Abstract/Summary
Src kinases are activated and relocalize to the cytoplasm during mitosis, but their mitotic function has remained elusive. We describe here a novel mitotic substrate of src kinases. Trask (Transmembrane and Associated with Src Kinases) is a 140kd type I transmembrane glycoprotein unrelated to currently known protein families. Src kinases phosphorylate Trask in vitro and mediate its mitotic hyperphosphorylation in vivo. Trask associates with both yes and src, is localized to the cell membrane during interphase, and undergoes cytoplasmic relocalization during mitosis. Overexpression of Trask leads to cell rounding and a loss of adhesion phenotype. Consistent with a function in cell adhesion, Trask interacts with a number of adhesion and matrix proteins including cadherins, syndecans, and the membrane-type serine protease 1 (MT-SP1), and is proteolytically cleaved by MT-SP1. Trask is unique among cell adhesion molecules in that it is under cell cycle regulation and thus links src kinases with the mitotic regulation of cell adhesion. This suggests a potential pathway by which hyperactive src kinases in tumors can deregulate adhesion signaling and mediate the metastatic phenotype.
Introduction
Tyrosine kinases are important regulators of growth in multicellular organisms. Deregulation of tyrosine kinase signaling leads to abnormal growth and development, cancer, and metastasis. A fundamental challenge of molecular oncology is to understand the specific cellular and extracellular functions of tyrosine kinase families in order to understand their oncogenic functions. A potent oncogenic potential is inherent within members of the src family of non-receptor tyrosine kinases. When constitutively activated in experimental systems, src kinases are transforming in tissue culture and transgenic models (Jove and Hanafusa 1987; Guy et al.1994; Webster et al. 1995). The viral oncogenes v-src, v-yes, and v-fgr are activated homologs of cellular src proteins and are responsible for producing tumors in animal hosts. High src activity is characteristic of many common human tumors including colon cancers, breast cancers, lung cancers, sarcomas, and melanomas (Rosen et al. 1986; Cartwright et al. 1989; Irby and Yeatman 2000; Ottenhoff-Kalff et al. 1992). In a subset of these tumors this is due to mutational activation of src (Mao et al. 1997; Irby and Fujita 1999; Sugimura et al. 2000). The interest in determining a src signaling pathway has led to the identification of numerous src substrates involved in diverse cellular functions including growth factor receptor signaling, cytoskeletal organization, cell motility, and adhesion signaling (Erpel and Courtneidge 1995). Src function is also required for cell cycle progression as microinjection of anti-src/yes/fyn antibodies prevents G1/S transition and microinjection during G2 prevents G2/M progression (Roche et al. 1995a; Roche et al. 1995b). The G1/S activities of src are in part regulated through inhibition by RACK1 (Mamidipudi et al. 2004). Although the activities of src kinases are required for cell cycle progression in mammalian cells, the cycle-specific cellular functions which they perform have remained elusive.
Much indirect and direct evidence suggests that src is important in the mitotic phase of the cell cycle. Src is predominantly associated with the plasma membrane, however during G2/M is redistributed to the cytoplasmic compartment (David-Pfeuty and Nouvian-Dooghe 1990). Src kinase activity is increased during mitosis in association with amino-terminal serine and threonine phosphorylations as well as partial dephosphorylation of tyrosine 527 by RPTPalpha (Chackalaparampil and Shalloway 1988; Park and Cartwright 1995; Bagrodia et al. 1991; Mustelin and Hunter 2002). Dephosphorylation of tyrosine 527 leads to the release of intramolecular interactions and increased accessibility of the SH2 and SH3 domains for interaction with proteins specifically during mitosis (Bagrodia et al. 1994). Only one mitotic substrate of src, Sam68, has been identified so far (Taylor et al. 1995; Fumagalli et al. 1994). Sam68 is an RNA binding protein and its RNA binding activity is abolished when tyrosine phosphorylated by src kinases (Wang et al. 1995).The role of Sam68 phosphorylation with regards to the cell cycle and the RNA targets of Sam68 that mediate mitotic events remain to be defined (Lukong and Richard 2003).
Considerable evidence suggests that the transforming functions of src are mediated through alterations in cell migration and invasiveness rather than increased proliferative rate. Src transformed fibroblasts have a much higher metastatic potential compared to their ras transformed counterparts (Tatsuka et al. 1996). In other experimental xenograft models, src kinase activity is not required for tumorigenic growth in mice, but is essential for the development of metastases (Boyer et al. 2002). Consistent with a pro-metastatic function, elevated src activity is associated with alterations in cell adhesion, not increased proliferative rate (Jones et al. 2002).
Based on a hypothesis that cell cycle substrates of src kinases may be highly relevant to the cancer phenotype, we have used a human breast cancer model with activated src kinases to search for cell cycle phase specific substrates of the src family members src and yes, and report here the purification and identification of a novel mitotic substrate of src kinases. Experimental overexpression of this transmembrane glycoprotein reveals a function in cell adhesion and suggests that src kinases may signal the characteristic cell adhesion changes of epithelial cells undergoing mitosis.
Results
Purification of P85
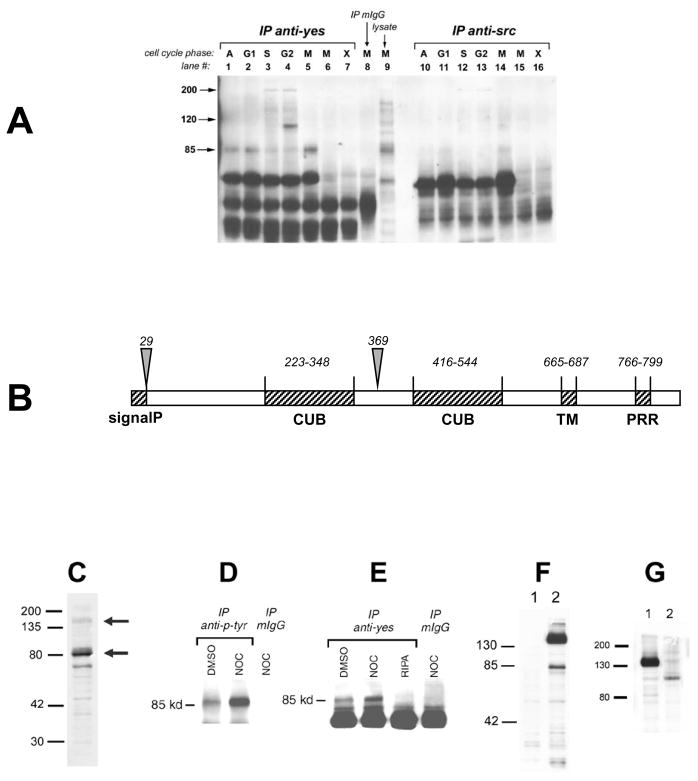
MDA-468 breast cancer cells have activated src and yes kinases (Moasser et al. 1999). Immunoprecipitation of src and yes from these cells during each of the phases of the cell cycle revealed the co-immunoprecipitation of an 85kd tyrosine phosphorylated protein predominantly within yes immune complexes (Figure 1A). This protein is hyperphosphorylated specifically in mitotic cells blocked with nocodazole but not in mitotic cells blocked with a src-selective tyrosine kinase inhibitor, leading us to suspect that it may be a mitotic substrate of src kinases (Figure 1, lanes 5 & 6). Immunoblot analyses failed to determine the identity of this 85kd protein therefore we sought to purify it. Purification was performed using anti-phosphotyrosine antibodies immobilized on solid support to purify p85 from mitotic lysates of MDA-468 cells. The purified p85 band was isolated by SDS-PAGE and subjected to trypsin digestion and two selected tryptic peptides were sequenced. The two sequences showed no homology with described proteins or cDNAs, however they matched with the same potential ORF of a human Est. The corresponding IMAGE clone contained a 1 kb partial cDNA with no identity within the cDNA and protein databases including NCBI, EMBL, or DDBJ (Japan). Northern blots probed with the radiolabeled partial cDNA revealed that p85 is encoded by a 6kb RNA message with increased expression in mitotic cells (shown in Figure 2B). The full length p85 cDNA sequence was then determined using 5′ and 3′ RACE PCR and this sequence information was then used to design primers and subsequently clone a 5kb cDNA by RT-PCR using RNA from mitotic MDA-468 cells.
