Abstract
The membrane proteins of peripheral light-harvesting complexes (LHCs) bind chlorophylls and carotenoids and transfer energy to the reaction centers for photosynthesis. LHCs of chlorophytes, chromophytes, dinophytes, and rhodophytes are similar in that they have three transmembrane regions and several highly conserved Chl-binding residues. All LHCs bind Chl a, but in specific taxa certain characteristic pigments accompany Chl a: Chl b and lutein in chlorophytes, Chl c and fucoxanthin in chromophytes, Chl c and peridinin in dinophytes, and zeaxanthin in rhodophytes. The specificity of pigment binding was examined by in vitro reconstitution of various pigments with a simple light-harvesting protein (LHCaR1), from a red alga (Porphyridium cruentum), that normally has eight Chl a and four zeaxanthin molecules. The pigments typical of a chlorophyte (Spinacea oleracea), a chromophyte (Thallasiosira fluviatilis), and a dinophyte (Prorocentrum micans) were found to functionally bind to this protein as evidenced by their participation in energy transfer to Chl a, the terminal pigment. This is a demonstration of a functional relatedness of rhodophyte and higher plant LHCs. The results suggest that eight Chl-binding sites per polypeptide are an ancestral trait, and that the flexibility to bind various Chl and carotenoid pigments may have been retained throughout the evolution of LHCs.
The gain of peripheral light-harvesting proteins for enhancement of light absorption and energy transfer to the photosynthetic reaction centers was a major advancement in the evolution of chloroplasts. In cyanobacteria, considered to be the ancestors of chloroplasts (1, 2), phycobilisomes serve as the primary light-harvesting antenna and feed energy to photosystem II. In contrast, most eukaryotic photosynthetic organisms contain membrane-intrinsic light-harvesting complexes (LHCs) associated with both photosystems I and II (PSI and PSII). In higher plants, possession of Chls a and b in association with LHCs and the carotenoid pigments of the xanthophyll cycle is regarded as an advantage for success on land (3). The increased absorbance in the range 450–550 nm by Chl c and fucoxanthin in chromophytes, and peridinin in dinophytes can enhance acclimation to varied light quality in marine environments (4).
The photosynthetic apparatus of red algae, as exemplified by Porphyridium cruentum, appears to represent a transitional state between cyanobacteria and chloroplasts of photosynthetic eukaryotes including higher plants. Rhodophytes are like cyanobacteria in their phycobilisome antenna, thylakoid architecture, and simplicity of pigmentation, with Chl a and zeaxanthin as the predominant thylakoid pigments (5, 6). However, rhodophytes like most other photosynthetic eukaryotes (5–9) have enhanced light-harvesting capacity by virtue of having LHCs associated with PSI. In P. cruentum, where phycobilisomes feed light energy to the reaction center of PSII, there are no intrinsic LHCs associated with this reaction center (5, 6), which is possibly indicative of a less advanced state.
The LHCs of the red alga P. cruentum (LhcaR1, LhcaR2) are similar in sequence and predicted structure to LHCs of green plants and of other algae (6, 10), but phylogenetic analyses suggest a closer phylogenetic affinity with LHCs of chromophytes and dinophytes than with those of chlorophytes (11). Three transmembrane helices are a common feature of most LHCs, with eight conserved Chl-binding sites in chromophytes, dinophytes, and rhodophytes (6, 10, 12). Whereas the PSII core antennas CP24, CP26, and CP29 of higher plants similarly possess 8 Chl-binding sites (13, 14), LHCII has 12 Chls. The 3.4 Å crystal structure established by Kühlbrandt et al. (15) shows that LHCII is also more complex and possesses a fourth helix with an additional Chl-binding residue (15). In green plants, these sites have been verified as Chl-binding sites by mutational analyses by showing loss of Chls and sometimes carotenoids (16–18).
Although LHC proteins are highly conserved across many taxa, they bind very different pigments in different taxonomic groups, e.g., Chls a and b and lutein in chlorophytes, Chls a and c and fucoxanthin in chromophytes; Chls a and c and peridinin in dinophytes; and Chl a and zeaxanthin in most rhodophytes. How did this diversity in pigment binding evolve? Were the pigment-binding sites inherent in the ancestral protein, or alternatively, did binding sites for specific pigments arise within the various taxa? In vitro reconstitution experiments showing that certain Chl-binding sites in higher plant LHCs are able to bind either Chl a or Chl b interchangeably and that functional LHCs can be assembled with a variety of higher plant carotenoid types and ratios (13, 16) indicate that a certain plasticity may be inherent in LHCs. The transitional nature of the Porphyridium chloroplast between cyanobacteria and photosynthetic eukaryotes and the simpler structure of its LHCs make this rhodophyte an ideal candidate for investigation of plasticity of pigment binding.
Analysis of primary amino acid sequences has been used to predict phylogenetic relatedness of LHCs (11). However, testing the relatedness, i.e., ascertaining the binding of heterologous pigments to individual LHC proteins, is presently beyond in vivo molecular genetic approaches. Biosynthetic pathways for most Chls and carotenoids generally follow phylogenetic lines, and the multigene nature of LHCs makes gene deletion in organisms difficult. In vitro binding of pigments to LHC proteins initially proved its feasibility (19) and has been extensively used for examining LHCs of higher plants (13, 20, 21), and in a recent report this was extended for the first time to the red algal LHCaR1 (22).
