Abstract
We describe a deep-sequencing procedure for tracking large numbers of transposon mutants of Pseudomonas aeruginosa. The procedure employs a new Tn-seq methodology based on the generation and amplification of single-strand circles carrying transposon junction sequences (the Tn-seq circle method), a method which can be used with virtually any transposon. The procedure reliably identified more than 100,000 transposon insertions in a single experiment, providing near-saturation coverage of the genome. To test the effectiveness of the procedure for mutant identification, we screened for mutations reducing intrinsic resistance to the aminoglycoside antibiotic tobramycin. Intrinsic tobramycin resistance had been previously analyzed at genome scale using mutant-by-mutant screening and thus provided a benchmark for evaluating the new method. The new Tn-seq procedure identified 117 tobramycin resistance genes, the majority of which were then verified with individual mutants. The group of genes with the strongest mutant phenotypes included nearly all (13 of 14) of those with strong mutant phenotypes identified in the previous screening, as well as a nearly equal number of new genes. The results thus show the effectiveness of the Tn-seq method in defining the genetic basis of a complex resistance trait of P. aeruginosa and indicate that it can be used to analyze a variety of growth-related processes.
IMPORTANCE
Research progress in microbiology is technology limited in the sense that the analytical methods available dictate how questions are experimentally addressed and, to some extent, what questions are asked. This report describes a new transposon tracking procedure for defining the genetic basis of growth-related processes in bacteria. The method employs next-generation sequencing to monitor the makeup of mutant populations (Tn-seq) and has several potential advantages over other Tn-seq methodologies. The new method was validated through the analysis of a clinically relevant antibiotic resistance trait in Pseudomonas aeruginosa, an important bacterial pathogen.
The process of defining the genetic basis of complex bacterial traits using mutant hunts has been greatly accelerated by the development of technologies for tracking individual strains in saturation level mixtures of transposon mutants. Several such procedures based on next-generation DNA sequencing have recently been described (1–4). The methods, referred to collectively here as Tn-seq (4), provide dramatic improvements in the specificity and dynamic range of mutant detection over techniques for tracking transposon mutants based on PCR or microarray hybridization. The Tn-seq procedures have employed several methods to capture and sequence transposon-genome junctions, including affinity purification of amplified junctions (1), ligation of adaptors into genome sequences distal to the transposon end with the aid of a specialized restriction site (2, 4), and selective amplification (3). Tn-seq has been used to characterize a variety of traits in several species, namely, pulmonary colonization in Haemophilus influenzae (1), in vivo fitness in the gut symbiont Bacteroides thetaiotaomicron (2), mutant interactions in Streptococcus pneumoniae (4), and bile tolerance genes in Salmonella enterica (3). In the work described here, we developed a new Tn-seq technology and used it to examine an antibiotic resistance trait in Pseudomonas aeruginosa.
The Gram-negative opportunistic pathogen P. aeruginosa exhibits unusually high intrinsic resistance to antibiotics and other toxic substances, properties which greatly limit the therapeutic options available to treat infections (5). The functions mediating resistance represent potential drug targets, since their inhibition could enhance the efficacy of antibiotics in combination therapies (6, 7). Intrinsic antibiotic resistance traits of P. aeruginosa and other Gram-negative species are complex, typically controlled by dozens of gene products, including efflux pumps, modifying enzymes, and elements of general stress responses. For example, in a genome-wide study of tobramycin resistance in P. aeruginosa, we identified mutations in over 100 genes that reduced intrinsic resistance to the antibiotic (7). The genes were identified by screening the members of a comprehensive, defined transposon mutant library one by one. The genes identified included several involved in a novel envelope stress response pathway which may help protect cells from tobramycin-induced mistranslation products which disrupt the membrane (7). In the study reported here, we employed intrinsic tobramycin resistance as a reference trait to validate the newly developed Tn-seq method of mutant identification.
RESULTS
A new Tn-seq methodology.
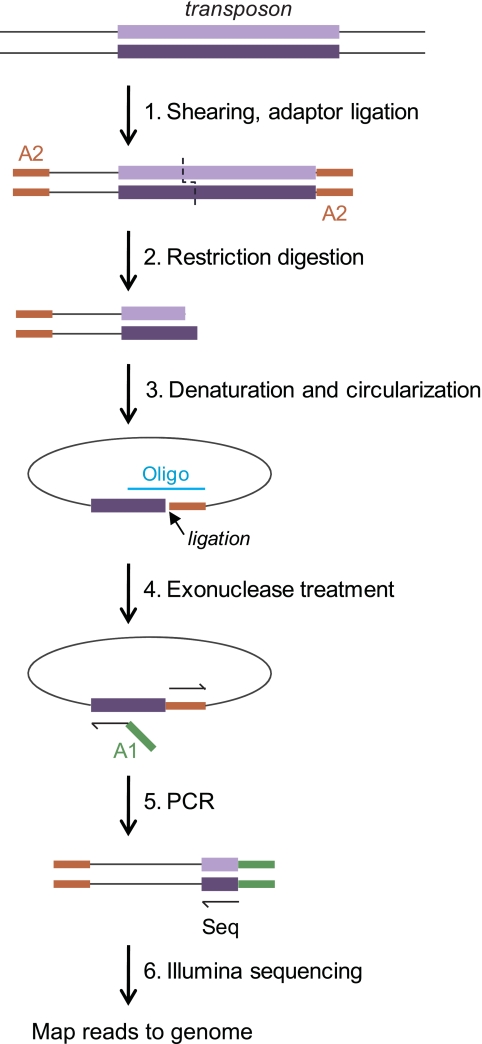
Using deep sequencing to track insertion mutants (Tn-seq) requires the selective sequencing of fragments which bear transposon-genome junctions. Three different methods for doing this have recently been described, all based on Illumina sequencing (1–4). We independently developed a fourth method, based on the generation of single-stranded DNA circles that carry transposon junctions, called the “circle method” (Fig. 1). The circles are protected from an exonuclease treatment which eliminates most genomic DNA, and they then serve as templates for quantitative amplification and sequencing of the junctions. The junction sequences are used to map the corresponding insertions in the genome of the mutagenized strain.