Figure 1. Identification and characterization of an 85kd mitotic phosphoprotein associated with yes.
A) An 85kd phosphoprotein was identified by anti-phosphotyrosine immunoblotting of anti-yes and anti-src immune complexes. Lanes correspond to asynchronous cells (1&10) or cells synchronized in G1 (2&11), S (3&12), G2(4&13), or M (5&14) phases of the cell cycle using lovastatin, aphidicolin, etoposide, and nocodazole respectively. Cells in lane 6 and 15 were blocked in mitosis by the src-selective tyrosine kinase inhibitor PD173955. Additional lanes include immunoprecipitates lacking cell lysates (7&16), or mitotic mIgG immunoprecipitates (8), or total lysate (9). B)The structural organization of Trask protein. Labels indicate the signal peptide (SignalP), two CUB domains, transmembrane domain (TM), and proline-rich region (PRR). Arrows indicate two known cleavage sites described in the text. C) Immunoblot of MDA-468 cells using anti-Trask 12F3 antibodies. D)Anti-phosphotyrosine immunoprecipitates from asynchronous and mitotic MDA-468 cells were immunoblotted with anti-Trask antibodies. E)Anti-yes immunoprecipitates from MDA-468 cells were immunoblotted with anti-Trask antibodies. Controls include immunoprecipitates from lysis buffer without cell lysate (3rdlane) and mIgG control (4thlane). F) MCF-7 cells were transfected with vector control (1) or pcDNA4-MycHis-Trask (2). An anti-myc immunoblot is shown here. Similar results were obtained by transfecting 293T or MDA-468 cells (not shown).G)MDA-468 cells were transiently transfected with pcDNA4-MycHis-Trask and treated with DMSO (1) or tunicamycin (2) for 24 hours. An anti-myc immunoblot is shown here.
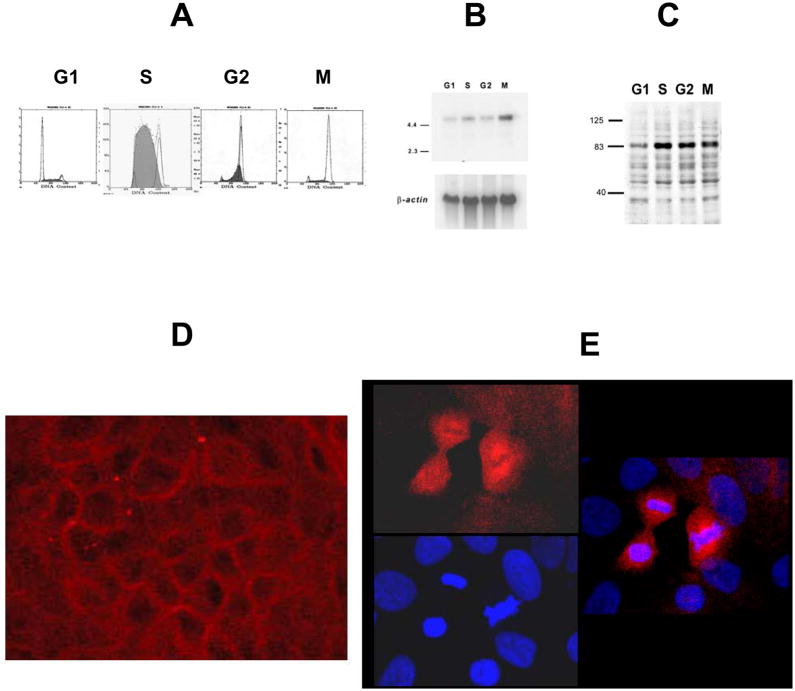
Figure 2. Cellular expression of Trask.
A) MDA-468 cells were synchronized in G1, S, G2, or M and cell cycle phase was verified. B)Northern blot analysis of polyA selected RNA from these cells was performed using a 2kb 32P labeled Trask cDNA probe. C) Immunoblot analysis of total lysates from these cells was performed using anti-Trask 12F3 monoclonal antibodies. D) Immunofluorescence microscopy of MCF10A breast epithelial cells using anti-Trask monoclonal antibodies labeled with Rhodamine-red. The majority of the cells show cell membrane staining as shown in this image. Mitotic cells were identified by their characteristic chromatin condensation and show cytoplasmic staining. A representative high power field is shown in image E.
P85 encodes a novel transmembrane protein
The open reading frame (ORF) of the p85 cDNA was determined from the purified peptide sequence information. A number of potential ATG start codons lie within the upstream sequences of this ORF of which the ATGs at positions 93, 669, and 651, in decreasing order comply best with Kozak consensus translational start sites (Kozak 1987). These regions identify coding regions of 2508, 1950, and 1932bps within the same ORF encoding proteins of 835, 649, and 643 amino acids with predicted MWs of 91, 73 and 72 kDs. This ORF is followed by a 3.5kb 3′ untranslated region. No other significant ORFs are found within the 6 kb p85 cDNA.
Comparison of the 835 AA p85 sequences against protein or cDNA databases does not reveal close homology with any currently known families of proteins. There are no homologs in prokaryotic or primitive eukaryotic organisms and close homologs of this protein are only found in more complex organisms. A transmembrane region is identified at AA position 665–686 with high probability using all major algorithms (PSORT, DAS, SMART, SOSUI, TMPRED) with an orientation that predicts a large extracellular N-terminal region and a smaller intracellular C-terminal region (Figure 1B). Consistent with a predicted extracellular localization, there are consensus signal peptide sequences at positions 1–29 of the 835 AA protein. Optimal Kozak translational sequences and 5′ sequences encoding a conserved signal peptide motif identify that the ATG start site at position 93 marks the correct translation initiation site and p85 is an 835 AA protein encoded by a 2508 bp coding region. AA positions 223–348 and 416–544 are identified with low scores as CUB domains (only one of the two is identified by SMART and the other identified by Pfam). There are five tyrosine residues within the intracellular domain and a proline rich region at 766–799 (identified by BLOCKS). This protein was named Trask (Transmembrane and Associated with Src Kinases) and is referred to as such from this point forward.