In exploring the plasticity of LHCaR1, we tested the in vitro binding of pigment extracts of spinach, a diatom, and a dinophyte. Predominant pigments in these groups, although not synthesized by P. cruentum, were shown to bind and transfer energy to Chl a. The data are consistent with the concept that flexibility in pigment binding was inherent in ancestral LHCs.
Materials and Methods
Purification of LHCaR1 Expressed in Escherichia coli.
LHCaR1 (10) was produced in E. coli and the inclusion bodies containing the polypeptide product were isolated according to the method of Paulsen et al. (20) as described in Grabowski et al. (22).
Pigment Preparations and LHC Reconstitution.
Unialgal cultures of the diatom Thallasiosira fluviatilis and the dinoflagellate Prorocentrum micans were grown in large-scale bioreactors and harvested at 5,000 × g for 5 min. Cell pellets were stored at −80°C, and pigments were extracted from pellets of T. fluviatilis and P. micans by a method modified from Svec (23). T. fluviatilis pigments were extracted in cold methanol, diethyl ether, and petroleum ether (5:2:1 vol/vol), and P. micans pigments were extracted in cold 100% methanol. Extractions of partially thawed cell pellets were done on ice and under dim light to minimize pigment degradation. Subsequently, the supernatant was partitioned into diethyl ether with the addition of 10% NaCl (wt/vol), evaporated, and stored under nitrogen at −20°C. Pigment extraction from fresh market spinach (Spinacea oleracea) leaves was with 100% acetone (23) and extracts were treated as above except the procedure was carried out at room temperature.
For reconstitution experiments, the total pigment extract from the diatom was resuspended in 75% diethyl ether and 25% absolute ethanol. Spinach pigment extracts were resuspended in 50% diethyl ether and 50% ethanol: Chl b (from Sigma) was dissolved in 100% diethyl ether. Reconstitution of LHCaR1 was according to Schmid et al. (21) as modified by Grabowski et al. (22). In the reconstitution with diatom pigments [Chl a/c ratio (mol/mol) 7.5], 50 μg of Chl a was incubated with 200 μg of inclusion body protein, whereas the spinach pigment mixture [Chl a/b ratio (mol/mol) 2.8] contained 60 μg of Chl and 200 μg of protein. Clearly resolved pigment complexes were collected from a discrete band in the upper third of sucrose gradients. Diatom and dinophyte pigments yielded a brownish-green band, and spinach pigments yielded a green band. Superimposition of pigment bands with Coomassie blue-stained bands, and absence of free pigments on nondenaturing PAGE gels (22) demonstrated adherence of pigments to proteins.
Reconstituted complexes with spinach and diatom pigment extracts corresponded to apparent dimers with a molecular mass of ca. 68 kDa, relative to molecular weight markers on sucrose gradients. Those with dinophyte pigments were larger and close to apparent trimers, as was also found with pigments from P. cruentum under the same conditions (22).
HPLC Analysis.
Pigments from LHCaR1 reconstituted with diatom and spinach pigments were extracted with ethyl acetate and analyzed by reverse-phase HPLC (Spherisorb ODS2 column) with a 30-min gradient of ethyl acetate (0–67%) in acetonitrile-water-triethylamine (9:1:0.01 vol/vol) at a flowrate of 1 ml/min. Pigments were detected at 450 nm with spectra recorded for peak fractions from 400 to 750 nm. Determinations of Chls a, b, c, and carotenoids were made according to standard compounds and published absorption coefficients for lutein, zeaxanthin, neoxanthin, violaxanthin, β-carotene (24), fucoxanthin, diadinoxanthin (25), and Chls a and b (26), and Chl c (27). Upon reconstitution, dinoflagellate pigments could not be reliably extracted from the protein and were not further quantified.
Spectroscopy.
Spectral measurements were made in 5 mM Tricine-NaOH (pH 7.8) at room temperature. Fluorescence emission spectra (ca. 6 μg/ml Chl a) were recorded for suitable excitation wavelengths: 438 nm (Chl a), 465 nm (Chl c), 467 nm (Chl b), and 495–508 nm (carotenoids). Excitation spectra were recorded with the emission wavelength set at 678 nm (Chl a). CD was determined (1 cm pathlength with 30–40 μg of Chl per ml) as described (22).
Protein and Chl Assay of LHC Preparations.
The protein content of inclusion body preparations and of the reconstitutions of LHCaR1 with diatom and spinach pigments were quantified by the bicinchoninic acid assay as described (22). Chl concentrations of diatom pigment extracts and reconstituted LHCs were determined in 90% acetone by using the extinction coefficients of Jeffrey and Humphrey (27). Chl and carotenoid concentrations of spinach pigments were determined in N,N-dimethylformamide as previously described (22).
Results
Reconstitution and Model of a Red Algal LHC.