FIG 1 .
Tn-seq circle method. The steps used to amplify and sequence transposon insertion junctions are illustrated, beginning with a DNA fragment carrying a transposon insertion (top). First, total DNA from a mutant pool is sheared and end repaired, and one Illumina adaptor (A2) is ligated to all free ends (step 1). The sample is then digested with a restriction enzyme that cuts near one transposon end (in this work, BamHI, which cuts 114 bp from the transposon’s left end) (step 2). Following a size selection step, single-strand fragments which include the transposon end are circularized by templated ligation (step 3). Oligo, oligonucleotide. Fragments which have not circularized (representing most of the DNA in the sample) are degraded in a subsequent exonuclease step (step 4). The transposon-genome junctions from the circularized fragments are then amplified by quantitative PCR in a step in which the second required Illumina adaptor (A1) is introduced (step 5). The products are sequenced on an Illumina flow cell using a sequencing primer corresponding to the transposon end (Seq), and each sequence read is then mapped to the genome (step 6).
Use of Tn-seq to profile pools of P. aeruginosa insertion mutants.
In a first test, we analyzed a pool of P. aeruginosa PAO1 mutants derived from a larger library (8) which included 3,625 strains with transposon insertions which had been mapped and confirmed by sequencing. The Tn-seq circle method identified 99.3% of these insertion locations (not shown), illustrating the effectiveness of the procedure. The result encouraged us to examine the performance of the procedure in profiling a much larger mutant pool.
For the second test, we created a pool of approximately 100,000 undefined ISlacZ/hah insertion mutants (nearly 20 insertions per gene), isolated DNA from the pool, and analyzed it in two independent Tn-seq runs. From the two technical replicates, 95,905 unique insertion locations were identified, in good agreement with the number of mutants pooled (Table 1). Approximately 91% of the unique insertion sites identified in each replicate were also identified in the other replicate, suggesting that the procedure primarily identified authentic insertion locations. Additional Tn-seq analysis of the same transposon mutant pool (Table 1) found that about 99% of the locations identified were detected in multiple Tn-seq runs and were thus presumably authentic. We assume that the irreproducible sites (about 1% of the total in each Tn-seq run) correspond to spurious amplification products. For subsequent analysis, we therefore included only locations that were detected in multiple independent Tn-seq runs (Table 1).
TABLE 1 .
Tn-seq analysis of P. aeruginosaa
| Growth condition | Total no. of reads | No. (%) of mapped reads | No. of locations identified | No. (%) of authentic locationsb |
|---|---|---|---|---|
| Initial poolc | 19,778,278 | 15,739,802 (80) | 95,905 | 94,531 (98.6) |
| Trial 1 | ||||
| Without tobramycin | 14,152,566 | 12,601,670 (89) | 96,745 | 96,185 (99.4) |
| With tobramycin | 10,699,404 | 9,691,469 (91) | 76,474 | 76,109 (99.5) |
| Trial 2 | ||||
| Without tobramycin | 12,660,378 | 11,081,531 (88) | 99,580 | 98,258 (98.7) |
| With tobramycin | 12,041,290 | 10,799,523 (90) | 98,243 | 96,925 (98.7) |
A pool of approximately 100,000 independent transposon mutants was analyzed by the Tn-seq circle method after growth under different conditions.
Authentic locations were defined as those identified in multiple Tn-seq assays of samples derived from the transposon mutant pool.
The values for the initial pool represent two replicate assays of a single DNA preparation.
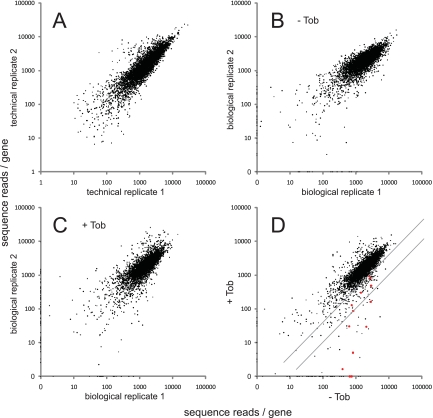
The number of sequence reads per individual location varied somewhat between technical replicates, but the total numbers of reads at all locations within each gene were highly correlated (Fig. 2A). The Tn-seq circle method thus appears to provide a reproducible and quantifiable readout of mutant representation at the gene level.
FIG 2 .
