kidneys and Tsc1 KO kidney tumors. 4 samples were analyzed in each setting. P > 0.05 for comparison between Tsc1 WT kidneys and Tsc1 KO polycystic kidneys for each gene. P < 0.01 for comparison between Tsc1 WT kidneys (or Tsc1 KO polycystic kidneys) and Tsc1 KO kidney tumors. (D) Box-plots showing Myc expression levels in ccRCC and normal kidney samples. (E) Positive correlation between expression levels of Myc and Myc signature genes in ccRCC and normal kidney samples. X axis: expression levels of Myc; Y axis: mean expression levels of Myc signature genes. Blue spots: normal kidney sample; Red spots: ccRCC sample. (F–H) The relative expression levels of Myc, Mxi1-SRa, mir-145 in 12 ccRCC samples and matched normal kidney samples. (I) FoxO inhibits Myc activity via transcriptional upregulation of Mxi1-SRα and downregulates Myc mRNA level via transcriptional upregulation of mir-145. Myc signaling is the key downstream effector of FoxO to suppress renal tumorigenesis. See also Figure S6.
Examination of Myc/Mxi1-SRα/mir-145 status in human RCCs in published human RCC transcriptome dataset (including 11 normal kidney samples and 59 ccRCC samples) (Beroukhim et al., 2009) revealed that Myc expression was significantly upregulated in ccRCCs compared with normal kidneys (Fig. 7D. P = 7.42 × 10−9), correlating with aforementioned data that FoxO1/3 are extinguished in the majority of human ccRCC samples. Further computational analyses revealed that the mean expression levels of Myc signature genes (the same signature genes used in our FoxO RCC transcriptome analysis in Fig. 5C) were significantly upregulated in ccRCCs compared with normal kidneys (Fig. S6A. P = 4.12 × 10−11) and that the expression levels of Myc positively correlated with the mean expression levels of Myc signature genes in ccRCC and normal kidney samples (Fig. 7E. Pearson correlation coefficient: 0.61, P = 2.03 × 10−8), strongly suggesting that Myc signaling is also upregulated in ccRCCs. We also performed RT-qPCR experiments to examine the expression levels of Myc, selected Myc targets (CCND2, GADD45A, LDHA, HK2 and ENO1), mir-145 and Mxi1-SRα in 12 human ccRCC and matched normal kidney samples. These experiments confirmed upregulation of Myc, CCND2, LDHA, HK2 and ENO1, but downregulation of mir-145, Mxi1-SRα and GADD45A (Myc repressed target) in ccRCC compared with matched normal kidney samples (Fig. 7F–7H, Fig. S6B–6F). Together, our data strongly suggests that FoxO regulation of Myc/Mxi1-SRα/mir-145 signaling network plays a role in suppressing renal tumorigenesis in Tsc1 renal tumor mouse model and human RCCs (Fig. 7I).
Discussion
The mTORC1- mediated negative feedback regulation of PI3K-AKT (Shaw and Cantley, 2006), cancer type-specific nature of PI3K-AKT pathway signaling in vivo, and the multiple AKT downstream effectors in cancer (Manning and Cantley, 2007) prompted us to define mTORC1 circuitry on the genetic, genomic and biological levels in RCC pathogenesis. By using genetically defined mice and human RCC cell line models, this study establishes that FoxOs play a critical role in an mTORC1-directed negative feedback circuit in the context of renal tumorigenesis. FoxO1 and FoxO3 are robustly activated in Tsc1 deficient benign polycystic neoplasias as well as Tsc1 KO MEFs, and consistently extinguished upon progression to renal adenomas and carcinomas. In addition, combined deletion of Tsc1 and FoxO1/3/4 provoked dramatic exacerbation of the renal tumor phenotypes, establishing activated FoxO signaling as a potent block in Tsc1-null renal cancer development.