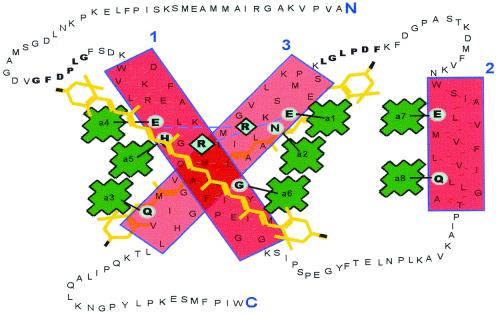
It was previously established that eight Chl a and up to four zeaxanthin molecules are incorporated into a rhodophyte LHCaR1 recombinant protein in vitro (22), with the resulting pigment–protein complex spectrally and functionally similar to the native multiprotein complex. The binding of eight Chl a molecules is consistent with the number of putative Chl-binding sites. However, the binding of four zeaxanthins per polypeptide indicates that carotenoid binding is proportionately greater than that in Chl a/b-binding proteins that have been examined. A model of LHCaR1 in Fig. 1 shows eight putative binding sites for Chl a. With a coding region of 194 aa residues deduced from the DNA sequence, the mature polypeptide (21.3 kDa) is smaller than most LHCs of higher plants (6). Like LHCaR2, another LHC from P. cruentum (10), it shares essential features with other LHCs: i.e., eight conserved Chl-binding residues, as well as conserved regions flanking helices 1 and 3 (GFDPLG and FDPLGL, respectively) that occur in many but not all LHCs. In helix 2, the putative Chl-binding sites are separated by 8 aa residues. Although a functional significance is obscure, such spacing is characteristic of LHCs of chromophytes and dinophytes (10, 12) rather than of chlorophytes, where a spacing of 7 aa residues occurs (6, 10). Probable Chl-binding residues in helix 1 are E56, H59, and G69; in helix 2, they are Q107 and E116; and in helix 3, they are E150, N153, and Q167. Binding of Chl via a glycine residue (G69) has been suggested (15, 28) to involve a water molecule as a second ligand to the Mg2+ of Chl. Like lutein in higher plants, at least two of the zeaxanthins likely provide structural stability for helices 1 and 3. Zeaxanthin, however, was not found to participate in excitation energy transfer (22).
Figure 1.
A model of LHCaR1 of the rhodophyte P. cruentum (based on the LHCII model of Kühlbrandt et al., ref. 15). The three boxed areas are the transmembrane helices 1–3 with putative Chl-binding sites encircled and connected by lines to porphyrin rings of Chl a (22). Two zeaxanthin molecules (of four) are shown in regions flanking helices 1 and 3 with possible carotenoid stabilization residues in bold text. Pairs of residues (E/R) thought to stabilize helices 1 and 3 are connected by a dashed line. The N and C termini are in large type.
Higher Plant Pigments Are Bound and Functionally Inserted.
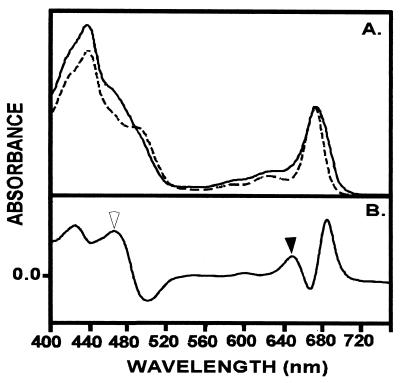
Spinach was selected as a representative of higher plants and its pigments were extracted for reconstitution experiments with the red algal LHC polypeptide. The incorporation of spinach pigment extracts (Chl a/b 2.8) into LHCaR1 showed a more complex absorption spectrum (Fig. 2A) than obtained with the P. cruentum pigment extracts. An increased absorption in the 640–650 nm region denotes binding of Chl b as is evident in the difference spectrum (Fig. 2B), and further supported by an enhanced absorbance in this region when additional Chl b (0.7 Chl a/b) is included in reconstitution experiments (not shown). The absorption characteristics of LHCaR1 reconstituted with spinach pigments are highly similar to those of higher plant LHCs (19, 28), where the spectrum includes a broadened absorption beyond the 672 nm maximum in the far-red region that is not observed for reconstitutions with P. cruentum pigments (Fig. 2).
Figure 2.
(A) An absorption spectrum of LHCaR1 reconstituted with spinach pigments (Chl a/b 2.8 mol/mol) (——) is compared with one for LHCaR1 reconstituted with P. cruentum pigments (Chl a) (– – – –). (B) A difference spectrum for the spectra in A illustrates the contribution of Chl b (solid arrowhead), and Chl b plus carotenoids (open arrowhead) and shows the increased absorption beyond 680 nm for reconstitution with spinach pigments. The spectra were normalized at 672 nm. (Measurements were at room temperature in 5 mM Tricine-NaOH, pH 7.8.)