Trask sequences were used to design peptide immunogens and two monoclonal antibodies were successfully raised against Trask sequences (clones 12F3 and 2G4). On immunoblot analyses of MDA-468 lysates these antibodies predominantly bind a 85kd protein that appears as a doublet consistent with phosphorylated and unphosphorylated forms (Figure 1C). Immunoprecipitation followed by immunoblot analyses confirm that Trask is associated with yes and to a smaller degree with src and is markedly tyrosine phosphorylated during the mitotic phase in MDA-468 cells (Figure 1D, E). These studies confirm that the purified protein is indeed the mitotic phosphoprotein associated with yes and not a background co-purification product. Transient transfection of a construct containing the full 2508bp ORF with a C-terminal myc tag (pcDNA4-MycHis-Trask) reveals that Traskis actually expressed as a 140kd protein and a secondary 85kd product (Figure 1F). A faint 140kd band is also seen in the endogenous Trask immunoblots of MDA-468 cells although it is not the predominant form of Trask in these cells (Figure 1C). The 85kd form of Trask is a product of proteolytic cleavage and is described further below. Treatment of MDA-468 cells with the glycosylation inhibitor tunicamycin decreases the molecular weight of myc-tagged Trask from 140kd to 100kd revealing that glycosylation accounts for a substantial increase in molecular weight above that predicted from its primary amino acid sequence (Figure 1G).
Cellular expression of Trask
Northern blot analysis using a 2kb coding region Trask cDNA probe shows low expression during most of interphase with a slight increase during S-phase, but a 4 fold induction of expression during the mitotic phase (Figure 2A, B). Aliquots of the same cells were lysed and studied by western blotting using anti-Trask 12F3 antibodies. This analysis shows a high level of Trask protein expression during the S, G2, and M phases compared to G1 (Figure 2A, C). The localization of Trask was studied by immunofluorescence microscopy using anti-Trask antibodies. Confocal microscopic analyses of a number of cell types including MCF10A, MDA-468, Du-145, and HT29 cells shows punctate and predominantly membrane staining as predicted by the encoded transmembrane domain (Figure 2D). This localization was seen using either 12F3 or 2G4 antibodies and using either paraformaldehyde or methanol fixation methods. Mitotic cells were identified within the asynchronous population by their characteristic chromatin condensation. Unlike interphase cells, many mitotic cells show predominantly cytoplasmic immunolocalization of Trask (Figure 2E). The punctate staining pattern and the cytoplasmic redistribution of Trask may reflect localization to the endosomal membrane, as is observed with src (Kaplan et al. 1992).
Src kinases phosphorylate Trask
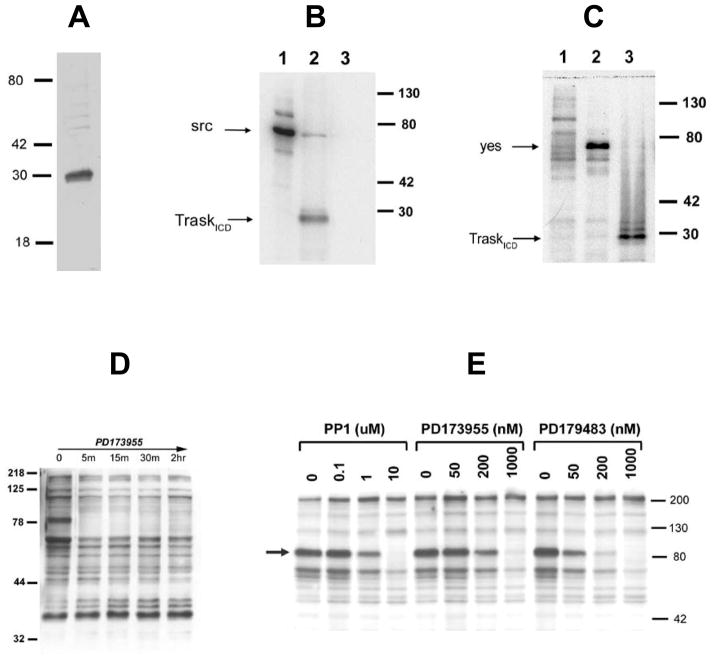
In order to determine whether Trask is a substrate of src kinases, recombinant Trask protein was synthesized. A 460 bp fragment encoding the entire intracellular domain (ICD) of Trask fused with carboxy terminal V5 and His tags was expressed in E.coli DH5α cells, purified over a charged nickel affinity column and subsequently dialyzed against PBS. Gel electrophoresis and immunoblot analysis of the purification products using anti-V5 antibodies confirms expression of a 30 kd recombinant fusion protein with a purity of approximately 80% by coomassie blue staining (Figure 3A). In vitro kinase reactions were performed with recombinant src and with immunopurified yes proteins using the purified recombinant Trask ICD as the substrate and [32P] labeled ATP. In this reaction both src and yes phosphorylate the Trask ICD (Figure 3B, C).
Figure 3. Src kinases phosphorylate Trask in vitro and in vivo.
A) The purity of recombinant Trask ICD was verified by SDS-PAGE separation and coomassie blue staining. B) In vitro src kinase reaction. Lanes 1–3 correspond to reactions containing src alone, src and TraskICD, and TraskICD alone. C) In vitro Yes kinase reaction. Lanes 1–3 correspond to reactions containing mIgG immunoprecipitates and TraskICD, α-yes immunoprecipitates alone, and α-yes immunoprecipitates and TraskICD. D)Asynchronous MDA-468 cells were treated with 1uM P173955 for the indicated times and lysates were immunoblotted using anti-phosphotyrosine antibodies. The 85 kd phosphotyrosine band indicated by the arrow is Trask, identified previously by purification. E)MDA-468 cells were treated for 20 hours with nocodazole, and mitotic shake-offs were washed and released into media containing the indicated concentrations of src-selective tyrosine kinase inhibitors for 30 minutes. Negative control lanes were treated with vehicle (DMSO) and are labeled as “0”. An anti-phosphotyrosine immunoblot is shown here. The 85kd band identified by the arrow is phosphorylated Trask, and disappears rapidly with increasing concentrations of src inhibitors.
To determine whether src kinases also phosphorylate Trask in vivo we treated MDA-468 cells with the tyrosine kinase inhibitor PD173955, which is selective for members of the src family. PD173955 is equally active against both src and yes in vitro(Moasser et al. 1999). Trask is dephosphorylated within 5 minutes of exposure to PD173955, suggesting that it is a substrate of src kinases in vivo (Figure 3D). Although Trask is tyrosine phosphorylated in interphase in MDA-468 cells, it undergoes significant hyperphosphorylation during mitosis. To determine whether src kinases also mediate this mitotic hyperphosphorylation, we released nocodazole-blocked MDA-468 cells for short times into media containing three different tyrosine kinase inhibitors selective for the src family (Kraker et al. 2000; Mizenina and Moasser 2004). Each of these compounds inhibits the mitotic hyperphosphorylation of Trask in a dose-dependent manner (Figure 3E). Although each of these tyrosine kinase inhibitors may have some non-specific activities and inhibit cellular kinases other than src and yes, all three inhibitors give similar results. These data suggest that the in vivo mitotic tyrosine phosphorylation of Trask is mediated through the src family kinases, of which src and yes are expressed in these cells.