In elucidating the biological mechanisms by which loss of FoxO serves to promote Tsc1-null renal cancer development, we provide evidence that knockdown of FoxO in RCC cells leads to increased Myc expression and enhanced cell proliferation, supporting a model that loss of FoxO and subsequent Myc activation function to promote cell cycling at the transition from premalignant polycystic kidney epithelial cells to adenoma in Tsc1 mice. In addition, while FoxO is a key progression factor, we acknowledge that loss of FoxO (and Tsc1 LOH) may represent one of several genetic networks commandeered in the course of kidney tumorigenesis, a view consistent with lengthy renal tumor latency in our Tsc1+/− FoxO−/− mice. Thus, this model system may serve as a foundation for identification of cooperative genetic events involved in renal tumorigenesis.
The FoxO family members serve as redundant tumor suppressors in vivo, and FoxO1 and FoxO3 exert the major tumor suppressor activity (Paik et al., 2007). Correspondingly, we observed that FoxO1 and FoxO3 are always co-extinguished in Tsc1 deficient mouse renal tumors and human renal tumors, suggesting that both FoxOs need to be eliminated in order to bypass FoxO-mediated checkpoint. Furthermore, activation of FoxO1 and FoxO3 in human renal cancer cells resulted in similar cellular phenotypes (apoptosis and cell cycle arrest) and transcriptomic alterations (such as inhibition of Myc expression). Thus, we conclude that FoxO1 and FoxO3 function as redundant tumor suppressors in renal tumorigenesis.
In this study, we found FoxO1 and FoxO3 levels are extinguished in the majority of the aggressive subtypes of human RCC, including clear cell and papillary tumors, but maintained or robustly activated in less aggressive carcinomas (i.e., chromophobe RCC) and benign tumors. The underlying mechanisms responsible for such differential expression/activation patterns of FoxOs in different kidney tumor types are not understood currently. It is worth noting that recent oncogenomic analysis identified frequent hemizygous deletions at 13q in human clear cell RCC samples, and FoxO1 was identified as a down-regulated gene in the minimal common region of the 13q deletion (Kojima et al.), suggesting that genomic alteration might be at least a contributing mechanism for the loss of FoxO1 staining in our analysis. We speculate that other mechanisms operating on the epigenetic and post-translational levels might be also involved in effecting extinction of FoxO expression/activity in these RCC samples.
Our integrated transcriptomic, computational and functional studies identified Myc-Mxi1 signaling as the key downstream effector of FoxOs in the regulation of renal cancer cell proliferation. This finding is particularly intriguing in light of our previous work showing that deletion of Mxi1 in mice led to the development of polycystic kidney disease (PKD) similar to the PKD observed in Tsc1 deficient mice (Schreiber-Agus et al., 1998). Notably, Myc transgenic mice also developed PKD (Trudel et al., 1991). These mouse genetic studies thus provide important complementary evidence to our computational and functional analyses in support of the critical function of Myc-Mxi1 signaling in mTORC1-FoxO-directed renal tumorigenesis.
Myc and Mxi1 are also highly relevant to human RCC. Recent oncogenomic profiles show that Myc is amplified in about 12% clear cell RCC samples and is the only gene in the 8q amplification peak region, confirming Myc as an oncogene in human kidney cancer (Beroukhim et al., 2009). Furthermore, Mxi1 locus is subjected to homozygous and hemizygous deletion in a subset of clear cell RCC samples (Beroukhim et al., 2009), suggesting that Mxi1 functions as a tumor suppressor in human kidney cancer. Mechanistically, it has been shown that Myc can crosstalk with HIF signaling to regulate cell cycle entry, metabolism and mitochondrial biogenesis in RCC cells (Dang et al., 2008; Gordan et al., 2007a; Gordan et al., 2007b; Zhang et al., 2007). Further study will be necessary to investigate whether FoxO can regulate cancer cell metabolism and mitochondrial biogenesis in the context of renal tumorigenesis via Myc signaling.