Identification of insertion mutants using Tn-seq. Pairs of Tn-seq assay results are compared, with the total number of reads per gene plotted. (A) Replicate Tn-seq analysis of a single DNA sample isolated from the 100,000-member transposon mutant pool. The total numbers of reads for genes were highly correlated (Pearson correlation coefficient = 87%). (B) Replicate analysis of DNA samples corresponding to two cultures of the 100,000-member mutant pool grown in the absence of tobramycin (Tob) (correlation coefficient = 80%). (C) Replicate analysis of DNA samples corresponding to two cultures of the mutant pool grown in the presence of a subinhibitory tobramycin concentration (0.36 µg/ml) (correlation coefficient = 71%). (D) Comparison of the growth of the mutant pool without tobramycin to its growth with tobramycin. Values represent average numbers of reads per gene from the pairs of biological replicates (panels B and C). Red points represent genes for which mutants were previously identified as being strongly tobramycin hypersensitive (7). The diagonal lines represent the thresholds for 2.5-fold and 10-fold negative selection, respectively. Points on the axes in panels B and C reflect genes represented in only one of the two biological replicates. In almost all cases, these represent genes with very few (fewer than three) total insertions. Most (79%) of these genes were excluded from the analysis of negative selection because of the low representation (see Materials and Methods). Read counts per gene were calculated and normalized as described in Materials and Methods, except for panel A, in which insertions at all positions within each gene were included and values were not normalized for gene length.
Identification of genes required for tobramycin resistance.
To examine whether the Tn-seq circle method could effectively identify genes required for a growth trait, we examined intrinsic resistance to tobramycin. Numerous genes associated with the trait are known (7), providing a reference set to help evaluate the new method. We grew the pool of 100,000 mutants characterized above in the presence or absence of a subinhibitory concentration of tobramycin (about one-half of the MIC) and used Tn-seq to determine which mutants were selectively lost in the treated culture. The selection and Tn-seq analysis were carried out twice, with highly reproducible results (Fig. 2B and C). Mutations affecting numerous candidate resistance genes were selectively lost in the presence of tobramycin (Fig. 2D).
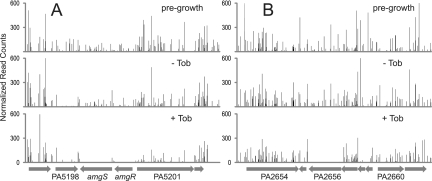
Loss of mutants in the presence of tobramycin was represented for each gene as the ratio of total mutant sequence reads with and without the antibiotic, with both biological replicates included (see Table S1 in the supplemental material). Reads corresponding to the 5′ and 3′ 10% of each open reading frame (ORF) were excluded to help eliminate insertions which were not inactivating, and the numbers of reads were normalized for gene length, total mapped reads in the corresponding Tn-seq run, and local read density (see Materials and Methods). Genes with very few mapped insertion locations or which showed a significant loss of mutant representation after growth in the absence of tobramycin were also excluded (see Materials and Methods). By these criteria, 117 genes showed greater than 2.5-fold negative selection (Fig. 2D; see Table S1 in the supplemental material), with 29 of them exhibiting 10-fold or greater negative selection. Several examples of genes exhibiting strong negative selection in the presence of tobramycin illustrate the loss of sequence reads relative to neighboring genes in which mutations were not negatively selected (Fig. 3).
FIG 3 .
Examples of negative selection revealed by Tn-seq. The graphs show the number of Tn-seq reads at each location from the sample prior to growth, after growth without tobramycin (Tob), and after growth with a subinhibitory concentration of tobramycin. Data are averaged from biological replicates and normalized as described in Materials and Methods. Regions of the genome corresponding to 8,000 bp are shown, with the predicted genes represented at the bottom of each panel. (A) The amgRS region. Insertions in both amgR and amgS are selected against strongly in the presence of tobramycin, whereas insertions in PA5198 are selected against whether or not tobramycin is present. (B) The PA2656 region. The graphs show strong negative selection of PA2656 mutants in the presence of tobramycin. PA2656 encodes a two-component regulator implicated in intrinsic tobramycin resistance by these results.
The 117 negatively selected genes included 13 of the 14 genes identified in the previous mutant-by-mutant screening as contributing strongly to tobramycin resistance (7) (Fig. 2D). The missing gene, PA4961, had been excluded because of very low Tn-seq representation after growth without tobramycin. The results show that the Tn-seq approach was relatively effective in identifying the known resistance functions.
Verification of sensitivity phenotypes using individual mutants.
Of the 117 genes associated with intrinsic tobramycin resistance by Tn-seq, 24 had been previously characterized using individual mutants (7). To test sensitivity phenotypes for newly identified genes, we recovered individual transposon mutants from the PAO1 mutant library (8) or constructed deletion mutants.
Transposon mutants corresponding to 58 of the 93 newly identified tobramycin resistance genes were assayed, with at least two different mutant alleles tested for most (42/58) (see Table S1 in the supplemental material). Each of the mutants was single colony purified and resequenced to confirm its identity prior to assaying sensitivity. Mutations in 47 of the 58 genes increased tobramycin sensitivity by MIC assay (see Table S1). The sensitivity phenotypes of 35 of the 47 were confirmed using multiple mutants.
For several of the tobramycin resistance genes identified by Tn-seq, there were either no (or few) transposon mutants available in the PAO1 library, the only mutants available corresponded to in-frame lacZ or phoA fusions (possibly causing sensitivity by a gain-of-function mechanism) (7), or different mutant alleles led to different tobramycin sensitivities. For seven such genes in the group displaying 10-fold or greater negative selection by Tn-seq, we created deletion mutants and examined their tobramycin sensitivities by MIC assays. All seven deletions increased tobramycin sensitivity (see Table S1 in the supplemental material).
Overall, of the 117 tobramycin resistance genes identified by Tn-seq, 85 were tested by assays of individual mutants in this and the previous study and 74 (87%) were confirmed to display at least a 2-fold decrease in the tobramycin MIC. The confirmed group included nearly all (26/27) of the genes with the strongest negative selection (10-fold or greater) identified by Tn-seq.