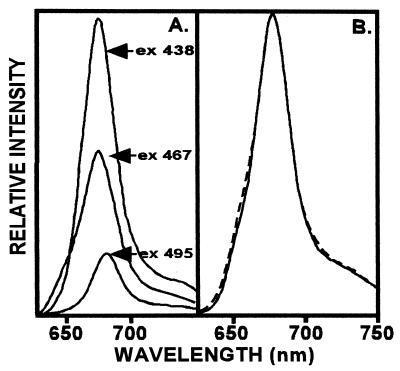
Fluorescence spectra (Fig. 3) indicate that the pigments are not only bound to the protein but they also participate in energy transfer. The participation of Chl b in energy transfer to Chl a is demonstrated upon excitation (467 nm) in the Chl b absorbing region with a resulting fluorescence from Chl a at 678 nm (Fig. 3A). Also, the 678 nm emission increased with 467 nm excitation for a reconstituted complex containing additional Chl b (0.7 Chl a/b) (not shown), which underscores participation of Chl b in transferring energy to Chl a. Interestingly, the fluorescence emission profile (Fig. 3B) did not reflect a broadened red-shift comparable to that seen in the absorption spectrum (Fig. 2A) and suggests that Chl b alone is not responsible for the long-wavelength absorption.
Figure 3.
(A) Fluorescence emission is at 678 nm for LHCaR1 reconstituted with spinach pigments (Chl a/b 2.8) whether excited at 438 nm (Chl a), 467 nm (Chl b region), or 495 nm (carotenoid region). (B) Normalized emission spectra at 678 nm (excitation at 476 nm) of samples with Chl a/b 2.8 (——) and Chl a/b 0.7 (– – – –). (Measurements were at room temperature in 5 mM Tricine-NaOH, pH 7.8/6 μg/ml Chl.)
Carotenoid participation in energy transfer to Chl a in LHCaR1 reconstituted with spinach pigments is evident in the fluorescence emission from Chl a (678 nm) with 495 nm excitation (Fig. 3A). It is well known that lutein is the major carotenoid in LHCs of higher plants and participates in energy transfer to Chl a. We conjecture that lutein is also the primary light-absorbing carotenoid in LHCaR1 reconstituted with spinach pigments that is responsible for exciting Chl a, although low concentrations of neoxanthin, violaxanthin and β-carotene were also present (Table 1).
Table 1.
Pigments per polypeptide of a rhodophyte LHCaR1 reconstituted with pigments of spinach, a diatom, and a red alga
| Pigment source | mol of pigment/mol of
polypeptide
|
Ratio
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chla | Chlb | Chlc | Zea | Fuco | Diadin | β-car | Lut | Neo | Vio | Chla/b | Car/Chl | |
| P. cruentum* | 8.2 ± 0.2 | — | — | 4.04 | — | — | 0.33 | — | — | — | — | 0.49 |
| Spinach† | 6.2 ± 0.8 | 1.8 ± 0.4 | — | — | — | — | 0.10 | 2.4 | 0.73 | 1.20 | 3.1 ± 0.2 | 0.54 |
| Spinach + Chlb‡ | 3.8 ± 1.1 | 3.7 ± 1.3 | — | — | — | — | 0.20 | 0.8 | 0.20 | 0.30 | 1.2 ± 0.3 | 0.17 |
| T. fluviatilis§ | 7.0 ± 0.4 | — | 1.0 ± 0.1 | — | 8.0 | 1.9 | — | — | — | — | — | 1.20 |
Values of mol of pigment/mol of polypeptide are means ± SD for three to six independent sample determinations.
As in Grabowski et al. (22).
Chl a/b in reconstitution mixture was 2.8 (mol/mol).
Chl a/b in reconstitution mixture was 0.7 (mol/mol).
Chl a/c in reconstitution mixture was 7.5 (mol/mol).
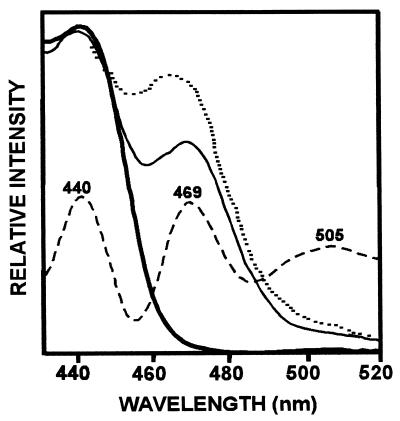
The contribution of Chl b and carotenoids (predominantly lutein) as accessory pigments to Chl a in the rhodophyte protein is further demonstrated by fluorescence excitation spectra (Fig. 4) for Chl a emission at 678 nm. A prominent shoulder in the region 465–475 nm resolves (by fourth derivative) into the Chl b maximum at ca. 469 nm and a carotenoid maximum at ca. 505 nm. In contrast, the excitation spectrum for the LHCaR1 reconstituted with P. cruentum pigments does not exhibit a contribution from zeaxanthin, as previously noted (22). Pigment organization in the protein as demonstrated by CD (Fig. 5) had signals at ca. 440 nm and ca. 683 nm reflective of Chl a, and carotenoids at ca. 500 nm similar, but not identical, to P. cruentum (figure 4B in ref. 22). With increased Chl b (Fig. 5, B) the Qy region exhibits a prominent shoulder at ca. 650 nm, which may be attributable to exciton interaction between Chls a and b (29). The Chl b enriched preparations also had discrete peaks at ca. 460 nm (−) and ca. 475 nm (+) and a more prominent shoulder at ca. 650 nm (Fig. 5, B).
