Abstract
The cancer stem cell (CSC) hypothesis postulates that tumors are maintained by a self‐renewing CSC population that is also capable of differentiating into non‐self‐renewing cell populations that constitute the bulk of the tumor. Although, the CSC hypothesis does not directly address the cell of origin of cancer, it is postulated that tissue‐resident stem or progenitor cells are the most common targets of transformation. Clinically, CSCs are predicted to mediate tumor recurrence after chemo‐ and radiation‐therapy due to the relative inability of these modalities to effectively target CSCs. If this is the case, then CSC must be efficiently targeted to achieve a true cure. Similarities between normal and malignant stem cells, at the levels of cell‐surface proteins, molecular pathways, cell cycle quiescence, and microRNA signaling present challenges in developing CSC‐specific therapeutics. Approaches to targeting CSCs include the development of agents targeting known stem cell regulatory pathways as well as unbiased high‐throughput siRNA or small molecule screening. Based on studies of pathways present in normal stem cells, recent work has identified potential “Achilles heals” of CSC, whereas unbiased screening provides opportunities to identify new pathways utilized by CSC as well as develop potential therapeutic agents. Here, we review both approaches and their potential to effectively target breast CSC.
Keywords: Cancer stem cell (CSC), Breast cancer, Novel therapeutics
1. What is a cancer stem cell?
The term “cancer stem cell” has led to a degree of confusion within the cancer research field; however, while the term is “imperfect” it is here to stay (Clarke et al., 2006). The existence of CSC does not imply that these cells are necessarily derived from normal tissue stem cells. Rather, the definition of “cancer stem cells” is based on three functional characteristics of these cells. These include the ability to initiate tumors in immunocompromised or syngeneic mice, self‐renewal capacity measured by tumor formation in secondary mice, and the capacity to differentiate into the non‐self‐renewing cells, which constitute the tumor bulk. Formal proof that a tumor follows the CSC paradigm is based on the ability to prospectively isolate fractions of cells with these properties as well as fractions depleted of these properties. On the other hand, the inability to fractionate cells with these properties suggests the tumor may not be hierarchically organized and each cell has equal tumor‐initiation potential determined by intrinsic and/or extrinsic factors (Dick, 2008). Given these properties, especially the ability to initiate tumors in model organisms, and confusion surrounding the term “cancer stem cell”, we suggest a more appropriate functional descriptor would be “tumor‐initiating cell”.
Stem cells can exhibit multiple fates: division, differentiation, quiescence, or death (Dick, 2008). A hallmark of stem cells is their ability to self renew whereby cell fate (i.e. tumor‐initiation) is segregated symmetrically or asymmetrically to daughter cells, which can be functionally defined by secondary transplantation (Dick, 2003; McKenzie et al., 2006). Asymmetric segregation of cell membranes, proteins, and even DNA impact cell fate, especially in stem cells (Neumuller and Knoblich, 2009). The existence of multiple stem cell populations with varying self‐renewal capacity has been demonstrated in the hematopoietic system (Majeti et al., 2007; McKenzie et al., 2006).
The third property of CSC is the ability to differentiate into the bulk, non‐tumorigenic cells. Work from the great pathologists over the last 100 years (Virchow, Maximov, etc) has shown tumors are morphologically heterogeneous, composed of relatively undifferentiated and differentiated cells. Seminal work by Pierce and colleagues demonstrated spontaneous differentiation of malignant teratocarcinoma cells into mature benign cells (Pierce et al., 1960). Similarities in the heterogeneity of tumors and normal tissue led to the proposal of cancer as a caricature of normal development, albeit with altered differentiation (Pierce and Speers, 1988). Cellular heterogeneity is typically measured using differential expression of protein differentiation markers (i.e. CD44, CD24, CD34, CD38, etc) and is generated in xenograft models upon serial transplantation. The ability to harness this differentiation process as a therapy has been successfully demonstrated by the treatment of acute promyelocytic leukemia with all‐trans retinoic acid (Nowak et al., 2009).
2. Brief history of CSCs
While the formal description of cancer stem cells is a relatively recent development, the concept is an old one related to tissue‐resident stem cells, especially in the hematopoietic system (Dick, 2008). After the demonstration by Furth and Kahn in 1937 that a single murine leukemia cell could initiate a tumor in mice (Furth and Kahn, 1937), work over the last 70 years has shown a wide variation of tumor‐initiation frequency. Surprisingly, human studies where single cell suspensions of cancers were autotransplanted back into the patient showed tumor‐initiation was rare and required more than 106 cells (Southam et al., 1962).
In the 1960s, a major conceptual development was the demonstration that the three major lineages of the blood system are derived from one common precursor stem cell (McCulloch and Till, 1960), tumors exhibited functional heterogeneity (Clarkson et al., 1970), and a small subset of acute myeloid leukemia (AML) cells proliferate slower than the mature blast cells (Clarkson, 1969). These studies laid the groundwork for the seminal discovery of AML stem cells by John Dick and colleagues in 1994 (Lapidot et al., 1994). Technical advances, including the development of NOD/SCID (non‐obese diabetic/severe combined immune deficiency) immunocompromised mouse models (Shultz et al., 2007, 1995), monoclonal antibody production (Spangrude et al., 1988), and fluorescent activated cell sorting (FACS) (Bonner et al., 1972) provided the tools needed to demonstrate that engraftment of primary AML cells in immunocompromised mice is mediated by a rare cell that can be prospectively isolated. Further discoveries showed AML is clonally derived, hierarchically organized, and can be serially passaged in immunocompromised mice (Bonnet and Dick, 1997; Hope et al., 2004).
Until the identification of CSCs in human breast cancer in 2003 (Al‐Hajj et al., 2003), the applicability of these observations to solid cancers remained hotly debated. In the following years, CSCs have been identified in most solid cancers, including brain, colon, pancreatic, head and neck, and others (Li et al., 2007; O'Brien et al., 2007; Prince et al., 2007; Singh et al., 2004). However, xenotransplantation studies have inherent problems, including the foreign microenvironment, incompatible growth factor and cytokine signals, and the lack of immune cells in these models. This raises very important points that must be addressed. Indeed, some human cancers, such as melanoma, may not follow the CSC paradigm with an inability to prospectively isolate tumor‐initiating cells and a high frequency of tumor‐initiation (Kelly et al., 2007; Quintana et al., 2008). Recently, Roesch et al. suggest that in melanoma a slow‐cycling subpopulation may play an important role in tumor “maintenance” rather than initiation and that this may represent a “dynamic” rather than static state (Roesch et al., 2010). However, it has been shown in syngeneic studies in human (Southam et al., 1962) and mice (Cho et al., 2008; Huntly et al., 2004; Yilmaz et al., 2006; Zhang et al., 2008) that not all cancer cells can reinitiate the tumor suggesting that all the work from xenotransplantation studies cannot be summarily disregarded. Importantly, the CSC paradigm does not depend solely on the frequency of tumor‐initiating cells, but rather the evidence of functional heterogeneity and hierarchal organization (Dick, 2008; Kennedy et al., 2007).
3. Hierarchal organization of the human breast
The human breast is composed of a branching network of ducts terminating in small ductal structures termed terminal ductal lobular units (TDLU). The ducts contain luminal epithelial and myoepithelial cells while fibroblast, endothelial, adipocytes, and hematopoietic cells constitute the remaining stromal component. The luminal lineage is divided into ductal and alveolar cells that line the ducts or the alveolar units present during pregnancy, whereas the myoepithelial cells are basal, contractile cells surrounding the ducts that force secreted milk proteins through the ductal network to the nipple during lactation. The large expansion of breast epithelium during puberty and multiple pregnancies suggests the existence of cells with extensive proliferative and regenerative properties typically associated with stem cells (Visvader, 2009).
Observational studies have provided additional lines of evidence for breast stem cells. Women exposed to radiation as teenagers, such as atomic bomb survivors, have an increased risk of developing breast cancer later in life, suggesting the presence of long‐lived cells (Land and McGregor, 1979). Examination of X‐linked inactivation and loss of heterozygosity in normal and adjacent breast cancer tissue showed large tracts of epithelial cells sharing similar chromosomal alterations suggesting they are derived from the same stem cell (Deng et al., 1996; Tsai et al., 1996).
The seminal development of the in vivo mouse mammary fat pad transplant system by DeOme et al. provided an assay to study breast stem cell function and remains the “gold standard” (DeOme et al., 1959). In this assay, the epithelial portion in the pre‐pubertal mammary fat pad is surgically removed leaving the stromal portion. Donor cells are transplanted into this “cleared fat pad” and outgrowths are quantified. Using this assay, it was demonstrated that outgrowths could be generated from different regions of the mouse mammary gland, young and old virgin or pregnant mice, and the outgrowths could be serially transplanted for multiple generations (Daniel et al., 1968; Daniel and Young, 1971; Smith and Medina, 1988; Young et al., 1971). Retroviral marking of mammary cells conclusively proved mammary outgrowths could be derived from a single cell (Kordon and Smith, 1998). Kuperwasser et al. further improved the transplant assay for human breast epithelial cells by “humanizing” the fat pad via injection of irradiated human fibroblasts, which generated a stromal environment that more closely simulates human breast tissue (Kuperwasser et al., 2004). Recently, Eirew et al. developed an alternative in vivo method for quantifying human breast stem cell activity based on transplantation of cells embedded in collagen gels into the renal capsule of mice supplemented with estrogen and progesterone (Eirew et al., 2008).
The development of these in vivo transplant assays has facilitated the prospective isolation of mouse and human breast stem and progenitor cells based on various cell‐surface markers. Mouse stem cells are enriched in the CD49fhiCD29hiCD24+Sca1− fraction (Shackleton et al., 2006; Stingl et al., 2006). Human breast stem cells have been found to be contained in the CD49fhiEpCAM− (Eirew et al., 2008; Lim et al., 2009) and aldehyde dehydrogenase (ALDH)+ fractions (Ginestier et al., 2007). Breast cancer stem cells are found in the CD44+CD24− fraction with as few as 200 cells able to initiate tumors (Al‐Hajj et al., 2003). Luminal progenitor cells within the human breast have been identified using in vitro differentiation assays and exhibit a CD49f+EpCAM+ phenotype (Eirew et al., 2008; Lim et al., 2009). Myoepithelial progenitor cells were concentrated in the CD49f+EpCAM−/low fraction along with bi‐potent progenitors (Eirew et al., 2008). Basal cells are marked by CD49f+EpCAM− expression whereas mature luminal cells are mainly CD49f−EpCAM+ (Lim et al., 2009). Additionally, recent studies have suggested that ductal and alveolar structures may originate from different stem cells in the mouse breast (Jeselsohn et al., 2010). Additionally, it has been shown using in vitro suspension culture that mammosphere formation is a property of breast progenitor cells (Charafe‐Jauffret et al., 2009; Dontu et al., 2003; Ginestier et al., 2007; Liu et al., 2006).
4. What is the cell of origin for breast cancer?
The cell of origin represents an important question in cancer research and has implications for prevention, early detection, and treatment. The quiescent, hence relatively long‐lived, nature of stem cells has been suggested as important in allowing these cells to acquire the multiple genetic lesions required for tumorigenesis (Reya et al., 2001). The cell‐of‐origin hypothesis postulates that cells within a tissue acquire some initial hit(s), somatic or germ‐line, but only the stem cell survives by virtue of its quiescent program and eventually passes the mutation(s) to progeny cells when it self‐renews. On the other hand, progenitor cells, with limited self‐renewal capacity, may generate cancers through acquisition of self‐renewal capacity. Experimental evidence for both possibilities has been demonstrated by the generation of murine leukemias after introduction of leukemogenic oncogenes into purified hematopoietic stem cells or progenitor cells (Cozzio et al., 2003; Krivtsov et al., 2006; Somervaille and Cleary, 2006). Armstrong and colleagues showed reactivation of self‐renewal pathways in normal progenitors led to transformation (Krivtsov et al., 2006). However, in these studies, stem cells were more efficiently transformed and, thus, the most common target may be the stem cell.