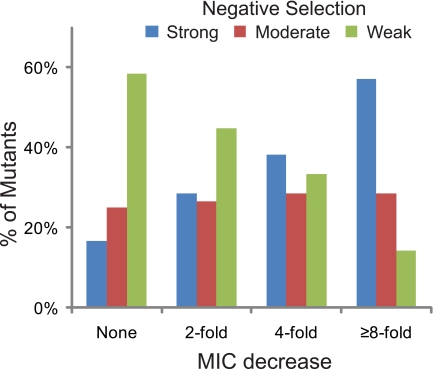
Mutations in 28 genes led to at least 4-fold decreases in the tobramycin MIC (Table 2). This group included 13 previously identified and 15 new genes. The importance of large-scale verification with individual mutants is illustrated by the fact that some of the mutations leading to large reductions in the tobramycin MIC were not in the top group negatively selected in the Tn-seq experiments (Table 2). Nevertheless, there was a correlation between the magnitude of the negative selection in the Tn-seq analysis and the MICs of the corresponding individual mutants (Fig. 4).
TABLE 2 .
Genes identified by Tn-seq exhibiting strong mutant hypersensitivity to tobramycina
| Locus | Gene | Function | No. of normalized reads (no. of hits with reads)b |
Selection ratiob | Previously identifiedc | No. of mutants testedd | MICe | ||
|---|---|---|---|---|---|---|---|---|---|
| Pregrowth | Growth without Tob | Growth with Tob | |||||||
| None (PAO1) | 1.0 | ||||||||
| PA3303 (control) | 1,888 (17) | 2,062 (20) | 1,703 (20) | 0.83 | 1 | 1.0 | |||
| PA0392 | Conserved hypothetical | 644 (5) | 1,374 (5) | 0 (0) | 0 | No | 2f | 0.25 | |
| PA4077 | Transcriptional regulator | 1,572 (3) | 1,230 (3) | 0 (0) | 0 | No | 1f | 0.25 | |
| PA5199 | amgS | Two-component sensor | 764 (18) | 663 (20) | 0 (0) | 0 | Yes | 0.125 | |
| PA5366 | pstB | Phosphate transport | 1,748 (8) | 761 (8) | 0 (0) | 0 | Yes | 0.25 | |
| PA5200 | amgR | Two-component response regulator | 752 (9) | 396 (12) | 2 (1) | 0.00 | Yes | 0.063 | |
| PA0016 | trkA | Potassium uptake | 1,089 (16) | 823 (16) | 5 (2) | 0.01 | Yes | 0.25 | |
| PA4942 | hflK | Protease subunit | 2,436 (14) | 2,104 (16) | 30 (2) | 0.01 | Yes | 0.25 | |
| PA3014 | faoA | Fatty acid oxidation | 1,465 (23) | 1,227 (27) | 56 (5) | 0.05 | No | 5 | 0.25 |
| PA4398 | Two-component sensor | 862 (19) | 633 (19) | 30 (2) | 0.05 | Yes | 0.25 | ||
| PA5528 | Hypothetical | 2,363 (12) | 2,839 (13) | 161 (4) | 0.06 | Yes | 0.125 | ||
| PA1805 | ppiD | Peptidyl-prolyl isomerase | 3,920 (29) | 4,583 (29) | 397 (12) | 0.09 | No | 5 | 0.25 |
| PA3016 | Hypothetical | 1,630 (9) | 840 (8) | 87 (1) | 0.10 | Yes | 0.125 | ||
| PA4223 | Transport | 638 (19) | 790 (19) | 106 (10) | 0.13 | No | 3 | 0.25 | |
| PA4960 | Amino acid metabolism | 916 (15) | 507 (17) | 80 (2) | 0.16 | No | 2 | 0.25 | |
| PA0374 | ftsE | Cell division | 4,583 (8) | 4,366 (8) | 705 (4) | 0.16 | No | 3 | 0.125g |
| PA3013 | foaB | Fatty acid oxidation | 3,632 (29) | 2,925 (32) | 485 (14) | 0.17 | Yes | 0.25 | |
| PA5471 | Hypothetical | 655 (8) | 755 (8) | 129 (4) | 0.17 | Yes | 0.25 | ||
| PA0502 | Biotin biosynthesis | 1,131 (9) | 627 (9) | 108 (3) | 0.17 | No | 1 | 0.25 | |
| PA3194 | edd | Carbohydrate metabolism | 253 (10) | 161 (11) | 29 (5) | 0.18 | No | 1 | 0.125 |
| PA1775 | cmpX | Cytoplasmic membrane protein | 1,670 (13) | 747 (11) | 134 (7) | 0.18 | No | 3 | 0.25 |
| PA0427 | oprM | Multidrug efflux | 1,877 (15) | 1,427 (15) | 298 (6) | 0.21 | Yes | 0.25 | |
| PA4222 | Transport | 551 (16) | 333 (18) | 74 (6) | 0.22 | No | 2 | 0.25 | |
| PA5369 | pstS | Phosphate transport | 1,037 (10) | 631 (9) | 164 (3) | 0.26 | No | 3 | 0.25g |
| PA2018 | mexY | Multidrug efflux | 3,025 (45) | 2,761 (46) | 811 (40) | 0.29 | Yes | 0.25 | |
| PA5285 | Hypothetical | 1,293 (4) | 1,559 (4) | 466 (4) | 0.30 | No | 4 | 0.25 | |
| PA2604 | Conserved hypothetical | 2,736 (8) | 2,162 (9) | 649 (8) | 0.30 | No | 3 | 0.25 | |
| PA2019 | mexX | Multidrug efflux | 2,403 (20) | 2,488 (21) | 930 (18) | 0.37 | Yes | 0.25 | |
| PA4050 | pgpA | Phospholipid biosynthesis | 1,931 (6) | 1,214 (7) | 483 (4) | 0.40 | No | 3 | 0.125 |
Genes whose mutation caused a 4-fold or greater MIC change are listed.