Trask is an adhesion protein
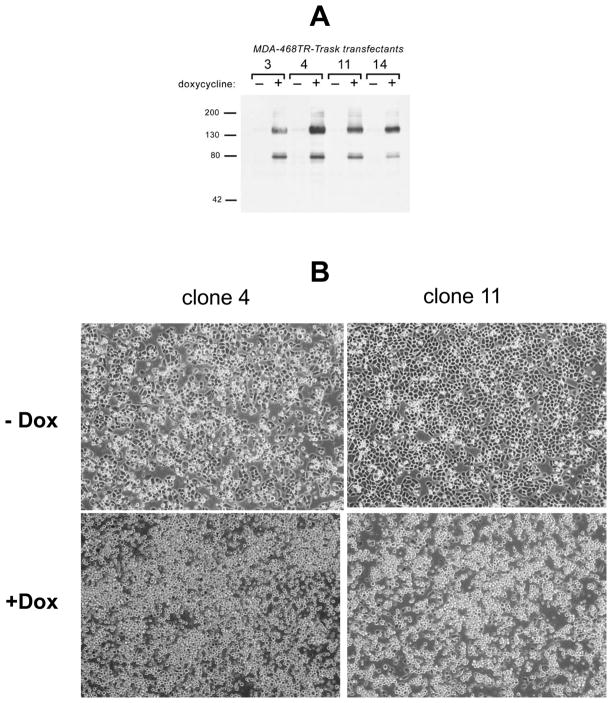
In order to study the cellular functions of Trask, we established an inducible expression system to overexpress it in MDA-468 cells. MDA-468 cells were first stably transfected with the pcDNA6/TR vector to express the tet-repressor protein and subsequently transfected with a pcDNA4-TO-MycHis vector containing the full length Trask cDNA in frame with C-terminal myc and His tags. Induction of these transfectants (MDA-468TR-Trask) with doxycycline leads to overexpression of p140Trask as well as p85Trask, again confirming that the two forms are products of the same transcript (Figure 4A). Overexpression of Trask has no effect on the proliferative rate of MDA-468 cells measured in assays of monolayer growth or anchorage independent growth (data not shown). However, overexpression of Trask has a profound effect on cell shape and adhesion, easily apparent under light microscopy. When induced to overexpress Trask, MDA-468 cells develop a rounded shape, lose their flattened adherent morphology, and grow in a loosely adherent suspension (Figure 4B). This effect is seen in all transfectants that overexpress Trask and not in doxycycline-treated vector controls (not shown).
Figure 4. Trask over-expression in MDA-468 cells.
A) MDA-468TR cells were transfected with pcDNA4-TO-MycHis-Trask and stable transfectants (named MDA-468TR-Trask) selected in Zeocin. Inducible expression of the myc-tagged Trask protein is shown here by anti-myc immunoblots of 4 clones. Induction of expression of full length Trask cDNA results in expression of a 140kd full length Trask protein as well as an 85kd cleaved product. B)MDA-468TR-Trask cells were grown in the absence or presence of 100ng/ml doxycycline and inspected under inverted light microscopy. Induction with doxycycline results in cell rounding and continued growth in a loosely adherent semi-suspended state morphology. Clones 4 and 11 are shown here and are typical of all transfected clones. Doxycyline treatment of vector transfected controls has no visible effects on cell morphology (not shown).
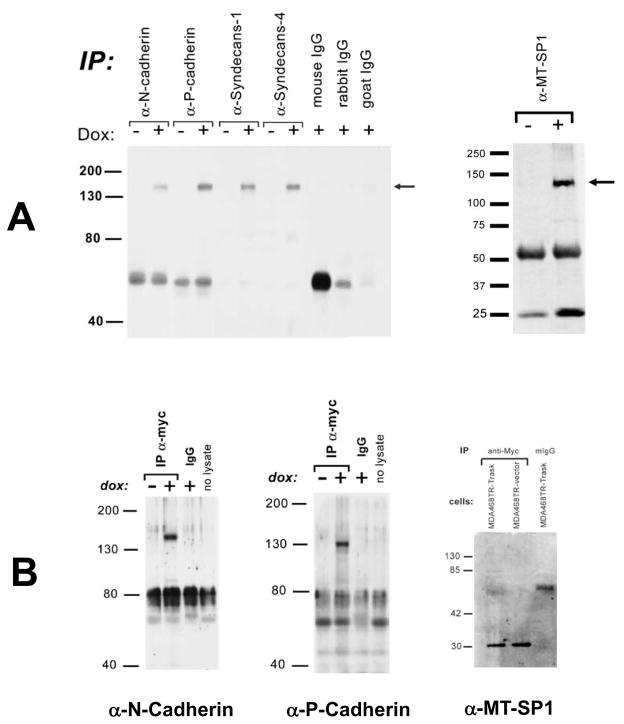
Since the transfection data suggests a function in cell adhesion, we explored Trask interactions with other proteins involved in cell adhesion and cell matrix association. MDA-468TR-Trask cells were induced to overexpress Trask and a number of candidate membrane, matrix, and adhesion proteins were immunoprecipitated with specific antibodies. We looked for the co-immunoprecipitation of Trask in these immune complexes by anti-myc immunoblotting. These studies identify that Trask interacts with the adhesion proteins N-cadherin and P-cadherin, the matrix proteins syndecans 1 and 4, and the membrane serine protease MT-SP1 (Figure 5A). The interaction between Trask and MT-SP1 may be mediated by the CUB domains in both proteins as CUB domains are thought to play a role in protein-protein interactions in the extracellular environment. These studies were also repeated by anti-myc immunoprecipitations which further confirm the interactions with N-cadherin and P-cadherin and MT-SP1 (Figure 5B) Thus far we have not been able to detect an interaction between Trask and selected members of the integrin family.
Figure 5. Trask interactions with membrane and matrix proteins.
A) MDA-468TR-Trask cells were induced with doxycyline to express Trask and cell lysates were immunoprecipitated with the indicated specific antibodies. Immune complexes were separated by SDS-PAGE and immunoblotted using anti-myc antibodies to detect the presence of myc-tagged Trask in the complexes. Controls include immune complexes using non-immune IgGs and immune complexes from un-induced cells that do not express the construct. B)The same experiments were performed in reverse order such that lysates were immunoprecipitated with anti-myc antibodies and immunoblotted with the indicated specific antibodies.