Parallel studies comparing the transformation efficiency of mouse mammary progenitor and stem cells with mammary oncogenes, such as Wnt‐1, have not been possible until the recent description and isolation of these subsets (Eirew et al., 2008; Lim et al., 2009; Shackleton et al., 2006; Stingl et al., 2006). Ectopic Wnt‐1 expression has been shown to increase mammary stem and progenitor cells in pre‐neoplastic tissue suggesting Wnt‐1 may alter the epithelial hierarchy and MMTV‐Wnt‐1 tumors contain cells capable of multi‐lineage differentiation whereas MMTV‐Neu tumors are composed of luminal committed cells suggesting these tumors originate from different cells in the mammary hierarchy (Liu et al., 2004; Vaillant et al., 2008). However, these descriptive studies cannot distinguish if Wnt‐1 more efficiently transforms stem or progenitor cells. In the mouse, Bouras et al. demonstrated constitutive Notch activation in the stem or luminal progenitor cells induced similar pre‐neoplastic lesions at comparable frequency suggesting Notch signaling can transform either population (Bouras et al., 2008).
Breast cancer is a histologically, molecularly and epidemiologically heterogeneous disease. There are six molecular subtypes based on gene expression analysis, which include normal breast‐like, luminal A and B, basal‐like, claudin‐low, and HER2/ERBB2 over‐expressing (Herschkowitz et al., 2008, 2007, 2000, 2001, 2006). The molecular heterogeneity between breast cancers has been suggested to result from different targets of transformation. Lim and colleagues examined the different epithelial populations in normal and pre‐neoplastic tissue from BRCA1 mutation carriers (Lim et al., 2009). In women heterozygous for BRCA1 mutations, they reported an increase in luminal progenitor cells and these tumors were molecularly more similar to luminal progenitors than the stem cell enriched population. These results seemingly contrast with an earlier study showing an expansion of breast stem cells after siRNA‐mediated knockdown of BRCA1 in normal mammary epithelial cells (Liu et al., 2008). However, these observations suggest a critical role of BRCA1 dosage since BRCA1 expression is lower in stem cells compared to luminal progenitors and differentiated luminal cells. Taken together, these data suggest expansion of the stem and/or luminal progenitor population may be a target for transformation in BRCA1 mutation carriers (Ginestier et al., 2009).
5. Resistance to chemotherapy and radiotherapy
While the existence of CSC in multiple human tumors has been firmly established, the functional and clinical significance of these cells remains hotly debated (Shipitsin and Polyak, 2008). The idea that CSC might be resistant to treatment had foundations in seminal leukemia studies that predicated relapse and ultimate failure of chemotherapy was due to the inability to eradicate CSC (Clarkson and Fried, 1971). Indeed, work from Craig Jordan's lab showed AML LSC are resistant to the chemotherapeutic cytarabine, which is part of standard induction therapy (Guzman et al., 2001, 2005). Recent studies have highlighted the chemo‐ and radio‐resistance of CSCs in solid tumors as a possible mechanism to explain why current treatment modalities fail to cure most patients (Bao et al., 2006; Creighton et al., 2009; Li et al., 2008; Phillips et al., 2006; Yu et al., 2007). NOTCH and Wnt signaling, pathways utilized in normal stem cell self‐renewal, were found to mediate radio‐resistance in glioblastoma and breast cancer (Phillips et al., 2006; Wang et al., 2010; Woodward et al., 2007).
CSCs have been invoked as the seed for distant metastases, which typically are responsible for end‐stage disease and ultimately death. The epithelial–mesenchymal transition (EMT) process is critical for metastasis. Numerous reports suggest a link between EMT and CSC (Mani et al., 2008), and recently the concept of “migrating cancer stem cells” has been proposed (Brabletz et al., 2005). Interestingly, the basal fraction (CD49fhiEpCAM−) containing normal human stem cells and myoepithelial progenitor cells express the common EMT marker vimentin suggesting some cells may be undergoing EMT (Eirew et al., 2008; Lim et al., 2009). However, since this fraction is not purely stem cells, it remains to be determined if most or all stem cells express EMT markers. Nonetheless, given the congruence between EMT factors and stem cell markers in invasive cancer cells, strategies to target CSC must also focus these “migrating cancer stem cells.”
6. The realities of targeting CSC
The development of CSC‐specific agents presents new challenges for evaluating clinical efficacy (Wang, 2007). The commonly accepted criteria for clinical efficacy is overall survival, but this can only be demonstrated in phase III trials requiring large patient numbers and long‐term follow‐up. Phase II trials are designed to assess a drug's activity using surrogate endpoints to determine whether a phase III trial is warranted. The typical endpoint used in phase II trials is tumor shrinkage using criteria defined by RECIST (Eisenhauer et al., 2009), which can be measured over weeks or months. The premise is that tumor regression equates with clinical benefit. Although tumor shrinkage may temporarily relieve symptoms related to tumor burden, a correlation between tumor response and overall survival has rarely been shown for patients with hematological and solid cancers (Huff et al., 2006). For example, there is only a modest overall survival advantage for pancreatic, prostate and metastatic breast cancer patients treated with combination chemotherapy even with tumor response (Abratt et al., 2004; Chung et al., 2008; Rocha Lima et al., 2004). This paradox between response and survival may be explained by an inability of chemotherapy to effectively target the CSC. The problems associated with correlation between tumor response and survival in phase II/III clinical trials highlight a need for novel clinical trial design to test anti‐CSC efficacy. These trials may utilize surrogate intermediate endpoints evaluating cells expressing CSCs markers.
One such approach is the use of neoadjuvant trials where CSC populations from tumor tissue obtained pre‐ and post‐therapy are assayed in vitro and in vivo (Creighton et al., 2009). Additionally, there are inherent differences when testing potential anti‐CSC therapeutics in the adjuvant and advanced settings. Agents causing tumor regression in the advanced setting likely reflect effects on the bulk tumor population, but may have minimal effect on the CSC population. In contrast, a CSC‐specific therapeutic would show modest effects on tumor growth when tested in the advanced setting, but may have substantial clinical benefit in the adjuvant setting.
An ideal CSC‐specific therapeutic would target the CSC and bulk tumor cells with minimal adverse effects. There are more than 500 kinds of adverse effects associated with cancer therapy and range from minor to life threatening or death (Trotti et al., 2003). Chemotherapy‐induced myelosuppression is a major dose‐limiting toxicity (Daniel and Crawford, 2006; Nirenberg, 2003) and is not limited to chemotherapy regimens; treatment with the molecularly targeted BCR‐ABL inhibitor imatinib (Gleevec) led to myelosuppression in patients with chronic myeloid leukemia (CML) or solid tumors (Agis et al., 2006). Additionally, total body irradiation was shown to selectively induce senescence in mouse hematopoietic stem cells, as compared to progenitor cells (Wang et al., 2006b). The relationship between chemotherapy‐induced toxicity in other tissues, especially the gastrointestinal, and normal stem cells has been demonstrated for numerous regimens (Dekaney et al., 2009, 1983, 1987). However, within human skin, the CD49+CD71low stem cells were more radio‐resistant than the CD49+CD71+ progenitor cells (Rachidi et al., 2007). Strategies to eradicate breast CSC do not necessarily need to spare normal breast stem cells since most women diagnosed with breast cancer are post‐menopausal and do not need the ability to lactate. However, these therapies should not target other normal stem cells, such as hematopoietic, gastrointestinal, skin, and neuronal, which are critical for patient survival.
7. Pathways utilized by solid CSCs
In order to effectively target CSC, a detailed understanding of the pathways regulating the growth, survival, and self‐renewal of CSC is needed. For example, seminal studies from Craig Jordan's lab first demonstrated a reliance on NF‐κB signaling in AML LSC (Guzman et al., 2001), which could be exploited using parthenolide, a natural product sesquiterpene lactone from the medicinal plant feverfew (Guzman et al., 2005). Parthenolide induced apoptosis in the LSC in vitro and was able to inhibit leukemia engraftment in NOD/SCID mice after a brief (16 h) treatment in vitro. More importantly, parthenolide had little effect on normal HSC suggesting parthenolide treatment in AML patients may spare normal blood cell production. An orally bioavailable parthenolide analog with similar anti‐LSC activity is currently in phase I clinical trials (Guzman et al., 2007). While NF‐κB signaling and sensitivity to parthenolide may not apply to all CSC, these studies present a model for rational drug development for anti‐CSC therapeutics.
7.1. New role for an old tumor suppressor: p53
Since the discovery of the p53 tumor suppressor (Hollstein et al., 1991), there have been thousands of articles detailing numerous roles of p53 in cancer. A recent article highlights a newly discovered role for p53 in normal and cancer stem cell biology. The genetic pathways regulating stem cell self‐renewal via symmetric or asymmetric cell divisions has been elegantly shown for Drosophila stem cells (Gonzalez, 2007; Morrison and Kimble, 2006; Neumuller and Knoblich, 2009); however, the machinery that controls asymmetric versus symmetric divisions in mammalian stem cells is poorly characterized. Cicalese and colleagues used the power of labeling cells with a cell‐surface dye, PKH26, to follow the fate of mouse breast stem cells (Cicalese et al., 2009). After labeling ∼99% of cells, they noticed only ∼1% of mammosphere cells retained high levels of PKH26. These PKH26high cells retained secondary mammosphere formation and outgrowth potential. By continuously imaging single PKH26high cells in vitro they found ∼80% of the first cell divisions were asymmetric. In contrast, the mammary cells of p53−/− mice contained a higher proportion of mammary stem cells, and in vitro these cells underwent a higher (∼75%) proportion of symmetric cell divisions. Over time, the number of stem cells increased at an apparent geometric rate. Similar results were obtained with ErbB2 over‐expressing cells. Restoration of p53 function with Nutlin3, a small molecule inhibitor of MDM2‐dependent p53 degradation, in ErbB2 over‐expressing cells switched cell divisions back to asymmetric in the absence of anti‐proliferative effects. Finally, Nutlin3 treatment reduced mammosphere and tumor formation in ErbB2 over‐expressing cells indicating increased self‐renewal contributes to tumor growth. These studies suggest that the balance between symmetric and asymmetric CSC self‐renewal may be a determinant of CSC frequency. Therapeutic agents developed to modulate these pathways may show efficacy in reducing CSC populations.
7.2. “Akt”ing as a central node in CSC signaling
Akt (protein kinase B) is a central regulator in the Wnt and PI3K signaling pathways and is critical in energy homeostasis. Upstream of Akt is the PTEN tumor suppressor, which encodes a lipid and protein phosphatase, and is frequently mutated in human cancer (Keniry and Parsons, 2008). PTEN dephosphorylates PIP3, a product of PI3K. Loss of PTEN results in accumulation of PIP3, thereby activating a signaling cascade, including AKT, S6 kinase, mTOR, Rac1, Cdc42, and phosphatidylinositol‐dependent kinases (Hill and Wu, 2009). AKT activation results in cell cycle progression via p27 down‐regulation and leads to down‐regulation of pro‐apoptotic factors, including BAD and caspase‐9. Various PTEN mouse models have shown the PTEN/PI3K/AKT pathway controls stem cell homeostasis and malignancies in numerous tissues (Guo et al., 2008; Hill and Wu, 2009; Wang et al., 2006a; Yilmaz et al., 2006; Zhang et al., 2006).
A role for PTEN in breast cancer and stem cells was shown when it was observed that PTEN deletion in mammary epithelial cells led to precocious mammary gland development and breast cancer (Li et al., 2002). Germ‐line mutation of the PTEN gene is associated with Cowden's disease, an autosomal dominant multi‐neoplasia syndrome, which predisposes men and women to an increased risk of breast cancer (Eng, 2003). In normal human breast tissue, knockdown of PTEN resulted in increased mammosphere formation, Akt activity, and activation of the Wnt/β‐catenin pathway via GSK3β (Korkaya et al., 2009). It was observed that PTEN knockdown in human mammary cells generated disorganized hyperplastic lesions when these cells were introduced into the humanized mammary fat pads of NOD/SCID mice. Pharmacological inhibition of AKT with perifosine, but not the mTOR inhibitor rapamycin, resulted in fewer mammospheres and completely inhibited formation of hyperplastic lesions. PTEN knockdown in breast cancer cells lead to increased mammosphere formation in vitro and tumor‐initiation in NOD/SCID mice. Accordingly, treatment with the AKT inhibitor perifosine reduced tumor growth and secondary tumor formation. Previous studies have shown oncogenic activation of Wnt/β‐catenin signaling increased mammary stem and progenitor cells (Li et al., 2003; Liu et al., 2004). In agreement, activation of Wnt signaling increased human mammosphere formation (Korkaya et al., 2009). Taken together, these results suggest that the PTEN/AKT/Wnt signaling axis regulates normal and malignant breast stem cells.