See text for method of calculation. Tob, tobramycin.
Reference 7.
Number of independent mutants tested. Thirty-four of the 42 mutants examined were confirmed by resequencing, including at least one representative for each gene.
MIC values (in micrograms per milliliter) for representative mutants as measured in this study or in reference 7.
Includes one deletion mutant.
Phenotype possibly due to expression of hybrid proteins.
FIG 4 .
Tobramycin sensitivities of individual mutants of genes identified by Tn-seq. The negative selection classes corresponding to genes showing different mutant tobramycin MIC reductions are shown. Mutants include insertion and deletion strains tested in this and a previous study (7). The Tn-seq negative selection classes are strong (≥10-fold depleted), moderate (5-fold to 10-fold depleted), and weak (2.5-fold to 5-fold depleted).
DISCUSSION
This report describes the development and validation of a procedure to identify the genetic basis of growth-related traits in bacteria. The procedure (the Tn-seq circle method) employs second-generation DNA sequencing to track changes in the makeup of saturation-level pools of transposon mutants grown under selection. The procedure was validated by the analysis of intrinsic P. aeruginosa resistance to the aminoglycoside antibiotic tobramycin, a trait which had been analyzed previously by genome-scale mutant-by-mutant screening (7). The new procedure identified both the most significant tobramycin resistance genes established in the previous study and a number of new genes.
The Tn-seq circle method differs from several other procedures employing deep sequencing to track transposon mutants in that it employs a single-strand circularization step to selectively protect transposon-genome junction fragments from exonuclease degradation prior to amplification and sequencing. Other methods enrich for junction fragment sequences by employing a Mariner transposon carrying an MmeI restriction site at one or both ends to allow the addition of adaptors to genome sequences 16 bp from the junction (2, 4) by selective amplification following DNA fragmentation and adaptor ligation using a transposon-specific primer alone (3) or followed by affinity purification of amplified junctions (1). The method we developed has several potential advantages. First, it does not require the use of a specific transposon carrying an MmeI site. Rather, the presence of a recognition site for almost any 6- or 8-bp cutter near (within ~500 bp) either transposon end is sufficient; it can thus be used with most transposons. This versatility may be particularly valuable for the analysis of bacteria like P. aeruginosa with GC-rich DNA, since Mariner requires a TA sequence at its insertion sites. Second, our method yields a high proportion of sequence reads from transposon-genome junctions. In most assays, approximately 90% of the reads mapped to the PAO1 genome and greater than 98% of the mapped positions appeared to be authentic transposon insertion locations (Table 1). By comparison, at most 67% of the reads from the selective amplification method bore transposon-identifying tags and mapped to the target genome (3). For the affinity purification method (1), the proportion of mapped reads was not reported, but the total number of reads which mapped to nonrepetitive chromosomal regions was about 15-fold lower than that obtained by the circle method. The nucleolytic elimination of unwanted genome sequences in the Tn-seq circle method may reduce the background of irrelevant sequence reads relative to the other methods. A direct comparison of the different methods using common test samples should help reveal their relative merits more definitively.
For genes identified through genomic-scale mutant screens, validation of phenotypes using individual mutants is generally a limiting step (9, 10). However, studies with P. aeruginosa benefit from the availability of near-saturation defined transposon mutant libraries from which individual mutants can easily be retrieved and tested (8, 11). Thus, in this study, we assayed the tobramycin sensitivity of individual insertion mutants affected in more than half (>50) of the genes newly identified using Tn-seq. For most of the genes, two or more different mutants were assayed to provide confirmation of phenotypes. In several cases, we also created and tested deletion mutants. Altogether, of 117 intrinsic resistance genes identified by Tn-seq, 85 were examined in this and the previous study (7) and 74 of the 85 were found to decrease the tobramycin MIC.
We observed only a partial correlation between mutant loss from the pool exposed to tobramycin and the MIC change observed in assays of individual strains (Fig. 4). For example, there were striking cases of strong negative selection in the pool-based screens which were not reflected in correspondingly large MIC changes with individual mutants (e.g., PA2656 in Fig. 3B; see Table S1 in the supplemental material). The result suggests that there may have been undefined differences in the selective pressures under the two assay conditions. However, attempts to show altered sensitivities for specific mutants when mixed with excess wild-type cells were not successful (data not shown). Overall, the findings illustrate the importance of large-scale validation of Tn-seq results using individual strains to identify the subsets of mutants with the strongest phenotypes.
These studies identified several new tobramycin resistance functions whose inactivation greatly increased sensitivity, including PA1805 (ppiD), PA4077, and PA0392 (Table 2). PA1805 encodes a peptidyl-prolyl cis-trans isomerase and is thought to facilitate the folding of membrane and exported proteins (12–14). Since misfolded proteins resulting from aminoglycoside-induced translation errors are thought to compromise the membrane barrier and contribute to bacterial killing (7), the loss of functions promoting proper folding may enhance sensitivity. Mutations inactivating two other putative peptidyl-prolyl isomerases (PA1996 and PA1800) also increased sensitivity to tobramycin (see Table S1 in the supplemental material).