Figure 4.
Fluorescence excitation spectra (for emission at 678 nm) of LHCaR1 reconstituted with P. cruentum pigments (thick solid line), or with spinach pigments of Chl a/b 2.8 (thin solid line) and Chl a/b 0.7 (- - - - -). Conditions were as in Fig. 3. Chl b (469 nm), carotenoids (505 nm), and Chl a (440 nm) regions are resolved in the fourth derivative spectrum (– – – –) of the Chl a/b 2.8 sample.
Figure 5.
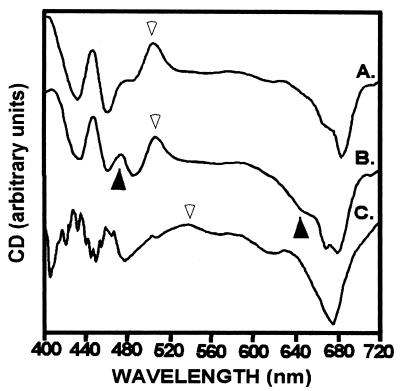
CD spectra of LHCaR1 reconstituted with spinach pigments Chl a/b 2.8 (A), Chl a/b 0.7 (B), and diatom pigments (C). The increased signals at ca. 470 and 650 nm (▴) correlate with higher Chl b content (B). Signals at ca. 505 nm (A and B) and ca. 540 nm (C) are reflective of the carotenoid region (▵). (Measurements at room temperature were made at Chl concentrations of 30–40 μg/ml (1 cm pathlength). An average of four scans (100 nm/min) were corrected for buffer.
Binding and Functional Insertion of Diatom Pigments.
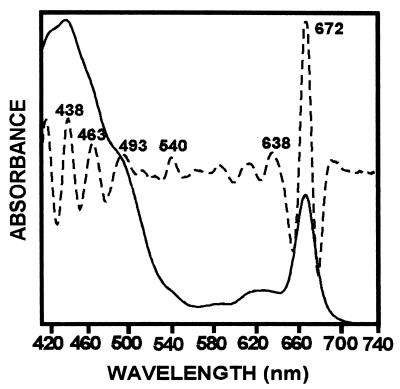
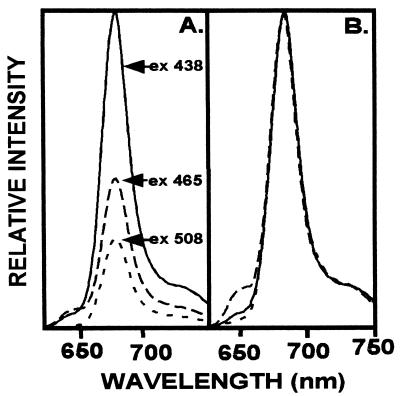
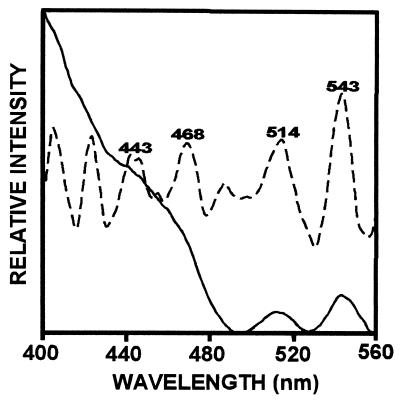
Pigment extracts of T. fluviatilis containing Chls a and c, fucoxanthin, diadinoxanthin and β-carotene, when reconstituted with the LHCaR1 rhodophyte protein, were recovered in a brown band on sucrose gradients with an absorption spectrum (Fig. 6) similar to those of Chl a/c LHCs of other chromophytes (30–33). Absorption maxima of individual pigments are resolved for Chl a at 438 and 672 nm, Chl c at 463 nm and 638 nm, and fucoxanthin at 493 and 540 nm by fourth derivative analysis. Participation in energy transfer of Chl c, fucoxanthin, and possibly diadinoxanthin is shown by the fluorescence emission at 678 nm with respective excitation at 465 nm and 508 nm (Fig. 7A). Upon normalization of the emission spectra (Fig. 7B) a small emission shoulder at 650 nm (excitation at 465 nm) is seen and may indicate that some Chl c is not optimally oriented for energy transfer to Chl a. However, the functional participation of Chl c is certainly evident from the excitation spectrum (Fig. 8) with a prominent shoulder at 450–480 nm, which is resolved into a peak at 468 nm by fourth derivative analysis. The contribution of fucoxanthin is indicated by the prominent peaks at 514 and 543 nm in the fourth derivative spectrum (Fig. 8) in agreement with measurements on diatom complexes (31).
Figure 6.
Absorption of LHCaR1 reconstituted with pigments of the diatom T. fluviatilis (——). Absorption maxima of Chl a (438/672 nm), Chl c (463/638 nm), and fucoxanthin (493/540 nm) are resolved by the fourth derivative spectrum (– – – –).
Figure 7.
Fluorescence spectra for LHCaR1 reconstituted with diatom pigments. (A) Emission at 678 nm results upon excitation at 438 nm Chl a (——), 465 nm Chl c (– – – –), and 508 nm fucoxanthin (- - - - -) indicating energy transfer to Chl a. (B) Normalization of the emission spectra was at 678 nm.