7.3. Paracrine and autocrine Hedgehog signaling
Aberrant activation of the Hedgehog (Hh) pathway is common in basal cell carcinoma, medulloblastoma, a tumor of cerebellar granule neuron progenitor cells, and rhabdomyosarcoma, a muscle tumor. The Hh pathway represents an attractive target for drug development and has shown promise in Phase I clinical trials of advanced basal cell carcinoma and medulloblastoma with GDC‐0449, an Hh pathway inhibitor (Rudin et al., 2009; Von Hoff et al., 2009). Hh is known to signal through autocrine, juxtacrine, and paracrine mechanisms (Rubin and de Sauvage, 2006; Theunissen and de Sauvage, 2009). Secreted Hh (Sonic, Indian, or Dessert) signals through the 12 trans‐membrane Patched 1 (PTCH) receptor, which de‐represses the 7 trans‐membrane Smoothened (SMOH) protein. Translocation of activated SMOH to cilia initiates a signaling cascade resulting in nuclear translocation of active GLI transcription family members and up‐regulation of target genes including GLI1 and PTCH1.
Given the limited evidence of activating mutations of the Hh pathway, the role of activated Hh signaling in other solid tumors had been poorly characterized. Watkins et al. reported that ∼25% of small cell lung cancer (SCLC) samples displayed high expression of Sonic Hh ligand and GLI1. Inhibition of Hh signaling with cyclopamine, a natural product SMOH inhibitor derived from corn lilies, or an anti‐Hh antibody blocked growth of SCLC cell lines in vitro. Furthermore, cyclopamine prevented growth of SCLC in immunocompromised mice (Watkins et al., 2003). Hh pathway activation has also been demonstrated in pancreatic cancer. Thayer et al. found altered Sonic Hh, PTCH1, and SMOH expression in the epithelial and stromal compartments of human tumors suggesting potential autocrine and paracrine signaling (Thayer et al., 2003). Treatment with cyclopamine prevented growth of pancreatic cell lines in vitro and, more importantly, in vivo.
Given the role of Hh signaling in regulating cell proliferation and differentiation during development, it is not surprising Hh also regulates normal and malignant stem cells in both Drosophila and mammalian systems (Jiang and Hui, 2008). Hh pathway activation in mouse primitive hematopoietic cells induced cycling and expansion, but eventual exhaustion of hematopoietic stem cells (Trowbridge et al., 2006). A role for Hh signaling in CSC was demonstrated by specific deletion of Smoothened in BCR‐ABL positive chronic myeloid leukemia stem cells, which prevented tumor‐initiation, and treatment with cyclopamine increased survival of mice transplanted with BCR‐ABL leukemia cells (Dierks et al., 2008).
Hh signaling also has been shown to play an important role in normal and malignant breast stem cells. Liu et al. showed Indian Hh, PTHC1, SMOH, GLI1, and GLI2 are expressed in stem and progenitor cells when cultured as mammospheres. Expression of the Hh pathway components was substantially reduced when these cells underwent differentiation (Liu et al., 2006). Exogenous Hh ligand increased mammosphere formation whereas cyclopamine treatment inhibited mammosphere formation. Hh signaling induced BMI1 expression and reduction of BMI1 led to decreased mammosphere formation. Furthermore, PTCH1, GLI1, GLI2, and BMI1 were expressed at higher levels in CD44+CD24− breast CSC compared to bulk cancer cells indicating Hh signaling has a role in normal and malignant breast stem cells.
7.4. Another notch in CSC defenses?
Notch signaling is activated by binding of Notch receptors (Notch 1–4) with ligands (Delta, Delta‐like, and Jagged1 and Jagged2) on adjacent cells. Binding initiates a cascade of proteolytic cleavages mediated by members of the ADAM metalloprotease family as well as internal cleavage by γ‐secretase. Therapeutic inhibition of the Notch pathway can be achieved by γ‐secretase inhibitors (GSI), which are currently in clinical trials for Alzheimer's disease, T‐cell acute lymphoblastic leukemia (T‐ALL), and breast cancer. Cleavage results in the translocation of the Notch intracellular domain (NICD) to the nucleus where it cooperates with the CSL DNA‐binding protein CBF1/RBPkJ forming a trimeric complex with CBF1/RBPkJ and the co‐activator Mastermind. This induces expression of Notch target genes including Hes, Hey, c‐Myc, cyclin D1, and p21/Waf1. NICD activity is regulated by CDK8‐dependent phosphorylation thereby resulting in ubiquitination of NICD. In the absence of NICD, co‐repressors, such as CtBP are recruited. Additionally, Notch activity is inhibited by Numb through an endocytotic mechanism. Interestingly, Numb is asymmetrically segregated into one of two daughter cells in numerous cell types, and is inhibited by Musashi at the level of Numb mRNA translation (Bray, 2006).
A role for Notch signaling in cancer was demonstrated by the identification of activating Notch1 mutations in more than 50% of T‐ALL (Weng et al., 2004) and retroviral insertions into Notch4 in mouse mammary cancers (Jhappan et al., 1992). Analysis of normal human breast stem cells showed activation of Notch signaling using soluble Delta ligand promoted stem cell self‐renewal and differentiation of progenitor cells with negligible effects on mature differentiated epithelial cells (Dontu et al., 2004). Additionally, it was shown that Musashi1 and Notch1 regulate human breast stem cells (Clarke et al., 2003, 2005). In the mouse mammary gland, Notch signaling was shown to regulate the expansion of stem cells and differentiation to luminal progenitor cells (Bouras et al., 2008). Recently, Harrison and colleagues showed Notch4 activity was increased in breast CSC, and that inhibition of Notch4 signaling reduced breast CSCs and completely inhibited tumor‐initiation (Harrison et al., 2010). Interestingly, Notch1 activity was lower in breast CSC compared to more differentiated progenitor cells. This suggests that there is specificity of different Notch receptors in the regulation of breast stem and progenitor cells. If this is the case, then selective inhibition of Notch4 may be more efficacious and potentially less toxic than Notch1 inhibitors or γ‐secretase inhibitors that inhibit all Notch receptors. GSI are currently being evaluated in early phase clinical trials for advanced breast cancer.
7.5. Cytokine signaling and the tumor microenvironment
The view of solid tumors as a homogenous sheet of epithelial cells, such as in vitro culture, is clearly too simplistic. Rather, tumors are composed of epithelial cells, fibroblasts, endothelial, hematopoietic, and other cells that communicate with each in a complex network of growth factors and cytokines and the cognate receptors. The ability to interfere with this network represents a growing interest in drug discovery. Indeed, utilization of monoclonal antibodies (i.e. CD44, CD47, CD123) to inhibit signaling between leukemia stem cells (LSC) and supporting cells has been very effective at eradicating AML LSC in NOD/SCID mice (Jin et al., 2006, 2009, 2009).
Several cytokines have been implicated in regulating breast CSCs, including IL‐6 (Sansone et al., 2007) and IL‐8 and the cognate CXCR1 receptor. CXCR1 expression is higher in ALDH+ cells from numerous breast cancer cells lines and exogenous IL‐8 increased the CSC population (Charafe‐Jauffret et al., 2009). This suggested that inhibition of IL‐8/CXCR1 signaling could target breast CSC. Ginestier and colleagues showed that <2% of breast cancer cells expressed CXCR1, which overlapped with the ALDH+ fraction (Ginestier et al., 2010). Interestingly, repertaxin, a small molecule inhibitor of CXCR1, was able to decrease viability of the whole population even though only a small fraction expressed CXCR1. Repertaxin mediated cell killing in the bulk cell population through a bystander effect via secretion of FAS ligand. CXCR1 is known to signal through AKT thereby inhibiting FOXO3A localization, and FAS ligand expression. Repertaxin treatment inhibited AKT signaling resulting in nuclear FOXO3A and FAS ligand expression, but, interestingly, was ineffective in PTEN knockdown cells. Chemotherapy is known to kill cells via a bystander effect through FAS ligand, but also induces IL‐8 and thereby protects CSC from FAS ligand. This suggested that repertaxin might block this effect and target the CSC population. Repertaxin reduced the CSC population in vitro and in tumor xenografts. As a single agent, repertaxin had marginal effect on tumor growth, but significantly reduced tumor volume in combination with the chemotherapy drug docetaxel. In addition, repertaxin reduced metastatic burden and secondary tumor formation. Taken together, these results suggest that repertaxin sensitizes CSC to the bystander effect via FAS ligand and that CXCR1 blockade may represent a new approach to targeting and eliminating breast CSCs.
7.6. Nothing “micro” about microRNA in CSC signaling
In the 10 years since RNA interference (RNAi) was first described by Fire et al. (1998), the field of small RNAs has virtually exploded. While small‐interfering RNA (siRNA) has been extremely useful as a laboratory tool for silencing target gene expression (Jackson and Linsley, 2010), the class of microRNAs (miRNAs) has garnered considerable interest based on the wide range of possible targets (∼300) for each miRNAs (Bartel, 2009). As of December 2009, over 700 human miRNAs have been confirmed or predicted within the mirBase database (Griffiths‐Jones et al., 2008). miRNAs are endogenous non‐coding ∼20–23nt RNAs processed from larger hairpin structures that bind to complementary sequences in the 3′ untranslated regions (UTR) of mRNAs. Processed miRNAs are loaded into the Argonaute protein of the silencing complex (RISC) and pair with mRNAs. Binding is mediated primarily through a seed sequence of 7–8nt at the 5′ region of the miRNA via Watson–Crick pairing. Binding via the seed sequence leads to translational repression and/or mRNA destabilization, whereas extensive complementarity, such as through siRNA, can direct Argonaute‐catalyzed mRNA cleavage (Bartel, 2009).
The wide range of possible targets for each miRNA provides a simple mechanism for a stem cell to coordinately regulate a set of “stemness” genes. Deletion of miRNA processing proteins, such as DGCR8 or Dicer, has been shown to alter the G1‐S transition and proliferation of embryonic stem cells (ESC) (Murchison et al., 2005; Wang et al., 2008). More than 100 miRNAs are differentially expressed between human ESC and differentiated embryoid bodies (Morin et al., 2008). Indeed, miR‐145 is absent in self‐renewing ESCs and targets the pluripotency factors Sox2, Oct4, and KLf4 (Xu et al., 2009). These studies highlight the importance of miRNAs in regulating “stemness”.
A role for miRNAs in CSC has recently been shown for the let‐7 and miR‐200 families (Shimono et al., 2009; Wellner et al., 2009; Yu et al., 2007). Yu and colleagues compared miRNA expression in primary breast cancer cell lines, which were enriched for mammosphere formation and tumor‐initiation, to more differentiated cancer cells. Let‐7 family members were consistently reduced in the mammosphere cells. Forced expression of let‐7a reduced mammosphere formation in primary breast cancer and cell lines whereas reduction of let‐7a expression in differentiated cells increased mammosphere formation. Importantly, forced let‐7a expression suppressed tumor‐initiation and growth in NOD/SCID mice and in subsequent passages indicating let‐7a maintains breast CSCs.