Several new transcriptional regulators also contributed to intrinsic tobramycin resistance, including PA4077 and PA0762 (algU), implying a relatively complex regulation of aminoglycoside resistance. PA0762 (algU) encodes a homologue of a sigma factor (σE) that regulates an important envelope stress response in Escherichia coli (14–16), and mutations inactivating another component of this pathway (PA3649, mucP) also increased tobramycin sensitivity. We assume that, like peptidyl-prolyl isomerases, stress response functions controlled by AlgU contribute to intrinsic resistance by helping cells tolerate misfolded or aberrant membrane proteins (7). Functional redundancy of the AlgU response with other envelope-protective responses, including the AmgRS response (7), may account for the relatively modest increase in sensitivity which accompanies the inactivation of algU (see Table S1 in the supplemental material).
PA0392 encodes a conserved cytoplasmic membrane protein of unknown function. The E. coli ortholog of PA0392 (yggT) contributes to osmotic stress tolerance when potassium uptake is reduced (17). Since inactivating either of two genes of the Trk potassium uptake system (trkA and trkH) also increases tobramycin sensitivity in P. aeruginosa (see Table S1 in the supplemental material and reference 7), it is possible that PA0392 contributes to intrinsic resistance by promoting potassium homeostasis.
In summary, we have presented evidence that a combination of a new Tn-seq screening method and large-scale verification with individual mutants is an effective procedure for identifying the genes responsible for an intrinsic resistance trait in P. aeruginosa. The approach should facilitate the genetic dissection of a variety of growth-related processes in P. aeruginosa and other prokaryotes for which transposon mutagenesis is possible.
MATERIALS AND METHODS
Strains, media, and MIC assays.
The P. aeruginosa strain used was MPAO1 (18). E. coli SM10λpir bearing plasmid pIT2 (8) was used as a donor for transposon T8 (ISlacZhah-tc) mutagenesis. Insertion mutants used for validation tests were obtained from a defined mutant library (8). In-frame deletion mutants were generated by amplification and joining of ~800-bp regions flanking each deletion, followed by recombination into MPAO1 using a pEX19Tc derivative carrying the deletion allele (7, 19). The sequences of the primers used for creating the deletions are listed in Table S2 in the supplemental material. Growth media included LB (10 g/liter tryptone, 5 g/liter yeast extract, 8 g/liter NaCl) and TYE (10 g/liter tryptone, 5 g/liter yeast extract, 8 g/liter NaCl, 15 g/liter agar). MIC assays were performed as previously described (7).
Mutant pool.
To create the ~100,000-member undefined transposon mutant pool, MPAO1 was mutagenized with the Tn5-based transposon T8 (ISlacZhah-tc) by conjugation as previously described (8). Mutagenesis reaction mixtures were frozen in aliquots and later thawed and plated on 2× TYE agar (1× salt) with 60 µg/ml tetracycline (to select for insertion mutants) and 10 µg/ml chloramphenicol (to select against the E. coli conjugal donor) in QTrays (Genetix) at a density appropriate to obtain 15,000 to 20,000 insertion mutant colonies per QTray. After incubation for 25 h at 37°C, the colonies in each QTray were collected into 30 ml LB with 5% dimethyl sulfoxide, frozen rapidly by immersion in ethanol and dry ice, and then stored at −80°C. Equal volumes of the pools from seven QTrays were later combined to create the final pool, containing approximately 105 unique strains. The pool was frozen rapidly in aliquots and stored at −80°C. A portion was used for DNA isolation as the “pregrowth” sample (Qiagen DNeasy Blood & Tissue Kit).
Negative selection.
To carry out selection against tobramycin-hypersensitive strains, approximately 4 × 107 CFU from the complex mutant pool were plated on TYE (solidified with 1.5% agarose) in QTrays with either no antibiotic or 0.36 µg/ml tobramycin (Sigma-Aldrich) and incubated at 37°C for seven to nine doublings (6.5 to 9 h). Preliminary experiments showed that growth for 7 to 10 generations on solid media with a subinhibitory concentration of tobramycin provided robust negative selection, whereas growth for 16 or more generations substantially diminished the complexity of the pool. The level of tobramycin (about half of the MIC) was chosen in order to identify mutants with even weak tobramycin sensitivity phenotypes. Following incubation, the cells were collected into LB and DNA was isolated (Qiagen DNeasy Blood & Tissue Kit). The total numbers of CFU in the inoculating and harvested cultures were determined by serial dilution and plating. Selection (with and without tobramycin) was carried out twice independently.
Tn-seq circle method.