Trask is a serine protease substrate
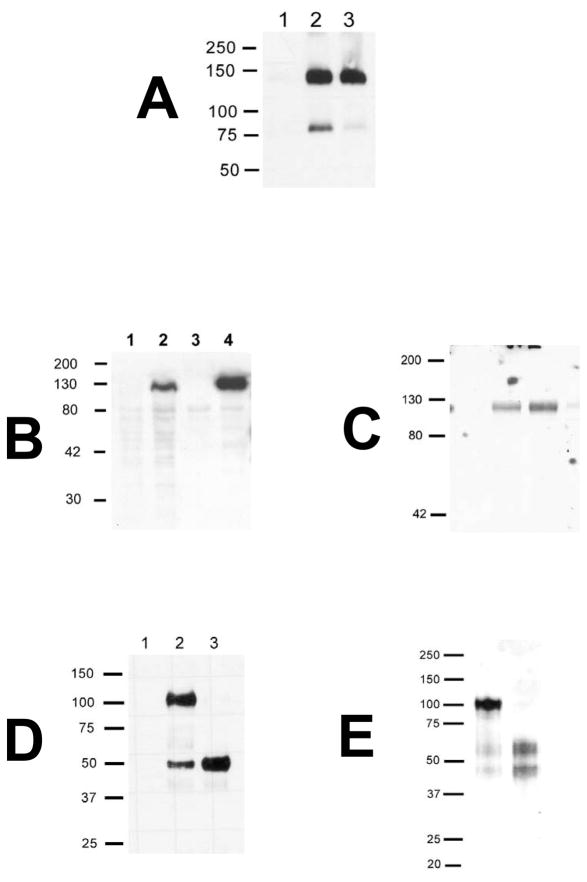
The expression of 140kd and 85kd forms of Trask from the same cDNA raise the possibility that p85Trask may be a cleavage product of p140Trask. Its interaction with the membrane serine protease MT-SP1 further strengthens that hypothesis. The induction of Trask expression in MDA-468TR-Trask cells in the presence of broad serine protease inhibitors results in a reduction in the amount of the 85kd product. More specifically, the trypsin-fold specific macromolecular serine protease inhibitor, ecotin, abrogates cleavage of p140 suggesting that an extracellular trypsin fold serine protease induces Trask processing (Figure 6A). To determine whether MT-SP1 cleaves Trask we generated recombinant extracellular domain of Trask (rTraskECD) to test as a substrate for MT-SP1 in vitro. The entire Trask extracellular domain (ECD) was cloned into pcDNA4-TO-MycHis in frame with c-terminal Myc and His tags and transfected into 293T cells. Due to the presence of signal peptide sequences and the absence of a transmembrane domain, the expressed product is secreted into the conditioned media (Figure 6B). 293T-TraskECD cells were grown in serum-free media and rTraskECD was collected from the conditioned media, purified over a charged nickel column, and dialyzed against PBS (Figure 6C). rTraskECD was incubated with recombinant catalytic domain of MT-SP1 (rMT-SP1CD) in vitroand the reaction products were separated and identified by anti-myc immunoblots and silver staining (Figure 6D, 6E). In this reaction, rMT-SP1CD completely cleaves rTraskECD into two fragments. Our rTraskECD preparation prepared from 293T cells contains some amount of cleaved Trask as seen in both the silver stain and the myc immunoblots, however addition of rMT-SP1CD cleaves it completely. N-terminal amino-acid sequencing of the reaction products revealed a free N-terminal sequence of KFVPGCF, thereby identifying that MT-SP1 cleaves Trask after Arg369. MT-SP1 is well established as a trypsin-specificity protease cleaving after the basic amino acids arginine and lysine. Using a P1-fixed combinatorial peptide substrate library approach we previously reported the extended MT-SP1 cleavage site preferences to be K/R in P4, R/K/Q in P3, and S in P2 (Takeuchi et al. 2000). The sequence KQS at P4, P3, P2 of the Trask cleavage site matches precisely with these MT-SP1 substrate preferences. N-terminal sequencing of the N-terminal fragment of Trask after processing was FEIAL, consistent with cleavage of the signal peptide after Ala29.
Figure 6. Trask cleavage by MT-SP1.
A) MDA-468TR-Trask cells were left un-induced (lane 1) or induced to express Trask with 100ng/ml doxycycline (2–3). The induced cells were treated with no drug (2), or with the serine protease inhibitor 7uM ecotin (3) beginning 2 hours before doxycycline induction. 24 hours after doxycycline induction, cell lysates were harvested and immunoblotted using anti-myc antibodies. The expression of p85Trask is reduced by the protease inhibitor. B)293T cells were transfected with a myc/his-tagged TraskECD construct. Anti-myc immunoblots were performed to confirm the expression of TraskECD in cell lysates from controls (1) and transfectants (2), and the secretion of TraskECD in the media of controls (3) and transfectants (4). C) These transfected 293T cells were grown in serum free media and the soluble recombinant TraskECD was purified from conditioned media over a charged nickel column and dialyzed against PBS yielding a pure 120kd soluble TraskECD product shown here by coomassie staining. D)1ug rTraskECD was added to an in vitro protease reaction using recombinant catalytic domain of MT-SP1 (rMT-SP1CD). The mixture was incubated for 1hr at 37C in PBS, the reaction products were denatured, deglycosylated by PNGaseF treatment, separated on SDS-PAGE and immunoblotted using anti-myc antibodies. Lanes correspond to reactions containing rMT-SP1CD alone (1), rTraskECD alone (2), or rMT-SP1CD and rTraskECD (3). The rTraskECD construct is myc-tagged at the carboxy terminus, therefore these anti-myc immunoblots identify uncleaved ECD as well as the carboxy terminal cleavage product, but not the N-terminal cleavage product. E)Silver staining of rTraskECD before (1) and after (2) cleavage with rMT-SP1CD identifies both cleavage products. Our rTraskECD preparation from 293T cells contains some amount of cleaved Trask as seen in both the silver stain and the myc immunoblots here, however addition of rMT-SP1CD results in complete proteolysis. The precise site of the in vitro cleavage was determined by N-terminal sequencing of the reaction product, as described in the text.
Discussion
A potential mitotic function of src proteins has been the subject of speculation for more than a decade. However, direct analysis of this potential cell cycle function has been awaiting the identification of a mitotic substrate of src kinases. The first such substrate was Sam68, an RNA binding protein whose RNA binding activity is inhibited by tyrosine phosphorylation (Taylor and Shalloway 1994; Fumagalli et al. 1994). The role of this phosphorylation in regulating mitotic events remains to be defined. We present here a second mitotic substrate of src kinases, a transmembrane protein that functions in cell adhesion. The cell cycle and adhesion functions of this novel src substrate suggest the new hypothesis that src may regulate the mitotic changes in cell adhesion. Cell adhesion is generally presumed to be regulated during mitosis, particularly in epithelial cells which need to release themselves from their cell-matrix and cell-cell interactions transiently in order to allow cell duplication and separation of daughter cells. However the molecular mechanism that regulates this dynamic cell adhesion process is unknown. Using an inducible overexpression model, we show that Trask functions in cell adhesion. Trask is unique among currently described cell adhesion molecules in that it is under cell cycle regulation and is a candidate protein for regulating cell adhesion during mitosis.
The adhesion functions of Trask could be regulated through its phosphorylation. In our inducible Trask overexpression model, the induced Trask, unlike endogenous Trask, undergoes significant interphase tyrosine phosphorylation while producing a loss of adhesion phenotype. The excessive interphase phosphorylation of the induced construct may be due to the excessive nature of its expression in this model system. Whether the phosphorylation of Trask is the cause or the consequence of the loss of adhesion remains to be determined. Phosphorylation of Trask could mediate structural changes in its conformation leading to altered interaction of its extracellular domain, or could promote or inhibit its interaction with other transmembrane proteins involved in cell adhesion. Phosphorylation could also recruit cytoplasmic signaling proteins, in particular SH2 containing proteins involved in adhesion signaling. These are potential mechanisms by which Trask phosphorylation could influence interactions with other cell adhesion proteins and signal changes in cell adhesion.