Given the large number of miRNAs present in ESCs compared to differentiated cells (Morin et al., 2008), it is not surprising that additional miRNAs are differentially expressed in breast CSC. Shimono and colleagues identified 37 miRNAs up regulated or down regulated in CD44+CD24− breast CSC compared to bulk, non‐tumorigenic cells (Shimono et al., 2009). Three clusters of miRNAs were down regulated in the breast CSC, including miR‐200c‐141, miR‐200b‐200a‐429, and miRNA‐183‐96‐182. Similar seed sequences are shared between the miR‐200c‐141 and miR‐200b‐200a‐429 clusters indicating similar genes are targeted by these miRNAs. These miRNA clusters were also decreased in normal breast stem cells suggesting a link between normal and malignant breast stem cells. Among the numerous targets, Shimono et al. concentrated on BMI1 since it is known to regulate normal and CSCs (Lessard and Sauvageau, 2003; Molofsky et al., 2003; Park et al., 2003). Exogenous miR‐200c reduced BMI1 protein levels and suppressed growth of embryonal carcinoma cells. Expression of miR‐200c in normal mouse breast stem cells suppressed outgrowth formation in the mammary fat pad assay. Moreover, miR‐200c expression in human CD44+CD24− breast CSC also suppressed tumor‐initiation in NOD/SCID mice suggesting similar to let‐7 that miR‐200c also regulates breast CSC.
7.7. “Transitioning” from epithelial–mesenchymal cells
The epithelial–mesenchymal transition (EMT) is a developmental program typically activated in metastatic cancer. During the metastasis process, it is postulated cancer cells acquire some self‐renewal capability to give rise to macroscopic metastases. Mani and colleagues showed forced expression of Snail or Twist transcription factors, which induce EMT, increased the CD44+CD24− fraction in immortalized human breast epithelial cells suggesting a relationship between EMT and the CSC phenotype (Mani et al., 2008). Cells that had undergone EMT had increased mammosphere‐forming capacity and tumor‐initiation potential in NOD/SCID mice. The miR‐200 family is known to prevent EMT by suppressing ZEB1 and ZEB2 expression, which are transcriptional repressors of E‐cadherin (Gregory et al., 2008; Park et al., 2008; Wellner et al., 2009). ZEB1 is also known to inhibit expression of miR‐200 family members creating a double‐negative feedback loop (Bracken et al., 2008; Peter, 2010). Recently, Greene et al. showed miR‐205 is highly expressed in mouse mammary epithelial progenitors and targets PTEN as well as ZEB1 and ZEB2 (Greene et al., 2010).
This raises interesting questions about what prevents miR‐200 expression in stem cells. Wellner and colleagues demonstrated ZEB1 is expressed in the invasive front of pancreatic cancer samples (Wellner et al., 2009). Down‐regulation of ZEB1 reduced invasion and metastasis in NOD/SCID mice and inhibited tumor‐initiation and secondary tumor formation. Furthermore, knockdown of ZEB1 increased expression of miR‐203 and miR‐183, members of the miR‐200 family, and forced expression of miR‐203 and miR‐183 reduced pancreatic sphere formation. Examination of the BMI1 3′ UTR revealed binding sites for miR‐200, miR‐203, and miR‐183. It was also demonstrated that miR‐200 also repressed the pluripotency factors Sox2 and Klf4. Therefore, ZEB1 and the miR‐200 family are at the center of a complex network of miRNAs and stem cell regulators in multiple cancers, including breast cancer.
7.8. High‐throughput screening to identify novel targets and compounds
Until recently, most drug development targeting CSC was based on a priori understanding of pathways utilized by these cells. From a drug discovery standpoint, this strategy is attractive given the high costs (estimates range from $800 million to $2 billion), high attrition rate, and difficulties associated with moving a compound from target discovery to FDA approval (Booth and Zemmel, 2004; Munos, 2009).
Identification of the molecular target(s) for lead compounds has been extremely time consuming and difficult, but recent advances point to a future where target identification may become more routine. Chemical proteomics, a spectrometry‐based affinity chromatography approach, holds promise for directly identifying the target protein(s) bound by small molecules, further advancing drug development via new target proteins for subsequent discovery efforts (Rix and Superti‐Furga, 2009). Computational approaches provide a mechanism to predict molecular targets and potential side‐effects (Campillos et al., 2008; Keiser et al., 2009). Furthermore, development of databases of drugs (MDL Drug Data Report), known drug targets (DrugBank, Matador, SuperTarget), and small molecule gene expression signature profiles (i.e. Connectivity Map) can provide new insights into possible mechanisms of action for unknown compounds (Gunther et al., 2008; Lamb et al., 2006; Wei et al., 2006; Wishart et al., 2008). The promise of these approaches was demonstrated by in silico data mining the publicly available microarray datasets with a query signature from AML cells treated with parthenolide that identified new potential anti‐LSC agents, which were then validated in pre‐clinical models (Hassane et al., 2008).
The combination of high‐throughput RNAi screens with small molecule screens is another avenue to identify the molecular target(s) of lead compounds (Iorns et al., 2007; Perrimon et al., 2007; Whitehurst et al., 2007). Identification of new proteins and pathways in CSCs from RNAi screens also presents new opportunities for drug development, both in terms of using RNAi as a therapeutic and developing new targets for small molecule or antibody‐based screening. This approach was recently demonstrated by the screening of the human kinome in primary glioblastoma cells enriched for tumor‐initiation. Schultz and colleagues identified TRRAP, an adaptor protein with homology to phosphoinositide‐3‐OH‐kinase‐related (PIKK) proteins, as critical to self‐renewal and proliferation of brain CSC through effects on cell cycle progression and histone marks of cyclin A2. Importantly, silencing of TRRAP suppressed tumorigenicity of primary xenografts (Wurdak et al., 2010).
The applicability of these screens is not limited to RNAi, but has also been applied to small molecule libraries (Pollard et al., 2009). Dirks and colleagues utilized in vitro conditions that are able to sustain brain tumor‐initiating cells. A screen of 450 compounds that have passed phase I–III trials identified the serotonin and monoamine signaling pathways as potential targets for modulation in brain CSCs. These results mirror previous results showing sensitivity of mouse neurospheres, enriched for stem cell activity, to neurotransmitter pathway perturbation (Diamandis et al., 2007).
Application of these high‐throughput approaches and concepts to breast cancer was recently demonstrated and identified a previously unrecognized class of compounds with efficacy against breast CSCs (Gupta et al., 2009). Gupta and colleagues harnessed the interesting observation that cells undergoing EMT were enriched for and had characteristics of CSC (Mani et al., 2008). They used a growth inhibition screen with breast cells engineered to maintain an EMT state and the immortalized normal, parental cells. From a collection of ∼16,000 small molecules, only 32 targeted the EMT cells and not the parental cells, and 4 (salinomycin, etoposide, abamectin, and nigericin) were confirmed in follow‐up studies. Salinomycin, a potassium ionophore, showed the greatest effect in vitro by reducing mammosphere formation as well as the CD44+CD24‐ fraction, shown previously to include breast CSC (Al‐Hajj et al., 2003). Pre‐treatment of breast cancer cells in vitro with salinomycin dramatically reduced tumor‐initiation and lung metastases in NOD/SCID mice indicating salinomycin was able to target breast CSC. While the mechanism of action for salinomycin in CSC has not been established, it was shown recently this compound induced apoptosis in cancer cells regardless of p53, Bcl2, or multi‐drug resistance protein status and independent of cell cycle arrest, caspase activation, FAS/FAS ligand, and the proteasome (Fuchs et al., 2009). The success of these screens highlight the promise of identifying novel compounds with anti‐CSC activity as well as novel proteins and pathways functioning in CSC.
8. The future of targeting breast CSC
Currently, there are multiple potential anti‐CSC agents in pre‐clinical and clinical trials, including Hh, NOTCH, AKT, and CXCR1 inhibitors. While these are promising agents, the likelihood of clinical success will depend on many aspects, including safety, trial design, and rational endpoints. High‐throughput screens represent new avenues for drug discovery and identification of novel pathways regulating CSCs. Breast CSCs screens may be based on methods to enrich for CSCs, such as CD44+CD24−, ALDH+, or sphere formation from cell lines, primary breast cancer xenografts, or primary samples (Figure. 1A). These CSC‐enriched populations can be used to screen siRNA, lentiviral shRNA, or small molecule libraries using various cellular endpoints including growth inhibition (Gupta et al., 2009), flow cytometry (Krutzik and Nolan, 2006), sphere formation (Diamandis et al., 2007), or cell migration (Wurdak et al., 2010) (Figure. 1B).
Figure 1.
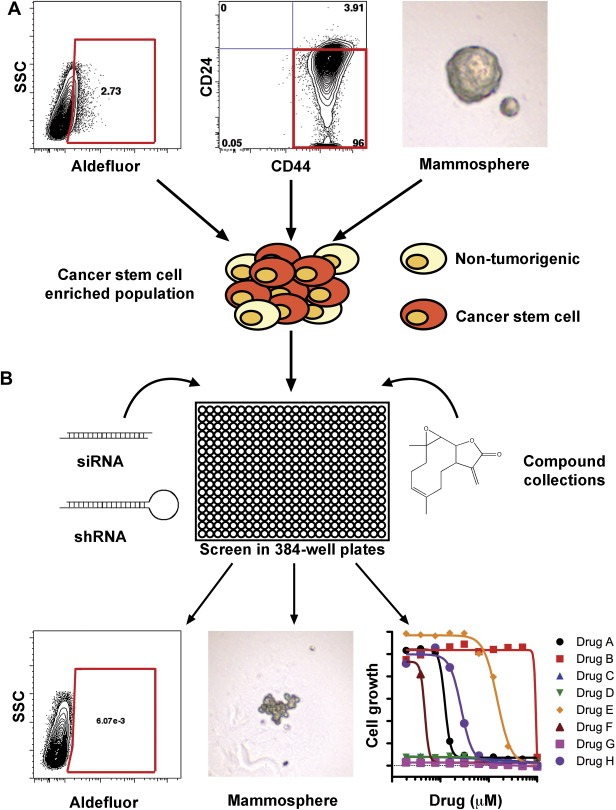
High‐throughput approaches to target breast CSC. A) Breast CSC can be isolated by sorting using the Aldefluor assay or CD44+CD24− populations (red box) or via culturing as mammospheres. Within these populations, the CSC frequency is enriched with some percentage of non‐tumorigenic bulk cells remaining. B) To screen the CSC population, cells are plated in 384‐well plates in conditions that maintain tumor‐initiation capacity, such as in serum‐free suspension (mammosphere) or in EGF‐ and FGF‐containing media on laminin‐coated plates (Pollard et al., 2009). Small molecule compound collections are added to plates at one compound/well, typically at high nanomolar or low micromolar concentrations. Additionally, the whole genome or selected gene sets (kinases, phosphatases, ‘druggable’) can be targeted using either siRNA or retroviral or lentiviral shRNA libraries with usually 4–5 constructs per gene. shRNA libraries can provide long‐term knockdown whereas siRNA knockdown is more transient. Cellular endpoints can include growth inhibition (Gupta et al., 2009), flow cytometry (Krutzik and Nolan, 2006), sphere formation (Diamandis et al., 2007), or cell migration (Wurdak et al., 2010) (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
High‐throughput screens provide a larger pool of compounds for testing in pre‐clinical and clinical models given the high attrition rate and decreasing productivity within the pharmaceutical industry (Booth and Zemmel, 2004; Munos, 2009). While some side‐effects from chemotherapy agents are poorly understood, it is increasing clear that others are due to toxic effects on normal stem cells in various tissues, such as hematopoietic, skin and gastrointestinal (see ‘The realities of targeting CSCs’ above). Thus, agents identified in primary high‐throughput screens need to be tested on normal stem cells to remove potentially cytotoxic agents with adverse side‐effects (Figure. 2A). These “confirmed” agents can then be tested in primary tumor xenograft mouse models, However, current models, based primarily on tumor growth from rapidly dividing progenitor cells, are poorly adept to identify anti‐CSC agents, which may have modest growth inhibition effects as a sole agent (Figure. 2B). Rather, testing agents in an early (i.e. adjuvant) setting would identify those agents with anti‐CSC activity since early tumor formation is stem cell dependent.
Figure 2.