The procedure depicted in Fig. 1 was carried out as follows. Six to 10 µg of DNA was sheared to an average fragment length of 400 bp using a Covaris E210 (a 10% duty cycle, an intensity of 5, 200 cycles per burst, and a duration of 100 s per tube) or a Bioruptor (Diagenode) (two tubes each with 300 µl DNA in TE, seven 15-min treatments, setting H). If the total volume of sheared DNA exceeded ~200 µl, it was concentrated with a QIAquick PCR purification kit (Qiagen). Sheared DNA was end repaired using either an End-It DNA End-Repair kit (Epicenter) or a NEBNext End Repair Module (NEB). The DNA was then purified using a QIAquick PCR purification kit (two columns per sample, elution with 91 µl of elution buffer [EB] per column, eluates combined after elution). A-tailing was carried out by incubating the end-repaired DNA for 20 min at 70°C in four 50-µl reaction mixtures each containing DNA with 1× PCR buffer with magnesium (Ambion), 0.5 mM dATP, and 0.25 µl Taq (Ambion). The DNA was purified using a MinElute PCR purification kit (Qiagen) (one column per A-tail reaction mixture, elution with 12.5 µl EB per column, eluates combined). Illumina adaptor no. 2 was ligated to the A-tailed DNA using a Quick Ligation kit (NEB) and a 20-fold molar excess of adaptor relative to the DNA fragment concentration (calculated based on NanoDrop measurement and predicted average fragment length). DNA was purified using a MinElute PCR purification kit (three columns per sample, eluates combined). The DNA was then digested overnight at 37°C with BamHI (NEB). Fragments of 200 to 400 bp were size selected by cutting from a precast 6% acrylamide TBE gel (Invitrogen), and the DNA was extracted from the crushed gel slices by incubation in TE at 65°C for several hours with periodic vortexing, followed by filtration using 0.2-µm Nanosep columns (Pall/VWR) and ethanol precipitation. DNA was resuspended in 10 to 30 µl of EB (Qiagen), and the concentration was measured by fluorometry or NanoDrop. Typical concentrations were 20 to 150 ng/µl. The steps described above reflect, with minor modifications, standard preparatory steps commonly used for Illumina sequencing.
The circularization and exonuclease steps were based on a protocol developed for selective and specific multiplex amplification of nucleic acids (20). The circularization reaction mixture, which included the size-selected DNA (1 to 4 pmol), an approximately 10-fold molar excess of collector probe (T8_COLLECT_1; see Table S2 in the supplemental material), 1× Ampligase buffer, and 0.5 µl Ampligase (Epicenter) in a 15-µl total reaction mixture volume, was cycled as follows: 95°C for 30 s, 15 cycles of 95°C for 30 s and 67°C for 3 min, 95°C for 2 min, and cooling to 10°C. Following thermocycling, a mixture of exonucleases (0.5 µl Exonuclease I [NEB, 20 U/µl], 0.75 µl LambdaExo [NEB, 5 U/µl], 0.75 µl T7 Gene 6 Exonuclease [USB, 50 U/µl]) was added and the sample was incubated at 37°C for 4 h and then at 85°C for 20 min. The DNA was purified using a MinElute PCR purification kit (one column per sample; elution with 11.5 µl EB). Amplification of transposon-genome junctions from the circularized fragments was carried out by real-time PCR using iTaq Supermix SBRG with ROX (Bio-Rad) in 50-µl reaction mixtures containing 3 µl of the purified DNA sample as the template and 250 nM each amplification primer (T8-SLXA_FOR_AMP2 and SLXA_REV_AMP; see Table S2), and cycling was as follows: 95°C for 2 h 30 min and then 35 cycles of 95°C for 25 s, 59°C for 25 s, and 72°C for 30 s. After an initial test reaction to determine the optimal number of cycles, the PCR was repeated and stopped at a cycle corresponding to approximately 50% maximum amplification (usually 24 to 26 cycles). The DNA was purified with a MinElute PCR purification kit (elution with 10 µl EB) and examined by gel electrophoresis to confirm the expected size range (controls were carried out for most samples which included a reaction mixture without the collector probe, a reaction mixture without Ampligase, and a reaction mixture without exonucleases). The samples were sequenced by standard Illumina sequencing (36-bp single-end runs) using sequencing primer T8-SEQ_STA or T8-SEQ_G (see Table S2 in the supplemental material).
To optimize the efficiency of the circularization step, we initially varied several parameters. For these tests, the amount of circularized product in each sample following the exonuclease step was determined by quantitative PCR using primers designed to amplify the ligated portion of the circles. We found that the procedure was robust when several parameters were varied, including the molar ratio of the template to the collector probe, ligation time and temperature, and the number of ligation cycles.
Data processing and analysis.
The standard Illumina preprocessing pipeline was carried out (21), and reads which passed default filtering were mapped using MAQ version 0.7.1 with default parameters (two mismatches per read permitted) (22). Typically 86 to 91% of the reads mapped to the PAO1 genome. Reads per insertion location were tallied using custom Perl scripts (available upon request). Reads which could not be mapped to an unambiguous location due to repetitive chromosomal sequences (MAQ mapping score of zero, fewer than 2% of mapped reads) were excluded from further analysis. In a few cases (fewer than 2% of locations), polymerase slippage during sequencing appeared to cause some reads to map to nucleotides adjacent to the insertion junction. These cases were recognizable as co-oriented reads mapping to adjacent nucleotides where the number of reads for one of the positions comprised a small fraction of the number of reads for the adjacent position. In such cases, all of the reads were considered to correspond to a single insertion and were assigned to the position with the largest number of reads.