Figure 8.
Fluorescence excitation spectrum for LHCaR1 reconstituted with diatom pigments (——). Fourth derivative analysis (– – – –) shows the individual peaks attributable to Chl a at 443 nm, Chl c at 468 nm, and fucoxanthin at 514 nm and 543 nm.
The CD spectrum for LHCaR1 reconstituted with diatom pigment extract (Fig. 5, C), unlike those with spinach (Fig. 5, A and B) and P. cruentum pigments (22), has a broad positive region at ca. 540 nm. The peak at ca. 674 nm, attributed to Chl a, and the 626 nm shoulder for Chl c are consistent with spectra of other chromophytes (32, 33). Unknown is the effect on CD spectra of apparent polypeptide dimers with spinach pigments, or apparent trimers with diatom pigments.
Functional Binding of Dinophyte Pigments.
Pigment extracts of the dinophyte P. micans, containing peridinin rather fucoxanthin as the major carotenoid pigment, were similarly reconstituted with LHCaR1 into a functional complex. Absorption, fluorescence (emission and excitation) and CD spectra showed binding and orientation for energy transfer (data not shown) largely similar to those of the LHCaR1 reconstituted with diatom pigments. An exception was a 495/509 nm absorption peak characteristic of peridinin, and a strong CD signal at ca. 480 nm (−). Reconstituted polypeptides with dinophyte pigments, although stable on “green gels” and suitable for spectral measurements, proved difficult for routine quantitative pigment/protein determination because extraction of Chl from the protein was sometimes incomplete.
Discussion
A Simple Red Algal LHC Protein Binds All Major Chlorophylls and Carotenoids of Plants and Algae.
The reconstitution experiments described here provide the first demonstration that an apparently “simple” LHC apoprotein from a rhodophyte can functionally bind the major types of Chls (a, b, and c) and carotenoids (lutein, fucoxanthin, and peridinin) known to function as photosynthetic accessory pigments. These results show that the rhodophyte LHC polypeptide retains a flexibility to functionally bind pigments foreign to its evolutionary history and that almost certainly evolved along separate phylogenetic lines, i.e., rhodophytes, chlorophytes, chromophytes, and dinophytes. The diversity in the pigment composition of LHCs of different taxa may not be reflective of protein diversity but rather of the ability of different photosynthetic organisms to synthesize pigments with different light-harvesting capabilities.
Insertion of Chl b, and especially Chl c, was unexpected. Binding of eight Chl/polypeptide (Table 1) is consistent with the eight conserved binding sites (Fig. 1) and previous results of the binding of eight Chl a/polypeptide (22). Our results suggest that many of these Chl-binding sites are not restricted to a specific type of Chl: it can be inferred that at least four of eight sites can be occupied by other Chl types. Competition for certain of the Chl-binding sites was observed when Chl b was increased, relative to Chl a, at a constant Chl/protein ratio. Reconstitution with extracts of market spinach leaves of lower Chl b content (2.8 Chl a/b) yielded six Chl a and two Chl b (Table 1) per polypeptide, but ca. four Chl a and four Chl b per polypeptide are bound with increased Chl b in the reconstitution mixture (Chl a/b 0.7). Competition for Chl-binding sites also occurred with Chl c. Whereas ca. seven Chl a and one Chl c per polypeptide was most commonly observed under our conditions, as many as two Chl c (with six Chl a) per polypeptide was sometimes attained.
Significantly, there was a decrease from 0.54 to 0.17 of the Car/Chl ratio with increased Chl b incorporation (Table 1). It appears that there is a spatial effect created by additional Chl b molecules in the polypeptide which interferes with carotenoid insertion. Such spatial hindrance would not be expected by viewing the LHCII crystal structure (15). However, changes in the Car/Chl ratios are also obtained in native complexes and with reconstituted chlorophyte LHCs (13, 34), thus decreases in Car/polypeptide with higher Chl b content are not likely attributable to in vitro reconstitution alone.
Are There Specific Carotenoid Binding Sites in LHCs?
Carotenoids clearly are required for the stability of the helices and for insertion of Chls into LHC apoproteins (19). Zeaxanthin in the P. cruentum polypeptide probably substitutes (Fig. 1) for the two molecules of lutein which, in the Kühlbrandt model (15) parallel helices 1 and 3. More unexpected is the insertion and proper orientation of foreign carotenoids into the rhodophyte polypeptide in proportions characteristic to those for native complexes of the phylogenetic taxa (9, 20, 31, 32) from which the pigments were derived (Figs. 2, 4, and 6).
Compared with those of chlorophytes and rhodophytes, the LHCs of chromophytes and dinophytes characteristically have a high Car/Chl ratio, as for example the 1.1 ratio of fucoxanthin/Chl (32, 35). In accord with pigment ratios reported for chromophyte LHCs, 10 carotenoid molecules were bound to the reconstituted red algal polypeptide (Table 1). This represents more than a 2-fold increase in Car/Chl incorporation and also the insertion of carotenoids that are not synthesized in the native organism. Substitution of related carotenoid types in chlorophyte plants (Chlamydomonas and Arabidopsis) has been shown for mutants (36). It has also been demonstrated in reconstitutions of LHCII (17), but our findings are the first demonstration that pigments of phylogenetically diverse taxa can form functional complexes.