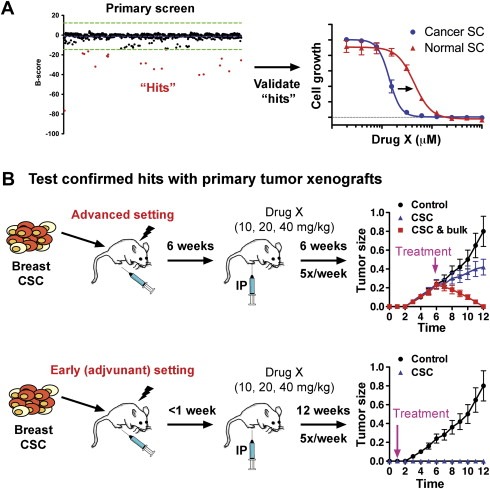
Validation of potential “hits” as anti‐CSC agents. A) Traditionally, “hits” are identified using a Z‐ or B‐score for each compound/RNA and at least 3 standard deviations (green line) from the mean (blue line). Hits (red dots) are “cherry‐picked” for validation studies (right) at additional doses to show specificity for CSC (blue circles) and not normal stem cells (red triangles). B) To confirm validated “hits” target CSCs, activity must be validated using primary tumor xenografts in immunocompromised mice. In the advanced setting (top), cells enriched for breast CSC are injected into mice, allowed to grow to a palpable size and then mice are treated with candidate drugs or siRNA/shRNA. A CSC‐specific agent (blue triangles) would have minimal effect in this assay since tumor growth is driven primarily by progenitors and not CSC. However, an agent that targets CSC and the bulk tumor cells (red squares) would show dramatic tumor reduction. This effect could be achieved using a CSC‐specific agent and a chemotherapeutic agent targeting the bulk population. In an early (i.e. adjuvant) setting (bottom), treatment is initiated soon after injection of CSC‐enriched cells. A selective anti‐CSC agent (blue triangles) would be predicted to have a much greater effect when administered in this setting (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
New therapies might come from traditional or existing medicines that were not tested for anti‐cancer efficacy and can be rapidly “repurposed” (Chen et al., 2008; Chong and Sullivan, 2007; Eberhard et al., 2009; Ginestier et al., 2010). The CXRC1 inhibitor repertaxin was developed to block organ transplant rejection, the gamma‐secretase inhibitors initially developed to treat Alzheimer's disease, and anti‐fungal ciclopirox olamine recently shown to target AML LSCs are examples of “repurposing” existing medicines.
Many proteins are considered “undruggable” based on cellular location, binding, and function (i.e. transcription factors) (Overington et al., 2006). The promise of RNAi is great considering the ability to target these “undruggable” proteins. However, RNAi has limitations, including off‐target effects, immune system modulation, and issues related to in vivo delivery (Jackson and Linsley, 2010; Whitehead et al., 2009). Systemic delivery, via intravenous injection, is likely required for treatment of disseminated disease and requires the ability to avoid non‐target tissues and efficient delivery to CSCs. The ability to bypass kidney filtration, phagocytosis, aggregation in serum, and enzymatic degradation is a key to successful RNAi therapy with numerous groups and companies actively trying to solve these issues. Indeed, RNAi therapeutics are currently in clinical trials for various diseases, such as age‐related macular degeneration, but most current trials rely on localized delivery instead of using delivery agents (Whitehead et al., 2009).
Much progress has been made in targeting CSC since the “original” discovery in AML (Lapidot et al., 1994) and the identification of parthenolide as an anti‐LSC agent (Guzman et al., 2005). Recent advances in understanding the pathways utilized by CSC, such as Hh, NOTCH, and Wnt, have led to exciting pre‐clinical and phase I clinical trials to test the clinical relevance of CSC. In addition, improved in vitro culture methods to maintain CSC activity from primary tumor samples open the arena of high‐throughput screening of small molecule and siRNA libraries. Initial results from these screens have provided new clues and leads in the fight against cancer. Taken together, these studies provide novel agents to target this critical cell population. It is hoped that successful targeting of CSC will significantly improve the outcome for patients with cancer.
McDermott Sean P., Wicha Max S., (2010), Targeting breast cancer stem cells, Molecular Oncology, 4, doi: 10.1016/j.molonc.2010.06.005.
References
- Abratt, R.P. , Brune, D. , Dimopoulos, M.A. , Kliment, J. , Breza, J. , Selvaggi, F.P. , Beuzeboc, P. , Demkow, T. , Oudard, S. , 2004. Randomised phase III study of intravenous vinorelbine plus hormone therapy versus hormone therapy alone in hormone-refractory prostate cancer. Ann. Oncol. 15, 1613–1621. [DOI] [PubMed] [Google Scholar]
- Agis, H. , Jaeger, E. , Doninger, B. , Sillaber, C. , Marosi, C. , Drach, J. , Schwarzinger, I. , Valent, P. , Oehler, L. , 2006. In vivo effects of imatinib mesylate on human haematopoietic progenitor cells. Eur. J. Clin. Invest. 36, 402–408. [DOI] [PubMed] [Google Scholar]
- Al-Hajj, M. , Wicha, M.S. , Benito-Hernandez, A. , Morrison, S.J. , Clarke, M.F. , 2003. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 100, 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, S. , Wu, Q. , McLendon, R.E. , Hao, Y. , Shi, Q. , Hjelmeland, A.B. , Dewhirst, M.W. , Bigner, D.D. , Rich, J.N. , 2006. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 444, 756–760. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. , 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner, W.A. , Hulett, H.R. , Sweet, R.G. , Herzenberg, L.A. , 1972. Fluorescence activated cell sorting. Rev. Sci. Instrum. 43, 404–409. [DOI] [PubMed] [Google Scholar]
- Bonnet, D. , Dick, J.E. , 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737. [DOI] [PubMed] [Google Scholar]
- Booth, B. , Zemmel, R. , 2004. Prospects for productivity. Nat. Rev. Drug Discov. 3, 451–456. [DOI] [PubMed] [Google Scholar]
- Bouras, T. , Pal, B. , Vaillant, F. , Harburg, G. , Asselin-Labat, M.L. , Oakes, S.R. , Lindeman, G.J. , Visvader, J.E. , 2008. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 3, 429–441. [DOI] [PubMed] [Google Scholar]
- Brabletz, T. , Jung, A. , Spaderna, S. , Hlubek, F. , Kirchner, T. , 2005. Opinion: migrating cancer stem cells – an integrated concept of malignant tumour progression. Nat. Rev. Cancer. 5, 744–749. [DOI] [PubMed] [Google Scholar]
- Bracken, C.P. , Gregory, P.A. , Kolesnikoff, N. , Bert, A.G. , Wang, J. , Shannon, M.F. , Goodall, G.J. , 2008. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial–mesenchymal transition. Cancer Res. 68, 7846–7854. [DOI] [PubMed] [Google Scholar]
- Bray, S.J. , 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689. [DOI] [PubMed] [Google Scholar]
- Campillos, M. , Kuhn, M. , Gavin, A.C. , Jensen, L.J. , Bork, P. , 2008. Drug target identification using side-effect similarity. Science. 321, 263–266. [DOI] [PubMed] [Google Scholar]
- Charafe-Jauffret, E. , Ginestier, C. , Iovino, F. , Wicinski, J. , Cervera, N. , Finetti, P. , Hur, M.H. , Diebel, M.E. , Monville, F. , Dutcher, J. , Brown, M. , Viens, P. , Xerri, L. , Bertucci, F. , Stassi, G. , Dontu, G. , Birnbaum, D. , Wicha, M.S. , 2009. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 69, 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S.T. , Dou, J. , Temple, R. , Agarwal, R. , Wu, K.M. , Walker, S. , 2008. New therapies from old medicines. Nat. Biotechnol. 26, 1077–1083. [DOI] [PubMed] [Google Scholar]
- Cho, R.W. , Wang, X. , Diehn, M. , Shedden, K. , Chen, G.Y. , Sherlock, G. , Gurney, A. , Lewicki, J. , Clarke, M.F. , 2008. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 26, 364–371. [DOI] [PubMed] [Google Scholar]
- Chong, C.R. , Sullivan, D.J. , 2007. New uses for old drugs. Nature. 448, 645–646. [DOI] [PubMed] [Google Scholar]
- Chung, N. , Zhang, X.D. , Kreamer, A. , Locco, L. , Kuan, P.F. , Bartz, S. , Linsley, P.S. , Ferrer, M. , Strulovici, B. , 2008. Median absolute deviation to improve hit selection for genome-scale RNAi screens. J. Biomol. Screen. 13, 149–158. [DOI] [PubMed] [Google Scholar]
- Cicalese, A. , Bonizzi, G. , Pasi, C.E. , Faretta, M. , Ronzoni, S. , Giulini, B. , Brisken, C. , Minucci, S. , Di Fiore, P.P. , Pelicci, P.G. , 2009. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 138, 1083–1095. [DOI] [PubMed] [Google Scholar]
- Clarke, M.F. , Dick, J.E. , Dirks, P.B. , Eaves, C.J. , Jamieson, C.H. , Jones, D.L. , Visvader, J. , Weissman, I.L. , Wahl, G.M. , 2006. Cancer stem cells–perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 66, 9339–9344. [DOI] [PubMed] [Google Scholar]
- Clarke, R.B. , Anderson, E. , Howell, A. , Potten, C.S. , 2003. Regulation of human breast epithelial stem cells. Cell Prolif. 36, (Suppl. 1) 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, R.B. , Spence, K. , Anderson, E. , Howell, A. , Okano, H. , Potten, C.S. , 2005. A putative human breast stem cell population is enriched for steroid receptor-positive cells. Dev. Biol. 277, 443–456. [DOI] [PubMed] [Google Scholar]
- Clarkson, B. , Fried, J. , Strife, A. , Sakai, Y. , Ota, K. , Okita, T. , 1970. Studies of cellular proliferation in human leukemia. 3. Behavior of leukemic cells in three adults with acute leukemia given continuous infusions of 3H-thymidine for 8 or 10 days. Cancer. 25, 1237–1260. [DOI] [PubMed] [Google Scholar]
- Clarkson, B.D. , 1969. Review of recent studies of cellular proliferation in acute leukemia. Natl. Cancer Inst. Monogr. 30, 81–120. [PubMed] [Google Scholar]
- Clarkson, B.D. , Fried, J. , 1971. Changing concepts of treatment in acute leukemia. Med. Clin. North Am. 55, 561–600. [DOI] [PubMed] [Google Scholar]
- Cozzio, A. , Passegue, E. , Ayton, P.M. , Karsunky, H. , Cleary, M.L. , Weissman, I.L. , 2003. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 17, 3029–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton, C.J. , Li, X. , Landis, M. , Dixon, J.M. , Neumeister, V.M. , Sjolund, A. , Rimm, D.L. , Wong, H. , Rodriguez, A. , Herschkowitz, J.I. , Fan, C. , Zhang, X. , He, X. , Pavlick, A. , Gutierrez, M.C. , Renshaw, L. , Larionov, A.A. , Faratian, D. , Hilsenbeck, S.G. , Perou, C.M. , Lewis, M.T. , Rosen, J.M. , Chang, J.C. , 2009. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl. Acad. Sci. USA. 106, 13820–13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, C.W. , De Ome, K.B. , Young, J.T. , Blair, P.B. , Faulkin, L.J. , 1968. The in vivo life span of normal and preneoplastic mouse mammary glands: a serial transplantation study. Proc. Natl. Acad. Sci. USA. 61, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, C.W. , Young, L.J. , 1971. Influence of cell division on an aging process. Life span of mouse mammary epithelium during serial propagation in vivo. Exp. Cell Res. 65, 27–32. [DOI] [PubMed] [Google Scholar]
- Daniel, D. , Crawford, J. , 2006. Myelotoxicity from chemotherapy. Semin. Oncol. 33, 74–85. [DOI] [PubMed] [Google Scholar]
- Dekaney, C.M. , Gulati, A.S. , Garrison, A.P. , Helmrath, M.A. , Henning, S.J. , 2009. Regeneration of intestinal stem/progenitor cells following doxorubicin treatment of mice. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G461–G470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, G. , Lu, Y. , Zlotnikov, G. , Thor, A.D. , Smith, H.S. , 1996. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 274, 2057–2059. [DOI] [PubMed] [Google Scholar]