The mitotic phosphorylation of Trask also potentially implicates src kinases in the regulation of Trask function and ultimately in the mitotic regulation of cell adhesion. In addition to MDA-468 cells, the mitotic hyperphosphorylation of Trask is evident in a number of other epithelial tumor cell lines with activated src kinases, but not readily apparent in an immortalized breast epithelial cell line or in fibroblasts (not shown).
In addition to its phosphorylation, the expression and localization of Trask are also regulated during the cell cycle. While the Trask RNA remains low during most of interphase with a small increase during S-phase, Trask protein expression increases during S, G2 and M phases. This is likely due to changes in mRNA stability and other post-transcriptional or post-translational mechanisms of regulation. Taken together, there is clear evidence of the cell cycle regulation of Trask at the level of expression, localization and post-translational modification by phosphorylation.
Trask function and expression may be important in human tumor progression. Consistent with this, Trask has also been identified by two other groups using global strategies to identify tumor-associated genes. Using cDNA chip hybridization techniques to search for genes preferentially expressed in solid tumors relative to normal tissues, Mostageer et al identified Est sequences corresponding to a cDNA identical to Trask, named CUB domain containing protein 1 (CDCP1), although the protein product was not identified in this study (Scherl-Mostageer et al. 2001). Of note, these authors use algorithms that predict a third CUB domain in the extracellular region of Trask, however in our analysis we find only two CUB domains and with low homology scores. The proposed third CUB domain falls below the threshold of homology of existing public domain algorithms for protein domain prediction. In addition, CUB domains are often only loosely related at the primary structural level making sequence-based predictions difficult. The ultimate characterization of the nature of these domains in Trask thus awaits tertiary structural analysis. In another approach to identify tumor-associated proteins, Hooper et al used subtractive immunization techniques to generate antibodies towards cell surface epitopes preferentially expressed by highly metastatic relative to non-metastatic carcinomas, and identified a cell surface glycoprotein named SIMA135, identical to Trask, and their analysis identifies two extracellular CUB domains (Hooper et al. 2003). We have independently cloned Trask as a cell cycle substrate of src kinases with a specific adhesion phenotype and interactions with adhesion proteins. We describe here the full length protein, its cell cycle expression pattern and its mitotic relocalization, its specific cleavage at Arg369by MT-SP1, its stable interaction and specific cell cycle phosphorylation by src and yes, and its interaction with matrix and adhesion proteins and with MT-SP1. Trask is the first adhesion molecule described that is under cell cycle regulation and its association and mitotic phosphorylation by src kinases, for the first time, implicates src in the regulation of adhesion in epithelial cells undergoing mitosis.
Proteolytic processing of Trask may a mechanism of regulation of its function. Trask is expressed predominantly in its cleaved 85kd form in MDA-468 breast cancer cells and we see no shifting between cleaved and uncleaved forms during cell cycle progression in these cells (data not shown). However there are substantial differences in the expression of cleaved and uncleaved Trask among various cancer cell lines, with predominant expression of cleaved Trask in most (not shown). Consistent with this, MDA-468 breast cancer cells, like many other epithelial cancer cell lines, exhibit characteristic upregulation of extracellular proteolytic pathways including activated MT-SP1. MT-SP1, which interacts with and cleaves Trask, is upregulated in many common human cancers including breast, ovarian, prostate, and colon cancers and may be responsible for Trask cleavage in most cancer cells (Bhatt et al. 2003; Santin et al. 2004). Additional serine proteases may also be able to cleave Trask. The addition of exogenous plasmin to human keratinocytes results in cleavage of gp140 to p80 and this protein is felt to be CDCP1/Trask based on anti-Fak immunoblots that seemingly cross react with the CDCP1/Trask protein (Brown et al. 2004) Although the direct significance of Trask cleavage has not yet been ascertained, it holds promise as an interesting mechanism of regulation of Trask activity.
The interaction of Trask with MT-SP1 may have specific functional implications. MT-SP1 has two extracellular CUB domains that may provide the structural basis for the interaction with Trask, since Trask also has CUB domains and CUB domains are known to be involved in homophilic interactions. MT-SP1 cleaves Trask and releases the N-terminal CUB domain. This would result in the release of binding partners of Trask that interact specifically with the N-terminal CUB domain. This may include adhesion molecules, extracellular peptide growth factors, soluble proteases, extracellular matrix components, matrix metalloproteases, or components of the plasminogen activation cascade. In addition to being a substrate of MT-SP1, Trask may play a role in regulating MT-SP1 activity or mediating the interaction of MT-SP1 with membrane and matrix proteins or with components of proteolytic cascades. MT-SP1 cleaves a number of cancer-associated proteins. In particular, MT-SP1 cleaves and activates urokinase-type plasminogen activator (uPA) which can trigger the plasminogen activation cascade (Takeuchi et al. 2000). Trask may provide a docking surface for MT-SP1 to interact with and activate uPA, leading to juxtamembrane activation of the plasminogen cascade, and increased invasiveness of cancer cells. MT-SP1 also cleaves pro-hepatocyte growth factor (pro-HGF) (Lee et al. 2000). Trask may also facilitate this interaction leading to increased HGF signaling which is associated with increased motility and invasiveness of tumor cells. Similarly, through a potential docking function, Trask may mediate the interaction of MT-SP1 with other soluble proteases involved in matrix remodeling such as matrix metalloproteases.
Trask is clearly a substrate of src family kinases within the cell and its function is likely modulated by phosphorylation of the intracellular domain. Although in MDA-468 cells Trask interacts predominantly with yes, it is difficult to draw any conclusions with regards to role of individual src family members in regulating Trask or mitotic processes, since src, yes, and fyn have largely overlapping functions. Yes, like src, is also activated in human cancers (Pena et al. 1995; Park et al. 1993). In 293 cells Trask interacts equally with both yes and fyn (data not shown). Src kinases are well-established players in oncogenesis and as more specific inhibitors of these kinases become available, a clearer delineation of the specificity and mechanism of Trask phosphorylation will be ascertained.
Trask is only the second mitotic substrate of src kinases identified to date and the first adhesion molecule found to be under cell cycle regulation. We have shown that Trask is phsophorylated in a cell cycle specific manner and that Trask represents a novel class of adhesion-related proteins. The phosphorylation of Trask represents a first possible method of regulation. Proteolytic cleavage of Trask is mediated by an extracellular proteolytic system and this may be yet another method of Trask regulation. This dual control and the identification of an adhesion-related, cell cycle substrate of src kinases presents a new paradigm to study the physiologic role of src kinases in mitosis and the pathologic role of src kinases in the mediating the invasive and metastatic properties of src driven tumors.