Read counts per gene were based on insertion locations in the central 80% of the ORF and were normalized to account for gene length, total mapped reads per Tn-seq run, and local read density. The overall correlation between gene length and number of insertion locations per gene was 83%, validating the use of gene length as a normalization factor. Local read density was calculated for each insertion location as the average number of reads for positions within an approximately 100-kb window surrounding that location whose read counts deviated from the overall average for the same window by no more than 2-fold. For each insertion position, the number of reads at that position was normalized according to its local read density relative to the average local read density for all positions. This normalization was necessary because some samples grown on tobramycin showed low relative density near the chromosomal replication origin and higher relative density near the terminus of replication, while samples grown without tobramycin showed higher relative density near the origin and lower relative density near the terminus. The overall correction from raw read numbers was, at most, 3-fold. The complete equation for normalization was as follows: normalized no. of reads per gene = Σ (for insertion locations in central 80% of ORF) [raw read counts × (average read density/local read density)] × 107/[gene length × total no. of mapped reads per run]. For determination of negative selection, genes with fewer than three mapped insertion locations in the middle 80% of the ORF were excluded, as were genes which showed ≥67% loss of representation after growth in the absence of tobramycin (based on read counts after growth relative to those before growth). Some genes (33/150) were also excluded if manual examination of the distribution of hits and reads within the ORF betrayed questionable evidence of negative selection, even though the strict numerical criteria were satisfied. Examples included genes with few (though at least three) insertion locations or genes with very low read numbers at almost all sites, such that excessive reads at a single location skewed the calculated degree of negative selection.
Nucleotide sequence accession numbers.
The Illumina sequencing reads listed in Table 1 have been deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRA026588.
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
This work was supported by grant U54AI057141 from the National Institutes of Health and by grant MANOIL08G0 from the Cystic Fibrosis Foundation.
We thank Emily Turner, Simon Fredriksson, Sam Lee, Aaron Hinz, Cho Li Lee, Kiara Held, Elizabeth Ramage, Metawee Thongdee, and Pradeep Singh for helpful discussions and technical assistance.
Footnotes
Citation Gallagher, L. A., J. Shendure, and C. Manoil. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2(1):e00315-10. doi:10.1128/mBio.00315-10.
REFERENCES
- 1. Gawronski J. D., Wong S. M., Giannoukos G., Ward D. V., Akerley B. J. 2009. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl. Acad. Sci. U. S. A. 106:16422–16427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goodman A. L., McNulty N. P., Zhao Y., Leip D., Mitra R. D., Lozupone C. A., Knight R., Gordon J. I. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Langridge G. C., Phan M. D., Turner D. J., Perkins T. T., Parts L., Haase J., Charles I., Maskell D. J., Peters S. E., Dougan G., Wain J., Parkhill J., Turner A. K. 2009. Simultaneous assay of every Salmonella typhi gene using one million transposon mutants. Genome Res. 19:2308–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Opijnen T., Bodi K. L., Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6:767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kerr K. G., Snelling A. M. 2009. Pseudomonas aeruginosa: a formidable and ever-present adversary. J. Hosp. Infect. 73:338–344 [DOI] [PubMed] [Google Scholar]
- 6. Cottarel G., Wierzbowski J. 2007. Combination drugs, an emerging option for antibacterial therapy. Trends Biotechnol. 25:547–555 [DOI] [PubMed] [Google Scholar]
- 7. Lee S., Hinz A., Bauerle E., Angermeyer A., Juhaszova K., Kaneko Y., Singh P. K., Manoil C. 2009. Targeting a bacterial stress response to enhance antibiotic action. Proc. Natl. Acad. Sci. U. S. A. 106:14570–14575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jacobs M. A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S., Will O., Kaul R., Raymond C., Levy R., Chun-Rong L., Guenthner D., Bovee D., Olson M. V., Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Autret N., Charbit A. 2005. Lessons from signature-tagged mutagenesis on the infectious mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 29:703–717 [DOI] [PubMed] [Google Scholar]
- 10. Chiang S. L., Mekalanos J. J., Holden D. W. 1999. In vivo genetic analysis of bacterial virulence. Annu. Rev. Microbiol. 53:129–154 [DOI] [PubMed] [Google Scholar]
- 11. Liberati N. T., Urbach J. M., Miyata S., Lee D. G., Drenkard E., Wu G., Villanueva J., Wei T., Ausubel F. M. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dartigalongue C., Raina S. 1998. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 17:3968–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duguay A. R., Silhavy T. J. 2004. Quality control in the bacterial periplasm. Biochim. Biophys. Acta 1694:121–134 [DOI] [PubMed] [Google Scholar]
- 14. Rowley G., Spector M., Kormanec J., Roberts M. 2006. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat. Rev. Microbiol. 4:383–394 [DOI] [PubMed] [Google Scholar]
- 15. Bazire A., Shioya K., Soum-Soutera E., Bouffartigues E., Ryder C., Guentas-Dombrowsky L., Hemery G., Linossier I., Chevalier S., Wozniak D. J., Lesouhaitier O., Dufour A. 2010. The sigma factor AlgU plays a key role in formation of robust biofilms by nonmucoid Pseudomonas aeruginosa. J. Bacteriol. 192:3001–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wood L. F., Ohman D. E. 2009. Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol. Microbiol. 72:183–201 [DOI] [PubMed] [Google Scholar]
- 17. Ito T., Uozumi N., Nakamura T., Takayama S., Matsuda N., Aiba H., Hemmi H., Yoshimura T. 2009. The implication of YggT of Escherichia coli in osmotic regulation. Biosci. Biotechnol. Biochem. 73:2698–2704 [DOI] [PubMed] [Google Scholar]
- 18. Gallagher L. A., McKnight S. L., Kuznetsova M. S., Pesci E. C., Manoil C. 2002. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184:6472–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 20. Fredriksson S., Baner J., Dahl F., Chu A., Ji H., Welch K., Davis R. W. 2007. Multiplex amplification of all coding sequences within 10 cancer genes by Gene-Collector. Nucleic Acids Res. 35:e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Illumina Inc 2009. CASAVA software version 1.7 user guide. Catalog no. SY-960-1701. Illumina, Inc., San Diego, CA [Google Scholar]
- 22. Li H., Ruan J., Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.