The sequence motifs GFDPLG/F in the stromal loop regions of helices 1 and 3 of LHCII, earlier were suggested as possible carotenoid-binding sites (37). These motifs are also present in the red algal LHCI polypeptides (Fig. 1) (6, 10). Heinemann and Paulsen (38) showed that a point mutation in such a domain of LHCII abolished in vitro reconstitution, suggesting that it is critical for assembly of a functional complex possibly because carotenoid binding cannot occur. However, conservation of the putative carotenoid binding sites is not consistent across taxa (11, 39). In any case, it is difficult to imagine how these putative carotenoid binding sites could accommodate eight or more carotenoid molecules in one LHC polypeptide. Thus, such motifs are either not essential for all carotenoids, or other additional sites exist in LHCs. For LHCIIb, multiple carotenoid binding sites have been proposed (17, 40). Neoxanthin binding has been suggested to require certain domains in helix 2 which, when modified, interfere with its insertion between helix 2 and 1/3 helix domains (17).
Whether or not the rhodophyte LHC alone retained the ability for functional insertion of various photosynthetic pigments remains to be ascertained. We would predict that considerable flexibility exists in other LHCs as well. Also interesting is the recent finding that Chl b was incorporated into the PSI reaction center of Synechocystis PCC6803 when transformed with chlorophyll a oxygenase (CAO) (41). The binding of different pigments in a flexible protein could have facilitated the radiation of photosynthetic eukaryotes into different aquatic and terrestrial light environments with subsequent protein modifications in specific groups of organisms. The results with the P. cruentum LHC polypeptides provide a simple in vitro model system by which the functional interaction of various pigments can be explored in the same protein environment thus advancing exploration of basic questions on pigment-protein interactions that were previously not possible to address.
Acknowledgments
We gratefully acknowledge receiving cells of Thallasiosira fluviatilis and Prorocentrum micans from Dr. Todd Kana (University of MD Center for Environmental Studies) and use of the CD spectropolarimeter by Dr. David Jollie. This work was supported in part by a grant from the National Science Foundation (MCB 9632157) and in part by funds from Monsanto, Quest International, and Zeneca.
Abbreviations
- Chl
chlorophyll
- LHC
light-harvesting complex
- LHCI and LHCII
LHC of photosystems I and II
- LHCaR1 and LHCaR2
the first and second cloned genes of rhodophyte LHCs associated with photosystem I
- PSI and PSII
photosystems I and II
Footnotes
See commentary on page 2119.
References
- 1.Delwiche C, Kuhsel M, Palmer J D. Mol Phylogeny Evol. 1995;4:110–128. doi: 10.1006/mpev.1995.1012. [DOI] [PubMed] [Google Scholar]
- 2.Moreira D, Le Guyader H, Phillipe H. Nature (London) 2000;405:69–72. doi: 10.1038/35011054. [DOI] [PubMed] [Google Scholar]
- 3.Demmig-Adams B, Gilmore A M, Adams W W., III FASEB J. 1996;10:403–412. doi: 10.1096/fasebj.10.4.8647339. [DOI] [PubMed] [Google Scholar]
- 4.Anderson J M, Barrett J. In: Photosynthesis III, Photosynthetic Membranes and Light Harvesting Systems. Staehelin L H, Arntzen C J, editors. Berlin: Springer; 1986. pp. 269–285. [Google Scholar]
- 5.Wolfe G R, Cunningham F X, Jr, Durnford D, Green B R, Gantt E. Nature (London) 1994;367:566–568. [Google Scholar]
- 6.Tan S, Cunningham F X, Jr, Gantt E. Plant Mol Biol. 1997;33:157–167. doi: 10.1023/a:1005715528297. [DOI] [PubMed] [Google Scholar]
- 7.Marquardt J, Rhiel E. Biochim Biophys Acta. 1997;1320:153–164. [Google Scholar]
- 8.Green B R, Durnford D G. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:685–714. doi: 10.1146/annurev.arplant.47.1.685. [DOI] [PubMed] [Google Scholar]
- 9.Larkum T, Howe C J. Adv Bot Res. 1997;25:257–330. [Google Scholar]
- 10.Tan S, Ducret A, Aebersold R, Gantt E. Photosynth Res. 1997;53:129–140. [Google Scholar]