Materials and Methods
Cell culture and immunoblotting
MDA-468 cells were grown in DME:F12 supplemented with 10% fetal bovine serum, glutamine, penicillin, streptomycin, at 37C in a humidified incubator with 5% CO2. For cell cycle synchronization cells were treated for 20 hours with 200 ng/ml nocodazole, 15 ug/ml lovastatin, 5ug/ml aphidicolin followed by a 2 hr release, or 2uM etoposide for synchronization in M, G1, mid-S, and G2 respectively. The cell cycle phase of synchronized cells was confirmed by FACS analysis of ethidium stained nuclei measuring DNA content. M cells were further differentiated from G2 cells because of their suspended state, their forward scatter characteristics, and their chromatin condensation. Lysates were prepared in RIPA buffer (10 mM Na phosphate pH 7.2, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% NP-40, 1% Na deoxycholate, and protease inhibitors). Immunoblots were performed using mouse monoclonal antibodies for myc and phosphotyrosine (Santa Cruz Biotechnology), Yes (Wako), Src (Calbiochem), rabbit polyclonal antibodies for N-Cadherin & P-Cadherin (Santa Cruz) and MT-SP1 (Takeuchi et al. 2000), goat polyclonal for Syndecans 1& 4 (Santa Cruz). PD173955 and PD179483 were obtained from Parke-Davis Pharmaceuticals. PP1 was from Calbiochem. Anti-Trask monoclonal antibodies were developed against the peptide sequences KERSGVVCQTGRAFMIIQEQRTC and CSPTSGKQLDLLFSVTLTPRT taken from the extracellular domain of Trask.
Purification of Trask
Trask was purified in its hyper-phosphorylated state from MDA-468 mitotic lysates. Cells were blocked in mitotis with 200ng/ml nocodazole and lysed in RIPA buffer. Lysates were boiled for 5 minutes and used in pilot experiments to screen a panel of anti-phosphotyrosine antibodies for the ability to immunopurify denatured Trask. The experiment was then scaled up and PY20 antibodies were bound to solid support used to immunopurify Trask from denatured mitotic lysates, eluted in phenyl phosphate, and separated by SDS-PAGE.
Amino acid sequence analysis
The purified gel-bound protein was digested with trypsin, peptides were partially fractionated, and were then analyzed by matrix-assisted laser-desorption/ionization reflectron time-of-flight (MALDI-reTOF) mass spectrometry (MS) (Erdjument-Bromage et al. 1998); matches of the masses were not found within the human segment of a protein non-redundant database. In a parallel analysis, two selected peptides from the pool were sequenced by nano-electrospray ionization (ESI) triple quadrupole MS/MS (Geromanos et al. 2000); spectra were inspected for y″ ion series and the information was taken to search dbEST using the PepFrag program. Matches were identified with a potential ORF of a human Est and verified by comparing the computer-generated fragment ion series of the predicted tryptic peptides with the experimental MS/MS data.
Northern blots
Poly A selected RNA was separated by formaldehyde gel electrophoresis and transferred to nylon membrane. The membrane was probed with a 2kb 32P labeled Trask cDNA probe hybridized in 50% formamide, 5xSSC, 5X Denhardt’s, 0.1%SDS, 100ug/ml Salmon Sperm DNA and washed in 0.2XSSC, 0.1%SDS at 65C and exposed to film overnight.
Transfections
Transfections were performed using Lipofectamine 2000 (Invitrogen). MDA-468 cells were stably transfected with pcDNA6/TR (Invitrogen), selected in Blasticidin and expression of the tet repressor confirmed in transient transfection assays. These cells were named MDA468TR. The 2.5kb human Trask cDNA was cloned by RT-PCR, the sequence was confirmed, and was then ligated into the pcDNA4/TO/MycHis vector (Invitrogen) under control of the CMV promoter containing tet operator sequences. The pcDNA4-Trask vector was stably transfected into MDA468TR cells, and transfectants were selected in Zeocin. In this expression system doxycycline induces gene expression.
Kinase assays
In vitro kinase reactions were performed in kinase buffer (50mM PIPES pH 7.0, 10mM MnCl2, 10mM DTT, 10μM ATP, 2μg acid-denatured enolase, and 5μCi of γ32P labelled ATP). Reactions were allowed to proceed at 30°C for 10 minutes, then immediately stopped by boiling in sample buffer, separated on a 10% SDS-PAGE gel, transferred to membrane and exposed to film. Src kinase assays were performed using 0.3ug of purified recombinant src protein (Panvera). Yes kinase assays were performed using yes immunopurified from 300ug cell lysates using anti-yes monoclonal antibodies (Wako).
Immunofluorescence studies
Cells were grown on glass cover slips, fixed in 4% paraformaldehyde, and permeabilized with 0.1 % Triton X-100. Fixed cells were stained initially with anti-Trask 12F3 monoclonal antibodies and subsequently with Rhodamine Red conjugated anti-mouse antibodies and counterstained with Hoechst stain. Localization of Trask was studied by confocal microscopy with appropriate filtering.
In vitro protease reactions
1ug of recombinant TraskECD was incubated with 100nM MT-SP1 in PBS for 1 hour at 37C. Laemmli buffer was added and the sample was boiled for 5 minutes to stop the cleavage reaction. The resulting sample was run on a 4–20% Tris-Glycine SDS-PAGE gel (Invitrogen). The gel was silver stained and and immunoblot analysis with anti-Myc antibodies was performed. Complete cleavage of Trask by MT-SP1 was observed. The C-terminal fragment was identified by immunoblot with an anti-Myc antibody (the myc-epitope is on the C-terminus of the protein) and the cleavage products were sequenced by Edman degradation at the UC Davis Molecular Structure Facility.
In vivo protease inhibition
MDA-468 cells were grown to 80% confluency and pre-treated with inhibitors 7uM Ecotin for 2 hours. Trask expression was induced with 100ng/mL doxycycline for 24 hours and cell lysates were separated by SDS-PAGE and immunoblotted using anti-myc antibodies.
Acknowledgments
This work was supported by the American Cancer Society RSG-02-139-01-CDD (MMM) and NIH CA 72006 (CSC). ASB is supported by the NIH medical scientist training grant and a fellowship from the ARCS foundation. The authors would like to thank Sami Mahrus and Chris Farady for reagents and helpful discussion regarding this manuscript.
Footnotes
The Trask sequence has been deposited in the NCBI database with genebank accession #AY167484.