Finally, our data also point to combination therapeutic regimens for improved treatment of renal cancer. Specifically, since our study demonstrated that FoxOs are extinguished in the majority of human RCC samples and serve as an critical tumor suppressor axis in renal cancer, it predicts that mTORC1 inhibition (or mTORC1/PI3K inhibition) would not affect the downstream oncogenic effect afforded by loss of FoxO, which explains, at least in part, why mTORC1 inhibition has relatively limited impact on kidney cancer patients. Our study further suggests combination therapy by mTORC1 inhibition and a FoxO re-stablization/activation and/or Myc de-stabilitization strategy might provide improved treatment in kidney cancer. As discussed above, currently we favor a model that FoxO extinction in human RCC might result from post-translational mechanism (such as phosphorylation-mediated protein degradation). Further understanding of how FoxOs are extinguished in human RCC might therefore provide treatment strategies in RCC, including such strategies as blocking Crm1-mediated nuclear export. Alternatively, our study suggests that combined inhibition of mTORC1 and Myc signaling might further improve the treatment of kidney cancer. Interference of Myc signaling may be provided by current efforts to target Myc heterodimerization with Max or by targeting factors regulating Myc protein degradation.
In summary, this integrated genetic, transcriptomic, computational and cell biological study has provided additional insights into the molecular basis of mTORC1-mediated negative feedback in the context of renal tumorigenesis. Our study identifies FoxO transcription factors as a critical checkpoint that functions to constrain mTORC1-mediated renal tumorigenesis and serves as a tumor suppressor axis in renal cancer. Further study of the FoxO network may illuminate additional points of therapeutic intervention and provide pharmacodynamic markers for guiding the development of agents targeting components of the PI3K pathway in RCC patients.
Experimental Procedures
Generation and analysis of mice
The Rosa26-CreERT2 mice (Vooijs et al., 2001) were mated with FoxO1/3/4L/L mice (Paik et al., 2007) to generate FoxO1/3/4L/L, Rosa26-CreERT2 mice, which were then crossed with Tsc1L/L mice (Kwiatkowski et al., 2002) to generate various mice lines with desired genotypes. All cohorts were in a FVB/n, C57Bl/6, and 129/Sv mixed genetic background. Littermates at 4–5 weeks of age were treated daily by intraperitoneal injection of tamoxifen in corn oil (0.13 mg/g body weight) for 5 consecutive days (Gan et al., 2008). The tamoxifen was intended to induce CreERT2 translocation into the nucleus, thereby leading to transient Cre-mediated recombination. Overall survival, as well as spontaneous formation of tumors, was monitored in various cohorts. Animals were autopsied and all tissues were examined regardless of their pathological status. All animal manipulations were performed with Harvard's and Dana-Farber Cancer Institute’s Institutional Animal Care and Use Committee (IACUC) approval.
Human samples
Human RCC TMA and RNA samples were obtained from Renal Cancer Tissue Acquisition, Pathology and Clinical Data Core (TAPCD) at Dana-Farber/Harvard Cancer Center (DF/HCC) Kidney Cancer SPORE directed by S. S.. All patients provided informed consent and the study was approved by the Institutional Review Boards of Dana-Farber Cancer Institute and DF/HCC Renal Cancer TAPCD Core Utilization Committee (RCTUC).
Antibodies for immunohistochemistry and western blot analysis
The following antibodies were used: Vinculin (Sigma), Myc (sc-42, Santa Cruz, to detect endogenous Myc), Myc (9E10, Santa Cruz, to detect Myc tag), S6K (Santa Cruz), TSC1, FoxO1 (C29H4), FoxO3 (75D8), FoxO4, Thr24 phospho-FoxO1/Thr32 FoxO3, AKT, Ser473 phospho AKT, S6, Ser240/244 phospho-S6, Thr389 phospho-S6K, PARP, cleaved-Caspase 3 (all from Cell Signaling Technology).
Constructs and reagents
FoxO1(TA) and FoxO3(TA) cDNAs were obtained from addgene, and then were fused with Myc-tagged ERT2 at C terminus and cloned in retroviral vector pBabe. Myc expression construct is described in our recent publication (Zheng et al., 2008). Lentiviral shRNA vectors targeting human Mxi1 and non-targeting control construct shGFP were obtained from the RNAi Consortium at the Dana-Farber/Broad Institute. Lentiviral mir-145 expression vector was purchased from System Biosciences. Anti-mir-145 and scrambled oligos were ordered from Exiqon.