- DeOme, K.B. , Fauklin, L.J. , Bern, H.A. , Blair, P.B. , 1959. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H. J. Natl. Cancer Inst. 78, 751–757. [PubMed] [Google Scholar]
- Diamandis, P. , Wildenhain, J. , Clarke, I.D. , Sacher, A.G. , Graham, J. , Bellows, D.S. , Ling, E.K. , Ward, R.J. , Jamieson, L.G. , Tyers, M. , Dirks, P.B. , 2007. Chemical genetics reveals a complex functional ground state of neural stem cells. Nat. Chem. Biol. 3, 268–273. [DOI] [PubMed] [Google Scholar]
- Dick, J.E. , 2003. Stem cells: self-renewal writ in blood. Nature. 423, 231–233. [DOI] [PubMed] [Google Scholar]
- Dick, J.E. , 2008. Stem cell concepts renew cancer research. Blood. 112, 4793–4807. [DOI] [PubMed] [Google Scholar]
- Dierks, C. , Beigi, R. , Guo, G.R. , Zirlik, K. , Stegert, M.R. , Manley, P. , Trussell, C. , Schmitt-Graeff, A. , Landwerlin, K. , Veelken, H. , Warmuth, M. , 2008. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 14, 238–249. [DOI] [PubMed] [Google Scholar]
- Dontu, G. , Abdallah, W.M. , Foley, J.M. , Jackson, K.W. , Clarke, M.F. , Kawamura, M.J. , Wicha, M.S. , 2003. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu, G. , Jackson, K.W. , McNicholas, E. , Kawamura, M.J. , Abdallah, W.M. , Wicha, M.S. , 2004. Role of notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 6, R605–R615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard, Y. , McDermott, S.P. , Wang, X. , Gronda, M. , Venugopal, A. , Wood, T.E. , Hurren, R. , Datti, A. , Batey, R.A. , Wrana, J. , Antholine, W.E. , Dick, J. , Schimmer, A.D. , 2009. Chelation of intracellular iron with the antifungal agent ciclopirox olamine induces cell death in leukemia and myeloma cells. Blood. 114, 3064–3073. [DOI] [PubMed] [Google Scholar]
- Eirew, P. , Stingl, J. , Raouf, A. , Turashvili, G. , Aparicio, S. , Emerman, J.T. , Eaves, C.J. , 2008. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat. Med. 14, 1384–1389. [DOI] [PubMed] [Google Scholar]
- Eisenhauer, E.A. , Therasse, P. , Bogaerts, J. , Schwartz, L.H. , Sargent, D. , Ford, R. , Dancey, J. , Arbuck, S. , Gwyther, S. , Mooney, M. , Rubinstein, L. , Shankar, L. , Dodd, L. , Kaplan, R. , Lacombe, D. , Verweij, J. , 2009. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer. 45, 228–247. [DOI] [PubMed] [Google Scholar]
- Eng, C. , 2003. PTEN: one gene, many syndromes. Hum. Mutat. 22, 183–198. [DOI] [PubMed] [Google Scholar]
- Fire, A. , Xu, S. , Montgomery, M.K. , Kostas, S.A. , Driver, S.E. , Mello, C.C. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Fuchs, D. , Heinold, A. , Opelz, G. , Daniel, V. , Naujokat, C. , 2009. Salinomycin induces apoptosis and overcomes apoptosis resistance in human cancer cells. Biochem. Biophys. Res. Commun. 390, 743–749. [DOI] [PubMed] [Google Scholar]
- Furth, J. , Kahn, M. , 1937. The transmission of leukemia of mice with a single cell. Am. J. Cancer. 31, 276–282. [Google Scholar]
- Ginestier, C. , Hur, M.H. , Charafe-Jauffret, E. , Monville, F. , Dutcher, J. , Brown, M. , Jacquemier, J. , Viens, P. , Kleer, C.G. , Liu, S. , Schott, A. , Hayes, D. , Birnbaum, D. , Wicha, M.S. , Dontu, G. , 2007. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 1, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier, C. , Liu, S. , Diebel, M.E. , Korkaya, H. , Luo, M. , Brown, M. , Wicinski, J. , Cabaud, O. , Charafe-Jauffret, E. , Birnbaum, D. , Guan, J.L. , Dontu, G. , Wicha, M.S. , 2010. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J. Clin. Invest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier, C. , Liu, S. , Wicha, M.S. , 2009. Getting to the root of BRCA1-deficient breast cancer. Cell Stem Cell. 5, 229–230. [DOI] [PubMed] [Google Scholar]
- Gonzalez, C. , 2007. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat. Rev. Genet. 8, 462–472. [DOI] [PubMed] [Google Scholar]
- Greene, S.B. , Gunaratne, P.H. , Hammond, S.M. , Rosen, J.M. , 2010. A putative role for microRNA-205 in mammary epithelial cell progenitors. J. Cell Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, P.A. , Bert, A.G. , Paterson, E.L. , Barry, S.C. , Tsykin, A. , Farshid, G. , Vadas, M.A. , Khew-Goodall, Y. , Goodall, G.J. , 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S. , Saini, H.K. , van Dongen, S. , Enright, A.J. , 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther, S. , Kuhn, M. , Dunkel, M. , Campillos, M. , Senger, C. , Petsalaki, E. , Ahmed, J. , Urdiales, E.G. , Gewiess, A. , Jensen, L.J. , Schneider, R. , Skoblo, R. , Russell, R.B. , Bourne, P.E. , Bork, P. , Preissner, R. , 2008. Super target and matador: resources for exploring drug–target relationships. Nucleic Acids Res. 36, D919–D922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W. , Lasky, J.L. , Chang, C.J. , Mosessian, S. , Lewis, X. , Xiao, Y. , Yeh, J.E. , Chen, J.Y. , Iruela-Arispe, M.L. , Varella-Garcia, M. , Wu, H. , 2008. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature. 453, 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, P.B. , Onder, T.T. , Jiang, G. , Tao, K. , Kuperwasser, C. , Weinberg, R.A. , Lander, E.S. , 2009. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 138, 645–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, M.L. , Neering, S.J. , Upchurch, D. , Grimes, B. , Howard, D.S. , Rizzieri, D.A. , Luger, S.M. , Jordan, C.T. , 2001. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood. 98, 2301–2307. [DOI] [PubMed] [Google Scholar]
- Guzman, M.L. , Rossi, R.M. , Karnischky, L. , Li, X. , Peterson, D.R. , Howard, D.S. , Jordan, C.T. , 2005. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood. 105, 4163–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, M.L. , Rossi, R.M. , Neelakantan, S. , Li, X. , Corbett, C.A. , Hassane, D.C. , Becker, M.W. , Bennett, J.M. , Sullivan, E. , Lachowicz, J.L. , Vaughan, A. , Sweeney, C.J. , Matthews, W. , Carroll, M. , Liesveld, J.L. , Crooks, P.A. , Jordan, C.T. , 2007. An orally bioavailable parthenolide analog selectively eradicates acute myelogenous leukemia stem and progenitor cells. Blood. 110, 4427–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, H. , Farnie, G. , Howell, S.J. , Rock, R.E. , Stylianou, S. , Brennan, K.R. , Bundred, N.J. , Clarke, R.B. , 2010. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 70, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassane, D.C. , Guzman, M.L. , Corbett, C. , Li, X. , Abboud, R. , Young, F. , Liesveld, J.L. , Carroll, M. , Jordan, C.T. , 2008. Discovery of agents that eradicate leukemia stem cells using an in silico screen of public gene expression data. Blood. 111, 5654–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz, J.I. , He, X. , Fan, C. , Perou, C.M. , 2008. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 10, R75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz, J.I. , Simin, K. , Weigman, V.J. , Mikaelian, I. , Usary, J. , Hu, Z. , Rasmussen, K.E. , Jones, L.P. , Assefnia, S. , Chandrasekharan, S. , Backlund, M.G. , Yin, Y. , Khramtsov, A.I. , Bastein, R. , Quackenbush, J. , Glazer, R.I. , Brown, P.H. , Green, J.E. , Kopelovich, L. , Furth, P.A. , Palazzo, J.P. , Olopade, O.I. , Bernard, P.S. , Churchill, G.A. , Van Dyke, T. , Perou, C.M. , 2007. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 8, R76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, R. , Wu, H. , 2009. PTEN, stem cells, and cancer stem cells. J. Biol. Chem. 284, 11755–11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein, M. , Sidransky, D. , Vogelstein, B. , Harris, C.C. , 1991. p53 mutations in human cancers. Science. 253, 49–53. [DOI] [PubMed] [Google Scholar]
- Hope, K.J. , Jin, L. , Dick, J.E. , 2004. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat. Immunol. 5, 738–743. [DOI] [PubMed] [Google Scholar]
- Huff, C.A. , Matsui, W. , Smith, B.D. , Jones, R.J. , 2006. The paradox of response and survival in cancer therapeutics. Blood. 107, 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntly, B.J.P. , Shigematsu, H. , Deguchi, K. , Lee, B.H. , Mizuno, S. , Duclos, N. , Rowan, R. , Amaral, S. , Curley, D. , Williams, I.R. , 2004. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 6, 587–596. [DOI] [PubMed] [Google Scholar]
- Ijiri, K. , Potten, C.S. , 1983. Response of intestinal cells of differing topographical and hierarchical status to ten cytotoxic drugs and five sources of radiation. Br. J. Cancer. 47, 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijiri, K. , Potten, C.S. , 1987. Further studies on the response of intestinal crypt cells of different hierarchical status to eighteen different cytotoxic agents. Br. J. Cancer. 55, 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorns, E. , Lord, C.J. , Turner, N. , Ashworth, A. , 2007. Utilizing RNA interference to enhance cancer drug discovery. Nat. Rev. Drug Discov. 6, 556–568. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L. , Linsley, P.S. , 2010. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 9, 57–67. [DOI] [PubMed] [Google Scholar]
- Jeselsohn, R., Brown, N.E., Arendt, L., Klebba, I., Hu, M.G., Kuperwasser, C., Hinds, P.W., 2010. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis 17, 65–76. [DOI] [PMC free article] [PubMed]
- Jhappan, C. , Gallahan, D. , Stahle, C. , Chu, E. , Smith, G.H. , Merlino, G. , Callahan, R. , 1992. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 6, 345–355. [DOI] [PubMed] [Google Scholar]
- Jiang, J. , Hui, C.C. , 2008. Hedgehog signaling in development and cancer. Dev. Cell. 15, 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L. , Hope, K.J. , Zhai, Q. , Smadja-Joffe, F. , Dick, J.E. , 2006. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat. Med. 12, 1167–1174. [DOI] [PubMed] [Google Scholar]
- Jin, L. , Lee, E.M. , Ramshaw, H.S. , Busfield, S.J. , Peoppl, A.G. , Wilkinson, L. , Guthridge, M.A. , Thomas, D. , Barry, E.F. , Boyd, A. , Gearing, D.P. , Vairo, G. , Lopez, A.F. , Dick, J.E. , Lock, R.B. , 2009. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell. 5, 31–42. [DOI] [PubMed] [Google Scholar]
- Keiser, M.J. , Setola, V. , Irwin, J.J. , Laggner, C. , Abbas, A.I. , Hufeisen, S.J. , Jensen, N.H. , Kuijer, M.B. , Matos, R.C. , Tran, T.B. , Whaley, R. , Glennon, R.A. , Hert, J. , Thomas, K.L. , Edwards, D.D. , Shoichet, B.K. , Roth, B.L. , 2009. Predicting new molecular targets for known drugs. Nature. 462, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, P.N. , Dakic, A. , Adams, J.M. , Nutt, S.L. , Strasser, A. , 2007. Tumor growth need not be driven by rare cancer stem cells. Science. 317, 337 [DOI] [PubMed] [Google Scholar]
- Keniry, M. , Parsons, R. , 2008. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 27, 5477–5485. [DOI] [PubMed] [Google Scholar]