References
- Bagrodia S, Chackalaparampil I, Kmiecik TE, Shalloway D. Nature. 1991;349:172–175. doi: 10.1038/349172a0. [DOI] [PubMed] [Google Scholar]
- Bagrodia S, laudano AP, Shalloway D. J Biol Chem. 1994;269:10247–10251. [PubMed] [Google Scholar]
- Bhatt AS, Takeuchi T, Ylstra B, Ginzinger D, Albertson D, Shuman MA, Craik CS. Biological Chemistry. 2003;384:257–266. doi: 10.1515/BC.2003.029. [DOI] [PubMed] [Google Scholar]
- Boyer B, Bourgeois Y, Poupon MF. Oncogene. 2002;21:2347–2356. doi: 10.1038/sj.onc.1205298. [DOI] [PubMed] [Google Scholar]
- Brown TA, Yang TM, Zaitsevskaia T, Xia Y, Dunn CA, Sigle RO, Knudsen B, Carter WG. J Biol Chem. 2004;279:14772–14783. doi: 10.1074/jbc.M309678200. [DOI] [PubMed] [Google Scholar]
- Cartwright CA, Kamps MP, Meisler AI, Pipas JM, Eckhart W. J Clin Inv. 1989;83:2025–2033. doi: 10.1172/JCI114113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chackalaparampil I, Shalloway D. Cell. 1988;52:801–810. doi: 10.1016/0092-8674(88)90422-9. [DOI] [PubMed] [Google Scholar]
- David-Pfeuty T, Nouvian-Dooghe Y. J Cell Biol. 1990;111:3097–3116. doi: 10.1083/jcb.111.6.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdjument-Bromage H, Lui M, Lacomis L, Grewal A, Annan RS, MacNulty DE, Carr SA, Tempst P. Journal of Chromatography. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- Erpel T, Courtneidge SA. Curr Opin Cell Biol. 1995;7:176–182. doi: 10.1016/0955-0674(95)80025-5. [DOI] [PubMed] [Google Scholar]
- Fumagalli S, Totty NF, Hsuan JJ, Courtneidge SA. Nature. 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- Geromanos S, Freckleton G, Tempst P. Anal Chem. 2000;72:777–790. doi: 10.1021/ac991071n. [DOI] [PubMed] [Google Scholar]
- Guy CT, Muthuswamy SK, Cardiff RD, Soriano P, Muller WJ. Genes & Development. 1994;8:23–32. doi: 10.1101/gad.8.1.23. [DOI] [PubMed] [Google Scholar]
- Hooper JD, Zijlstra A, Aimes RT, Liang H, Claassen GF, Tarin D, Testa JE, Quigley JP. Oncogene. 2003;22:1783–1794. doi: 10.1038/sj.onc.1206220. [DOI] [PubMed] [Google Scholar]
- Irby RB, Yeatman TJ. Oncogene. 2000;19:5636–5642. doi: 10.1038/sj.onc.1203912. [DOI] [PubMed] [Google Scholar]
- Irby RBM, Fujita DJJ. Nat Genet. 1999;21:187–190. doi: 10.1038/5971. [DOI] [PubMed] [Google Scholar]
- Jones RJ, Avizienyte E, Wyke AW, Owens DW, Brunton VG, Frame MC. Br J Cancer. 2002;87:1128–1135. doi: 10.1038/sj.bjc.6600594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jove R, Hanafusa H. Annu Rev Cell Biol. 1987;3:31–56. doi: 10.1146/annurev.cb.03.110187.000335. [DOI] [PubMed] [Google Scholar]
- Kaplan KB, Swedlow JR, Varmus HE, Morgan DO. J Cell Biol. 1992;118:321–333. doi: 10.1083/jcb.118.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAa. Nucleic Acids Research. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. Ref Type: Abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraker AJ, Hartl BG, Amar A, Barvian MR, Showalter HD, Moore CW. Biochem Pharmacol. 2000;60:885–898. doi: 10.1016/s0006-2952(00)00405-6. [DOI] [PubMed] [Google Scholar]
- Lee SL, Dickson RB, Lin CY. J Biol Chem. 2000;275:36720–36725. doi: 10.1074/jbc.M007802200. [DOI] [PubMed] [Google Scholar]
- Lukong KE, Richard S. Biochim Biophys Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Mamidipudi V, Zhang J, Lee KC, Cartwright CA. Molecular & Cellular Biology. 2004;24:6788–6798. doi: 10.1128/MCB.24.15.6788-6798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao W, Irby R, Coppola D, Fu L, Wloch M, Turner J, Yu H, Garcia R, Jove R, Yeatman TJ. Oncogene. 1997;15:3083–3090. doi: 10.1038/sj.onc.1201496. [DOI] [PubMed] [Google Scholar]
- Mizenina OA, Moasser MM. Cell Cycle. 2004;3:796–803. [PubMed] [Google Scholar]
- Moasser MM, Srethapakdi M, Sachar KS, Kraker AJ, Rosen N. Cancer Res. 1999;59:6145–6152. [PubMed] [Google Scholar]
- Mustelin T, Hunter T. Science’s Stke [Electronic Resource] Signal Transduction Knowledge Environment. 2002;115:PE3. doi: 10.1126/stke.2002.115.pe3. [DOI] [PubMed] [Google Scholar]
- Ottenhoff-Kalff AE, Rijksen G, van Beurden EA, Hennipman A, Michels AA, Staal GE. Cancer Res. 1992;52:4773–4778. [PubMed] [Google Scholar]
- Park J, Cartwright CA. Mol Cell Biol. 1995;15:2374–2382. doi: 10.1128/mcb.15.5.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Meisler AI, Cartwright CA. Oncogene. 1993;8:2627–2635. [PubMed] [Google Scholar]
- Pena SV, Melhem MF, Meisler AI, Cartwright CA. Gastroenterology. 1995;108:117–124. doi: 10.1016/0016-5085(95)90015-2. [DOI] [PubMed] [Google Scholar]
- Roche S, Fumagalli S, Courtneidge SA. Science. 1995a;269:1567–1569. doi: 10.1126/science.7545311. [DOI] [PubMed] [Google Scholar]
- Roche S, Koegl M, Barone MV, Roussel MF, Courtneidge SA. Mol Cell Biol. 1995b;15:1102–1109. doi: 10.1128/mcb.15.2.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen N, Bolen JB, Schwartz AM, Cohen P, DeSeau V, Israel MA. J Biol Chem. 1986;261:13754–13759. [PubMed] [Google Scholar]
- Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E, Anfossi S, Gokden M, Dunn D, Roman JJ, O’Brien TJ, Tian E, Cannon MJ, Shaughnessy J, Jr, Pecorelli S. Int J Cancer. 2004;112:14–25. doi: 10.1002/ijc.20408. [DOI] [PubMed] [Google Scholar]
- Scherl-Mostageer M, Sommergruber W, Abseher R, Hauptmann R, Ambros P, Schweifer N. Oncogene. 2001;20:4402–4408. doi: 10.1038/sj.onc.1204566. [DOI] [PubMed] [Google Scholar]
- Sugimura M, Kobayashi K, Sagae S, Nishioka Y, Ishioka S, Terasawa K, Tokino T, Kudo R. Jpn J Cancer Res. 2000;91:395–398. doi: 10.1111/j.1349-7006.2000.tb00958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Harris JL, Huang W, Yan KW, Coughlin SR, Craik CS. J Biol Chem. 2000;275:26333–26342. doi: 10.1074/jbc.M002941200. [DOI] [PubMed] [Google Scholar]
- Tatsuka M, Ota T, Yamagishi N, Kashihara Y, Wada M, Matsuda N, Mitsui H, Seiki M, Odashima S. Mol Carcinog. 1996;15:300–308. doi: 10.1002/(SICI)1098-2744(199604)15:4<300::AID-MC7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Taylor SJ, Anafi M, Pawson T, Shalloway D. J Biol Chem. 1995;270:10120–10124. doi: 10.1074/jbc.270.17.10120. [DOI] [PubMed] [Google Scholar]
- Taylor SJ, Shalloway D. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- Wang LL, Richard S, Shaw AS. J Biol Chem. 1995;270:2010–2013. doi: 10.1074/jbc.270.5.2010. [DOI] [PubMed] [Google Scholar]
- Webster MA, Cardiff RD, Muller WJ. Proc Natl Acad Sci USA. 1995;92:7849–7853. doi: 10.1073/pnas.92.17.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]