Cell culture studies
Immortalized human kidney cells (HK2), human kidney cancer cell lines used in this study, and human embryonic kidney cell line HEK293T were purchased from American Type Culture Collection (ATCC). Lentiviruses or retroviruses were produced in HEK293T cells with packing mix (ViraPower Lentiviral Expression System, Invitrogen) and used to infect target cells as per manufacturer’s instruction. To reactivate FoxO, FoxO1(TA)ERT2 or FoxO3(TA)ERT2 cells were treated with 100 nM 4OHT or vehicle for different periods of time as indicated and subjected to various analyses. Cell proliferation assays were performed in 12-well plates in triplicate with 10,000 cells per well. Cells were fixed in 10% formalin in PBS, and stained with crystal violet at different days as indicated. At the conclusion of the assay, crystal violet was extracted with 10% acetic acid and absorbance at 595nm was measured with 96-well plate reader.
ChIP analysis
Chip experiment was performed using EZ-Chip Kit (Chemicon) as per manufacturer’s instruction.
Expression profiling and quantitative real-time PCR analysis
Details of expression profiling and quantitative real-time PCR analysis are provided in supplemental experimental procedures.
Statistical Analysis
Tumor-free survivals were analyzed using Graphpad Prism4. Statistical analyses were performed using nonparametric Mann–Whitney test. Comparisons of cell growth and gene expression were performed using the unpaired Student's t-test. For all experiments with error bars, standard deviation was calculated to indicate the variation within each experiment and data, and values represent mean ± SEM.
Significance
The clinical response of RCC to mTORC1 inhibition has been modest. The clinical application of mTORC1 inhibitors may be improved by the development of mTORC1-directed mouse models of RCC and by illumination of the mTORC1 activation network in RCC pathogenesis. This study reports the engineering of a highly penetrant model of renal adenoma/carcinoma and identification of a FoxO-Myc network as integral regulators of renal tumorigenesis in mice and humans. Illumination of this circuit may provide strategies to understand the variable clinical response to mTORC1 inhibition in RCC patients and motivate the development of combination treatment by mTORC1 inhibitors and agents reactivating FoxO or targeting Myc in human RCC.
Highlight
FoxOs are activated in polycystic kidneys, but lost in renal tumors of Tsc1 KO mice.
Dual genetic inactivation of FoxO and Tsc1 drives renal tumor progression.
FoxOs are extinguished in aggressive, yet expressed in benign, human renal tumors.
Myc antagonists are FoxO effectors in suppressing renal tumorigenesis.
Supplementary Material
Acknowledgements
We are grateful to Anton Berns for providing Rosa26-CreERT2 mouse strain. We are also grateful to Shan (Julia) Zhou for the assistance in the animal facility. This research is supported by National Cancer Institute grant R21CA135057 (R. A. D. and B. G.), U01CA141508 (R. A. D.), 1P01CA120964 (D. J. K.), DF/HCC Kidney Cancer SPORE Career Development Grant (P50CA101942-06A1) and DOD TSCRP Career Transition Award (TS093049) (B. G.). B. G. is the Research Fellow of the Leukemia & Lymphoma Society. S. H. is supported by Damon Runyon Cancer Research Foundation Postdoctoral Fellowship. Y. A. W. is supported by Multiple Myeloma Research Foundation. R. A. D. was supported by an American Cancer Society Research Professorship and the Robert A. and Renee E. Belfer Foundation Institute for Innovative Cancer Science.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Number. Completed microarray data are deposited on the GEO website under super series accession number GSE23926.