- Kennedy, J.A. , Barabe, F. , Poeppl, A.G. , Wang, J.C. , Dick, J.E. , 2007. Comment on “Tumor growth need not be driven by rare cancer stem cells”. Science. 318, 1722 author reply 1722 [DOI] [PubMed] [Google Scholar]
- Kordon, E. , Smith, G. , 1998. An entire functional mammary gland may comprise the progeny from a single cell. Development. 125, 1921–1930. [DOI] [PubMed] [Google Scholar]
- Korkaya, H. , Paulson, A. , Charafe-Jauffret, E. , Ginestier, C. , Brown, M. , Dutcher, J. , Clouthier, S.G. , Wicha, M.S. , 2009. Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biol. 7, e1000121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov, A.V. , Twomey, D. , Feng, Z. , Stubbs, M.C. , Wang, Y. , Faber, J. , Levine, J.E. , Wang, J. , Hahn, W.C. , Gilliland, D.G. , Golub, T.R. , Armstrong, S.A. , 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 442, 818–822. [DOI] [PubMed] [Google Scholar]
- Krutzik, P.O. , Nolan, G.P. , 2006. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat. Methods. 3, 361–368. [DOI] [PubMed] [Google Scholar]
- Kuperwasser, C. , Chavarria, T. , Wu, M. , Magrane, G. , Gray, J.W. , Carey, L. , Richardson, A. , Weinberg, R.A. , 2004. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc. Natl. Acad. Sci. USA. 101, 4966–4971. Epub 2004 Mar 4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J. , Crawford, E.D. , Peck, D. , Modell, J.W. , Blat, I.C. , Wrobel, M.J. , Lerner, J. , Brunet, J.P. , Subramanian, A. , Ross, K.N. , Reich, M. , Hieronymus, H. , Wei, G. , Armstrong, S.A. , Haggarty, S.J. , Clemons, P.A. , Wei, R. , Carr, S.A. , Lander, E.S. , Golub, T.R. , 2006. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 313, 1929–1935. [DOI] [PubMed] [Google Scholar]
- Land, C.E. , McGregor, D.H. , 1979. Breast cancer incidence among atomic bomb survivors: implications for radiobiologic risk at low doses. J. Natl. Cancer Inst. 62, 17–21. [PubMed] [Google Scholar]
- Lapidot, T. , Sirard, C. , Vormoor, J. , Murdoch, B. , Hoang, T. , Caceres-Cortes, J. , Minden, M. , Paterson, B. , Caligiuri, M.A. , Dick, J.E. , 1994. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 367, 645–648. [DOI] [PubMed] [Google Scholar]
- Lessard, J. , Sauvageau, G. , 2003. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 423, 255–260. [DOI] [PubMed] [Google Scholar]
- Li, C. , Heidt, D.G. , Dalerba, P. , Burant, C.F. , Zhang, L. , Adsay, V. , Wicha, M. , Clarke, M.F. , Simeone, D.M. , 2007. Identification of pancreatic cancer stem cells. Cancer Res. 67, 1030–1037. [DOI] [PubMed] [Google Scholar]
- Li, G. , Robinson, G.W. , Lesche, R. , Martinez-Diaz, H. , Jiang, Z. , Rozengurt, N. , Wagner, K.U. , Wu, D.C. , Lane, T.F. , Liu, X. , Hennighausen, L. , Wu, H. , 2002. Conditional loss of PTEN leads to precocious development and neoplasia in the mammary gland. Development. 129, 4159–4170. [DOI] [PubMed] [Google Scholar]
- Li, X. , Lewis, M.T. , Huang, J. , Gutierrez, C. , Osborne, C.K. , Wu, M.F. , Hilsenbeck, S.G. , Pavlick, A. , Zhang, X. , Chamness, G.C. , Wong, H. , Rosen, J. , Chang, J.C. , 2008. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst. 100, 672–679. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Welm, B. , Podsypanina, K. , Huang, S. , Chamorro, M. , Zhang, X. , Rowlands, T. , Egeblad, M. , Cowin, P. , Werb, Z. , Tan, L.K. , Rosen, J.M. , Varmus, H.E. , 2003. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc. Natl. Acad. Sci. USA. 100, 15853–15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, E. , Vaillant, F. , Wu, D. , Forrest, N.C. , Pal, B. , Hart, A.H. , Asselin-Labat, M.L. , Gyorki, D.E. , Ward, T. , Partanen, A. , Feleppa, F. , Huschtscha, L.I. , Thorne, H.J. , Fox, S.B. , Yan, M. , French, J.D. , Brown, M.A. , Smyth, G.K. , Visvader, J.E. , Lindeman, G.J. , 2009. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 15, 907–913. [DOI] [PubMed] [Google Scholar]
- Liu, B.Y. , McDermott, S.P. , Khwaja, S.S. , Alexander, C.M. , 2004. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc. Natl. Acad. Sci. USA. 101, 4158–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Dontu, G. , Mantle, I.D. , Patel, S. , Ahn, N.S. , Jackson, K.W. , Suri, P. , Wicha, M.S. , 2006. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 66, 6063–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Ginestier, C. , Charafe-Jauffret, E. , Foco, H. , Kleer, C.G. , Merajver, S.D. , Dontu, G. , Wicha, M.S. , 2008. BRCA1 regulates human mammary stem/progenitor cell fate. Proc. Natl. Acad. Sci. USA. 105, 1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti, R. , Chao, M.P. , Alizadeh, A.A. , Pang, W.W. , Jaiswal, S. , Gibbs, K.D. , van Rooijen, N. , Weissman, I.L. , 2009. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 138, 286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti, R. , Park, C.Y. , Weissman, I.L. , 2007. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 1, 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani, S.A. , Guo, W. , Liao, M.J. , Eaton, E.N. , Ayyanan, A. , Zhou, A.Y. , Brooks, M. , Reinhard, F. , Zhang, C.C. , Shipitsin, M. , Campbell, L.L. , Polyak, K. , Brisken, C. , Yang, J. , Weinberg, R.A. , 2008. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell. 133, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch, E.A. , Till, J.E. , 1960. The radiation sensitivity of normal mouse bone marrow cells, determined by quantitative marrow transplantation into irradiated mice. Radiat. Res. 13, 115 [PubMed] [Google Scholar]
- McKenzie, J.L. , Gan, O.I. , Doedens, M. , Wang, J.C. , Dick, J.E. , 2006. Individual stem cells with highly variable proliferation and self-renewal properties comprise the human hematopoietic stem cell compartment. Nat. Immunol. 7, 1225–1233. [DOI] [PubMed] [Google Scholar]
- Molofsky, A.V. , Pardal, R. , Iwashita, T. , Park, I.K. , Clarke, M.F. , Morrison, S.J. , 2003. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 425, 962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, R.D. , O'Connor, M.D. , Griffith, M. , Kuchenbauer, F. , Delaney, A. , Prabhu, A.L. , Zhao, Y. , McDonald, H. , Zeng, T. , Hirst, M. , Eaves, C.J. , Marra, M.A. , 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 18, 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, S.J. , Kimble, J. , 2006. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 441, 1068–1074. [DOI] [PubMed] [Google Scholar]
- Munos, B. , 2009. Lessons from 60 years of pharmaceutical innovation. Nat. Rev. Drug Discov. 8, 959–968. [DOI] [PubMed] [Google Scholar]
- Murchison, E.P. , Partridge, J.F. , Tam, O.H. , Cheloufi, S. , Hannon, G.J. , 2005. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 102, 12135–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller, R.A. , Knoblich, J.A. , 2009. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 23, 2675–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg, A. , 2003. Managing hematologic toxicities: novel therapies. Cancer Nurs. 26, 32S–37S. [DOI] [PubMed] [Google Scholar]
- Nowak, D. , Stewart, D. , Koeffler, H.P. , 2009. Differentiation therapy of leukemia: 3 decades of development. Blood. 113, 3655–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, C.A. , Pollett, A. , Gallinger, S. , Dick, J.E. , 2007. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 445, 106–110. [DOI] [PubMed] [Google Scholar]
- Overington, J.P. , Al-Lazikani, B. , Hopkins, A.L. , 2006. How many drug targets are there?. Nat. Rev. Drug Discov. 5, 993 [DOI] [PubMed] [Google Scholar]
- Park, I.K. , Qian, D. , Kiel, M. , Becker, M.W. , Pihalja, M. , Weissman, I.L. , Morrison, S.J. , Clarke, M.F. , 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 423, 302–305. [DOI] [PubMed] [Google Scholar]
- Park, S.M. , Gaur, A.B. , Lengyel, E. , Peter, M.E. , 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou, C.M. , Sorlie, T. , Eisen, M.B. , van de Rijn, M. , Jeffrey, S.S. , Rees, C.A. , Pollack, J.R. , Ross, D.T. , Johnsen, H. , Akslen, L.A. , Fluge, O. , Pergamenschikov, A. , Williams, C. , Zhu, S.X. , Lonning, P.E. , Borresen-Dale, A.L. , Brown, P.O. , Botstein, D. , 2000. Molecular portraits of human breast tumours. Nature. 406, 747–752. [DOI] [PubMed] [Google Scholar]
- Perrimon, N. , Friedman, A. , Mathey-Prevot, B. , Eggert, U.S. , 2007. Drug-target identification in Drosophila cells: combining high-throughout RNAi and small-molecule screens. Drug Discov. Today. 12, 28–33. [DOI] [PubMed] [Google Scholar]
- Peter, M.E. , 2010. Regulating cancer stem cells the miR way. Cell Stem Cell. 6, 4–6. [DOI] [PubMed] [Google Scholar]
- Phillips, T.M. , McBride, W.H. , Pajonk, F. , 2006. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J. Natl. Cancer Inst. 98, 1777–1785. [DOI] [PubMed] [Google Scholar]
- Pierce, G.B. , Dixon, F.J. , Verney, E.L. , 1960. Teratocarcinogenic and tissue-forming potentials of the cell types comprising neoplastic embryoid bodies. Lab. Invest. 9, 583–602. [PubMed] [Google Scholar]
- Pierce, G.B. , Speers, W.C. , 1988. Tumors as caricatures of the process of tissue renewal: prospects for therapy by directing differentiation. Cancer Res. 48, 1996–2004. [PubMed] [Google Scholar]
- Pollard, S.M. , Yoshikawa, K. , Clarke, I.D. , Danovi, D. , Stricker, S. , Russell, R. , Bayani, J. , Head, R. , Lee, M. , Bernstein, M. , Squire, J.A. , Smith, A. , Dirks, P. , 2009. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 4, 568–580. [DOI] [PubMed] [Google Scholar]
- Prince, M.E. , Sivanandan, R. , Kaczorowski, A. , Wolf, G.T. , Kaplan, M.J. , Dalerba, P. , Weissman, I.L. , Clarke, M.F. , Ailles, L.E. , 2007. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana, E. , Shackleton, M. , Sabel, M.S. , Fullen, D.R. , Johnson, T.M. , Morrison, S.J. , 2008. Efficient tumour formation by single human melanoma cells. Nature. 456, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachidi, W. , Harfourche, G. , Lemaitre, G. , Amiot, F. , Vaigot, P. , Martin, M.T. , 2007. Sensing radiosensitivity of human epidermal stem cells. Radiother. Oncol. 83, 267–276. [DOI] [PubMed] [Google Scholar]
- Reya, T. , Morrison, S.J. , Clarke, M.F. , Weissman, I.L. , 2001. Stem cells, cancer, and cancer stem cells. Nature. 414, 105–111. [DOI] [PubMed] [Google Scholar]
- Rix, U. , Superti-Furga, G. , 2009. Target profiling of small molecules by chemical proteomics. Nat. Chem. Biol. 5, 616–624. [DOI] [PubMed] [Google Scholar]
- Rocha Lima, C.M. , Green, M.R. , Rotche, R. , Miller, W.H. , Jeffrey, G.M. , Cisar, L.A. , Morganti, A. , Orlando, N. , Gruia, G. , Miller, L.L. , 2004. Irinotecan plus gemcitabine results in no survival advantage compared with gemcitabine monotherapy in patients with locally advanced or metastatic pancreatic cancer despite increased tumor response rate. J. Clin. Oncol. 22, 3776–3783. [DOI] [PubMed] [Google Scholar]
- Roesch, A., Fukunaga-Kalabis, M., Schmidt, E.C., Zabierowski, S.E., Brafford, P.A., Vultur, A., Basu, D., Gimotty, P., Vogt, T., Herlyn, M., 2010. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. 141, 583–594. [DOI] [PMC free article] [PubMed]