- 11.Durnford D G, Deane J A, Tan S, McFadden G I, Gantt E, Green B R. J Mol Evol. 1999;48:59–68. doi: 10.1007/pl00006445. [DOI] [PubMed] [Google Scholar]
- 12.Passaquet C, Lichtlé C. Plant Mol Biol. 1995;29:135–148. doi: 10.1007/BF00019125. [DOI] [PubMed] [Google Scholar]
- 13.Bassi R, Croce R, Cugini D, Sandoná D. Proc Natl Acad Sci USA. 1999;96:10056–10061. doi: 10.1073/pnas.96.18.10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandonà D, Croce R, Pagano A, Crimi M, Bassi R. Biochim Biophys Acta. 1998;1365:207–214. doi: 10.1016/s0005-2728(98)00068-1. [DOI] [PubMed] [Google Scholar]
- 15.Kühlbrandt W, Wang D N, Fujiyoshi Y. Nature (London) 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- 16.Rogl H, Kühlbrandt W. Biochemistry. 1999;38:16214–16222. doi: 10.1021/bi990739p. [DOI] [PubMed] [Google Scholar]
- 17.Croce R, Weiss S, Bassi R. J Biol Chem. 1999;274:29613–29623. doi: 10.1074/jbc.274.42.29613. [DOI] [PubMed] [Google Scholar]
- 18.Remelli R, Varotto C, Sandoná D, Croce R, Bassi R. J Biol Chem. 1999;47:33510–33521. doi: 10.1074/jbc.274.47.33510. [DOI] [PubMed] [Google Scholar]
- 19.Plumley F G, Schmidt G W. Proc Natl Acad Sci USA. 1987;84:146–150. doi: 10.1073/pnas.84.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulsen H, Rumler U, Rudiger W. Planta. 1990;181:204–211. doi: 10.1007/BF02411539. [DOI] [PubMed] [Google Scholar]
- 21.Schmid V H R, Cammarata K V, Bruns B U, Schmidt G W. Proc Natl Acad Sci USA. 1997;94:7667–7672. doi: 10.1073/pnas.94.14.7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grabowski B, Tan S, Cunningham F X, Jr, Gantt E. Photosynth Res. 2000;63:85–96. doi: 10.1023/A:1006357107247. [DOI] [PubMed] [Google Scholar]
- 23.Svec W A. In: Chlorophylls. Scheer H, editor. Boca Raton, FL: CRC; 1991. pp. 89–102. [Google Scholar]
- 24.Davies B H. In: Chemistry & Biochemistry of Plant Pigments. 2nd Ed. Goodwin T W, editor. Vol. 2. New York: Academic; 1976. pp. 38–165. [Google Scholar]
- 25.Jeffrey S W. In: Phytoplankton Pigments in Oceanography: Guidelines to Modern Methods. Jeffrey S W, Mantoura R F C, Wright S W, editors. Paris: United Nations Educational and Scientific Organization; 1997. pp. 127–166. [Google Scholar]
- 26.Porra R J. In: Chlorophylls. Scheer H, editor. Boca Raton, FL: CRC; 1991. pp. 31–57. [Google Scholar]
- 27.Jeffrey S W, Humphrey G F. Biochem Physiol Pflanz. 1975;167S:191–194. [Google Scholar]
- 28.Walters R G, Horton P. Photosynth Res. 1999;61:77–89. [Google Scholar]
- 29.Gülen D, Knox R, Breton J. Photosynth Res. 1986;9:13–20. doi: 10.1007/BF00029727. [DOI] [PubMed] [Google Scholar]
- 30.Büchel C, Garab G. J Photochem Photobiol B Biol. 1997;37:118–124. [Google Scholar]
- 31.Alberte R S, Friedman A L, Gustafson D L, Rudnick M S, Lyman H. Biochim Biophys Acta. 1981;635:304–316. doi: 10.1016/0005-2728(81)90029-3. [DOI] [PubMed] [Google Scholar]
- 32.Pascal A A, Caron L, Rousseau B, Lapouge K, Duval J-C, Robert B. Biochemistry. 1998;37:2450–2457. doi: 10.1021/bi9719657. [DOI] [PubMed] [Google Scholar]
- 33.Mimuro M, Katoh T, Kawai H. Biochim Biophys Acta. 1990;1015:450–456. [Google Scholar]
- 34.Kleima F J, Hobe S, Calkoen F, Urbanus M L, Peterman E J G, van Grondelle R, Paulsen H, van Amerongen H. Biochemistry. 1999;38:6587–6596. doi: 10.1021/bi982823v. [DOI] [PubMed] [Google Scholar]
- 35.De Martino A, Douady D, Rousseau B, Duval J C, Caron L. Photochem Photobiol. 1997;66:190–197. [Google Scholar]
- 36.Niyogi K K. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- 37.Green B R, Kühlbrandt W. Photosynth Res. 1995;44:139–148. doi: 10.1007/BF00018304. [DOI] [PubMed] [Google Scholar]
- 38.Heinemann B, Paulsen H. Biochemistry. 1999;38:14088–14093. doi: 10.1021/bi991439a. [DOI] [PubMed] [Google Scholar]
- 39.Eppard M, Rhiel E. Mol Gen Genet. 1998;260:335–345. doi: 10.1007/s004380050902. [DOI] [PubMed] [Google Scholar]
- 40.Hobe S, Niemeier H, Bender N, Paulsen H. Eur J Biochem. 2000;267:616–624. doi: 10.1046/j.1432-1327.2000.01060.x. [DOI] [PubMed] [Google Scholar]
- 41.Satoh, S., Ikeuchi, M., Mimuro, M. & Tanaka, A. (November 9, 2000) J. Biol. Chem., 10.1074/jbc.M008238200.