References
- Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- Algaba F. Renal adenomas: pathological differential diagnosis with malignant tumors. Adv Urol. 2008 doi: 10.1155/2008/974848. 974848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Atkins MB, Hidalgo M, Stadler WM, Logan TF, Dutcher JP, Hudes GR, Park Y, Liou SH, Marshall B, Boni JP, et al. Randomized phase II study of multiple dose levels of CCI-779, a novel mammalian target of rapamycin kinase inhibitor, in patients with advanced refractory renal cell carcinoma. J Clin Oncol. 2004;22:909–918. doi: 10.1200/JCO.2004.08.185. [DOI] [PubMed] [Google Scholar]
- Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar PT, Hay N. The two TORCs and Akt. Dev Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Cai SL, Walker CL. TSC2, a key player in tumor suppression and cystic kidney disease. Nephrol Ther. 2006;2 Suppl. 2:S119–S122. [PubMed] [Google Scholar]
- Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353:2477–2490. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- Delpuech O, Griffiths B, East P, Essafi A, Lam EW, Burgering B, Downward J, Schulze A. Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Mol Cell Biol. 2007;27:4917–4930. doi: 10.1128/MCB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Sahin E, Jiang S, Sanchez-Aguilera A, Scott KL, Chin L, Williams DA, Kwiatkowski DJ, DePinho RA. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc Natl Acad Sci U S A. 2008;105:19384–19389. doi: 10.1073/pnas.0810584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007a;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007b;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Hanna SC, Heathcote SA, Kim WY. mTOR pathway in renal cell carcinoma. Expert Rev Anticancer Ther. 2008;8:283–292. doi: 10.1586/14737140.8.2.283. [DOI] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudes G, Carducci M, Tomczak P, Dutcher J, Figlin R, Kapoor A, Staroslawska E, Sosman J, McDermott D, Bodrogi I, et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med. 2007;356:2271–2281. doi: 10.1056/NEJMoa066838. [DOI] [PubMed] [Google Scholar]
- Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Minowa O, Sugitani Y, Takai S, Mitani H, Kobayashi E, Noda T, Hino O. A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proc Natl Acad Sci U S A. 2001;98:8762–8767. doi: 10.1073/pnas.151033798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T, Shimazui T, Horie R, Hinotsu S, Oikawa T, Kawai K, Suzuki H, Meno K, Akaza H, Uchida K. FOXO1 and TCF7L2 genes involved in metastasis and poor prognosis in clear cell renal cell carcinoma. Genes Chromosomes Cancer. 49:379–389. doi: 10.1002/gcc.20750. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Manning BD. Tuberous sclerosis: a GAP at the crossroads of multiple signaling pathways. Hum Mol Genet 14 Spec No. 2005;2:R251–R258. doi: 10.1093/hmg/ddi260. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Zhang H, Bandura JL, Heiberger KM, Glogauer M, el-Hashemite N, Onda H. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- Li Y, Corradetti MN, Inoki K, Guan KL. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem Sci. 2004;29:32–38. doi: 10.1016/j.tibs.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Linehan WM, Zbar B. Focus on kidney cancer. Cancer Cell. 2004;6:223–228. doi: 10.1016/j.ccr.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Lopez-Beltran A, Scarpelli M, Montironi R, Kirkali Z. 2004 WHO classification of the renal tumors of the adults. Eur Urol. 2006;49:798–805. doi: 10.1016/j.eururo.2005.11.035. [DOI] [PubMed] [Google Scholar]
- Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4:257–262. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- Manning BD. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J Cell Biol. 2004;167:399–403. doi: 10.1083/jcb.200408161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, Grunwald V, Thompson JA, Figlin RA, Hollaender N, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372:449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantuck AJ, Seligson DB, Klatte T, Yu H, Leppert JT, Moore L, O'Toole T, Gibbons J, Belldegrun AS, Figlin RA. Prognostic relevance of the mTOR pathway in renal cell carcinoma: implications for molecular patient selection for targeted therapy. Cancer. 2007;109:2257–2267. doi: 10.1002/cncr.22677. [DOI] [PubMed] [Google Scholar]
- Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- Robb VA, Karbowniczek M, Klein-Szanto AJ, Henske EP. Activation of the mTOR signaling pathway in renal clear cell carcinoma. J Urol. 2007;177:346–352. doi: 10.1016/j.juro.2006.08.076. [DOI] [PubMed] [Google Scholar]
- Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi AI, DePinho RA. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N, Meng Y, Hoang T, Hou H, Jr, Chen K, Greenberg R, Cordon-Cardo C, Lee HW, DePinho RA. Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature. 1998;393:483–487. doi: 10.1038/31008. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Trudel M, D'Agati V, Costantini F. C-myc as an inducer of polycystic kidney disease in transgenic mice. Kidney Int. 1991;39:665–671. doi: 10.1038/ki.1991.80. [DOI] [PubMed] [Google Scholar]
- Um SH, D'Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MJ, Stoneley M, Willis AE. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene. 1998;17:769–780. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.