- Rubin, L.L. , de Sauvage, F.J. , 2006. Targeting the Hedgehog pathway in cancer. Nat. Rev. Drug Discov. 5, 1026 [DOI] [PubMed] [Google Scholar]
- Rudin, C.M. , Hann, C.L. , Laterra, J. , Yauch, R.L. , Callahan, C.A. , Fu, L. , Holcomb, T. , Stinson, J. , Gould, S.E. , Coleman, B. , LoRusso, P.M. , Von Hoff, D.D. , de Sauvage, F.J. , Low, J.A. , 2009. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N. Engl. J. Med. 361, 1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone, P. , Storci, G. , Tavolari, S. , Guarnieri, T. , Giovannini, C. , Taffurelli, M. , Ceccarelli, C. , Santini, D. , Paterini, P. , Marcu, K.B. , Chieco, P. , Bonafe, M. , 2007. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J. Clin. Invest. 117, 3988–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton, M. , Vaillant, F. , Simpson, K.J. , Stingl, J. , Smyth, G.K. , Asselin-Labat, M.L. , Wu, L. , Lindeman, G.J. , Visvader, J.E. , 2006. Generation of a functional mammary gland from a single stem cell. Nature. 439, 84–88. [DOI] [PubMed] [Google Scholar]
- Shimono, Y. , Zabala, M. , Cho, R.W. , Lobo, N. , Dalerba, P. , Qian, D. , Diehn, M. , Liu, H. , Panula, S.P. , Chiao, E. , Dirbas, F.M. , Somlo, G. , Pera, R.A. , Lao, K. , Clarke, M.F. , 2009. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 138, 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin, M. , Polyak, K. , 2008. The cancer stem cell hypothesis: in search of definitions, markers, and relevance. Lab. Invest. 88, 459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz, L.D. , Ishikawa, F. , Greiner, D.L. , 2007. Humanized mice in translational biomedical research. Nat. Rev. Immunol. 7, 118–130. [DOI] [PubMed] [Google Scholar]
- Shultz, L.D. , Schweitzer, P.A. , Christianson, S.W. , Gott, B. , Schweitzer, I.B. , Tennent, B. , McKenna, S. , Mobraaten, L. , Rajan, T.V. , Greiner, D.L. , 1995. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J. Immunol. 154, 180–191. [PubMed] [Google Scholar]
- Singh, S.K. , Hawkins, C. , Clarke, I.D. , Squire, J.A. , Bayani, J. , Hide, T. , Henkelman, R.M. , Cusimano, M.D. , Dirks, P.B. , 2004. Identification of human brain tumour initiating cells. Nature. 432, 396–401. [DOI] [PubMed] [Google Scholar]
- Smith, G.H. , Medina, D. , 1988. A morphologically distinct candidate for an epithelial stem cell in mouse mammary gland. J. Cell Sci. 90, 173–183. [DOI] [PubMed] [Google Scholar]
- Somervaille, T.C. , Cleary, M.L. , 2006. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 10, 257–268. [DOI] [PubMed] [Google Scholar]
- Sorlie, T. , Perou, C.M. , Tibshirani, R. , Aas, T. , Geisler, S. , Johnsen, H. , Hastie, T. , Eisen, M.B. , van de Rijn, M. , Jeffrey, S.S. , Thorsen, T. , Quist, H. , Matese, J.C. , Brown, P.O. , Botstein, D. , Eystein Lonning, P. , Borresen-Dale, A.L. , 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA. 98, 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southam, C. , Brunschwig, A. , Dizon, Q. , 1962. Autologous and Homologous Transplantation of Human Cancer. Biological Interactions in Normal and Neoplastic Growth: a Contribution to the Tumor-Host Problem Little Brown; Boston, MA: pp. 723–738 [Google Scholar]
- Spangrude, G.J. , Heimfeld, S. , Weissman, I.L. , 1988. Purification and characterization of mouse hematopoietic stem cells. Science. 241, 58–62. (erratum appears in Science 1989 Jun 2. 244(4908), 1030) [DOI] [PubMed] [Google Scholar]
- Stingl, J. , Eirew, P. , Ricketson, I. , Shackleton, M. , Vaillant, F. , Choi, D. , Li, H.I. , Eaves, C.J. , 2006. Purification and unique properties of mammary epithelial stem cells. Nature. 439, 993–997. [DOI] [PubMed] [Google Scholar]
- Thayer, S.P. , di Magliano, M.P. , Heiser, P.W. , Nielsen, C.M. , Roberts, D.J. , Lauwers, G.Y. , Qi, Y.P. , Gysin, S. , Fernandez-del Castillo, C. , Yajnik, V. , Antoniu, B. , McMahon, M. , Warshaw, A.L. , Hebrok, M. , 2003. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 425, 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen, J.W. , de Sauvage, F.J. , 2009. Paracrine hedgehog signaling in cancer. Cancer Res. 69, 6007–6010. [DOI] [PubMed] [Google Scholar]
- Troester, M.A. , Herschkowitz, J.I. , Oh, D.S. , He, X. , Hoadley, K.A. , Barbier, C.S. , Perou, C.M. , 2006. Gene expression patterns associated with p53 status in breast cancer. BMC Cancer. 6, 276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotti, A. , Colevas, A.D. , Setser, A. , Rusch, V. , Jaques, D. , Budach, V. , Langer, C. , Murphy, B. , Cumberlin, R. , Coleman, C.N. , Rubin, P. , 2003. CTCAE v3.0: development of a comprehensive grading system for the adverse effects of cancer treatment. Semin. Radiat. Oncol. 13, 176–181. [DOI] [PubMed] [Google Scholar]
- Trowbridge, J.J. , Scott, M.P. , Bhatia, M. , 2006. Hedgehog modulates cell cycle regulators in stem cells to control hematopoietic regeneration. Proc. Natl. Acad. Sci. USA. 103, 14134–14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, Y.C. , Lu, Y. , Nichols, P.W. , Zlotnikov, G. , Jones, P.A. , Smith, H.S. , 1996. Contiguous patches of normal human mammary epithelium derived from a single stem cell: implications for breast carcinogenesis. Cancer Res. 56, 402–404. [PubMed] [Google Scholar]
- Vaillant, F. , Asselin-Labat, M.L. , Shackleton, M. , Forrest, N.C. , Lindeman, G.J. , Visvader, J.E. , 2008. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 68, 7711–7717. [DOI] [PubMed] [Google Scholar]
- Visvader, J.E. , 2009. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 23, 2563–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff, D.D. , LoRusso, P.M. , Rudin, C.M. , Reddy, J.C. , Yauch, R.L. , Tibes, R. , Weiss, G.J. , Borad, M.J. , Hann, C.L. , Brahmer, J.R. , Mackey, H.M. , Lum, B.L. , Darbonne, W.C. , Marsters, J.C. , de Sauvage, F.J. , Low, J.A. , 2009. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N. Engl. J. Med. 361, 1164–1172. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Wakeman, T.P. , Lathia, J.D. , Hjelmeland, A.B. , Wang, X.F. , White, R.R. , Rich, J.N. , Sullenger, B.A. , 2010. Notch promotes radioresistance of glioma stem cells. Stem Cells. 28, 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.C. , 2007. Evaluating therapeutic efficacy against cancer stem cells: new challenges posed by a new paradigm. Cell Stem Cell. 1, 497–501. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Garcia, A.J. , Wu, M. , Lawson, D.A. , Witte, O.N. , Wu, H. , 2006. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc. Natl. Acad. Sci. USA. 103, 1480–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Baskerville, S. , Shenoy, A. , Babiarz, J.E. , Baehner, L. , Blelloch, R. , 2008. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 40, 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Schulte, B.A. , LaRue, A.C. , Ogawa, M. , Zhou, D. , 2006. Total body irradiation selectively induces murine hematopoietic stem cell senescence. Blood. 107, 358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, D.N. , Berman, D.M. , Burkholder, S.G. , Wang, B. , Beachy, P.A. , Baylin, S.B. , 2003. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 422, 313–317. [DOI] [PubMed] [Google Scholar]
- Wei, G. , Twomey, D. , Lamb, J. , Schlis, K. , Agarwal, J. , Stam, R.W. , Opferman, J.T. , Sallan, S.E. , den Boer, M.L. , Pieters, R. , Golub, T.R. , Armstrong, S.A. , 2006. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 10, 331–342. [DOI] [PubMed] [Google Scholar]
- Wellner, U. , Schubert, J. , Burk, U.C. , Schmalhofer, O. , Zhu, F. , Sonntag, A. , Waldvogel, B. , Vannier, C. , Darling, D. , zur Hausen, A. , Brunton, V.G. , Morton, J. , Sansom, O. , Schuler, J. , Stemmler, M.P. , Herzberger, C. , Hopt, U. , Keck, T. , Brabletz, S. , Brabletz, T. , 2009. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11, 1487–1495. [DOI] [PubMed] [Google Scholar]
- Weng, A.P. , Ferrando, A.A. , Lee, W. , Morris, J.P.t. , Silverman, L.B. , Sanchez-Irizarry, C. , Blacklow, S.C. , Look, A.T. , Aster, J.C. , 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306, 269–271. [DOI] [PubMed] [Google Scholar]
- Whitehead, K.A. , Langer, R. , Anderson, D.G. , 2009. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 8, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehurst, A.W. , Bodemann, B.O. , Cardenas, J. , Ferguson, D. , Girard, L. , Peyton, M. , Minna, J.D. , Michnoff, C. , Hao, W. , Roth, M.G. , Xie, X.J. , White, M.A. , 2007. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 446, 815–819. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S. , Knox, C. , Guo, A.C. , Cheng, D. , Shrivastava, S. , Tzur, D. , Gautam, B. , Hassanali, M. , 2008. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36, D901–D906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, W.A. , Chen, M.S. , Behbod, F. , Alfaro, M.P. , Buchholz, T.A. , Rosen, J.M. , 2007. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc. Natl. Acad. Sci. USA. 104, 618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdak, H. , Zhu, S. , Romero, A. , Lorger, M. , Watson, J. , Chiang, C.Y. , Zhang, J. , Natu, V.S. , Lairson, L.L. , Walker, J.R. , Trussell, C.M. , Harsh, G.R. , Vogel, H. , Felding-Habermann, B. , Orth, A.P. , Miraglia, L.J. , Rines, D.R. , Skirboll, S.L. , Schultz, P.G. , 2010. An RNAi screen identifies TRRAP as a regulator of brain tumor-initiating cell differentiation. Cell Stem Cell. 6, 37–47. [DOI] [PubMed] [Google Scholar]
- Xu, N. , Papagiannakopoulos, T. , Pan, G. , Thomson, J.A. , Kosik, K.S. , 2009. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 137, 647–658. [DOI] [PubMed] [Google Scholar]
- Yilmaz, O.H. , Valdez, R. , Theisen, B.K. , Guo, W. , Ferguson, D.O. , Wu, H. , Morrison, S.J. , 2006. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 441, 475–482. [DOI] [PubMed] [Google Scholar]
- Young, L.J. , Medina, D. , DeOme, K.B. , Daniel, C.W. , 1971. The influence of host and tissue age on life span and growth rate of serially transplanted mouse mammary gland. Exp. Gerontol. 6, 49–56. [DOI] [PubMed] [Google Scholar]
- Yu, F. , Yao, H. , Zhu, P. , Zhang, X. , Pan, Q. , Gong, C. , Huang, Y. , Hu, X. , Su, F. , Lieberman, J. , Song, E. , 2007. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 131, 1109–1123. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Grindley, J.C. , Yin, T. , Jayasinghe, S. , He, X.C. , Ross, J.T. , Haug, J.S. , Rupp, D. , Porter-Westpfahl, K.S. , Wiedemann, L.M. , Wu, H. , Li, L. , 2006. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 441, 518–522. [DOI] [PubMed] [Google Scholar]
- Zhang, M. , Behbod, F. , Atkinson, R.L. , Landis, M.D. , Kittrell, F. , Edwards, D. , Medina, D. , Tsimelzon, A. , Hilsenbeck, S. , Green, J.E. , Michalowska, A.M. , Rosen, J.M. , 2008. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 68, 4674–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]


