Summary
Some superfamilies contain large numbers of protein domains with very different functions. The ability to refine the functional classification of domains within these superfamilies is necessary for better understanding the evolution of functions and to guide function prediction of new relatives. To achieve this, a suitable starting point is the detailed analysis of functional divisions and mechanisms of functional divergence in a single superfamily. Here we present such a detailed analysis in the superfamily of HUP domains. A biologically meaningful functional classification of HUP domains is obtained manually. Mechanisms of function diversification are investigated in detail using this classification. We observe that structural motifs play an important role in shaping broad functional divergence, whereas residue-level changes shape diversity at a more specific level. In parallel, we examine the ability of an automated protocol to capture the biologically meaningful classification, with a view to automatically extending this classification in the future.
Introduction
Proteins are made up of domains, which generally adopt well-defined globular 3D structures and perform specific functions, and are often considered to be fundamental units of protein evolution (Vogel et al., 2004). In the CATH database, domains are classified together in superfamilies when there is evidence that they are related by evolution, because they share high sequence identity, structural similarity, functional similarity or a combination thereof (Cuff et al., 2009b).
Most domain superfamilies consist of very few domains that all share the same function. In contrast, less than 5% of the total number of superfamilies contain large numbers of domains with highly diverse structures and functions (Goldstein, 2008;Chothia et al., 2009;Dessailly et al., 2009). Recent results suggest that such superfamilies account for at least 40% of predicted domains in genomes (Cuff et al., 2009a).
Because they consist of domains with diverse structures and functions, these superfamilies challenge the notion that homologous protein domains share similar structures and functions (Gerlt et al., 2001;Reeves et al., 2006;Dessailly et al., 2009). This is significant because most approaches for predicting function still rely on homology recognition (Lee et al., 2007).
These issues suggest a need to sub-classify domains according to function within superfamilies, and to recognise features that characterise the different sub-groups of functionally similar domains. The ability to classify and predict functions within diverse superfamilies relies in turn on a better understanding of the mechanisms of function diversification between homologous domains.
However, relatively few studies to date have been aimed at improving our understanding of the determinants of functional variation in these diverse superfamilies (Todd et al., 2001). Work by Babbitt and colleagues have concentrated on mechanistically diverse enzyme superfamilies, i.e. superfamilies of enzymes that catalyse different overall reactions with a common mechanistic step (Gerlt et al., 2001;Glasner et al., 2006). Six of these superfamilies are described in the Structure-Function Linkage Database (Pegg et al., 2006). Another recent study showed that the 294 largest superfamilies in CATH are very diverse structurally: domains in such superfamilies generally share a common structural core, but individual domains can display very large secondary structure embellishments relative to that superfamily core (Reeves et al., 2006). It was suggested that these embellishments may play a role in functional diversification, in line with other recent results indicating that indels can cause large length differences within domain superfamilies, and that they are often important for function (Sandhya et al., 2009). Finally, a large number of studies have focused on individual superfamilies, such as the Cupins (Agarwal et al., 2009) or the HAD superfamily (Burroughs et al., 2006).
One important conclusion from the extant body of work on the subject is that different superfamilies can use different strategies for achieving functional diversification. Whereas some superfamilies mainly achieve it via the exploration of different sets of residues or metal ions in their main active site (Glasner et al., 2006), emergence of diverse functions in others seem to be mainly due to structural variation within domains (Reeves et al., 2006), or to the recombination of domains with others (Dessailly et al., 2009).
Many function prediction algorithms have been developed and tested on large datasets of structures and sequences covering many superfamilies (Godzik et al., 2007;Redfern et al., 2008). However, it is likely that their performance will vary depending on the superfamily and the degree of structural and functional diversity captured within it. Therefore, as well as benchmarking over large sets of structures and sequences, it is also important to examine in detail the ability of these methods to capture functional divisions in diverse superfamilies.
Here, we describe a very detailed analysis of functional variation in a single large and diverse superfamily, and of the underlying mechanisms that generate such functional diversity. In parallel, we explore the possibility of using structure- and sequence-based automated methods to capture a biologically meaningful classification of domains within a superfamily.
HUP domains are chosen as a representative superfamily in which structural variation clearly plays a role in functional variation. This superfamily was selected because (a) it presents obvious structural variation with large embellishments; (b) it is very diverse functionally: the superfamily contains both enzymes and non-enzymes, enzymes that catalyse reactions with different mechanisms and different substrates, ligand types range from small ions to nucleic acids and proteins, and no common mechanistic step as observed in mechanistically diverse superfamilies is apparent; (c) a lot of structural data is available for this superfamily, notably because it has been specifically targeted by structural genomics projects; (d) a previous detailed evolutionary study of the superfamily by Aravind et al provides a good starting point for our functional analysis (Aravind et al., 2002).
Domains of known structure in the HUP superfamily are first manually classified based on their function and according to a biologically intuitive classification scheme. For this, HUP domains are first subdivided into manually curated functional sub-groups that group together domains with similar functional mechanisms but different specificities. Specific functions are further identified within these functional sub-groups. Both relevant literature and database annotations are exploited to obtain this multi-level and biologically intuitive classification, which is then used as a reference to refine our understanding of mechanisms underlying function diversification. Based on this analysis we derive guidelines towards an automated protocol to classify functions within this superfamily, and discuss the value of this protocol in classifying other very large diverse superfamilies where the size of the superfamily makes continuous manual curation intractable.
Results
HUP Core definition
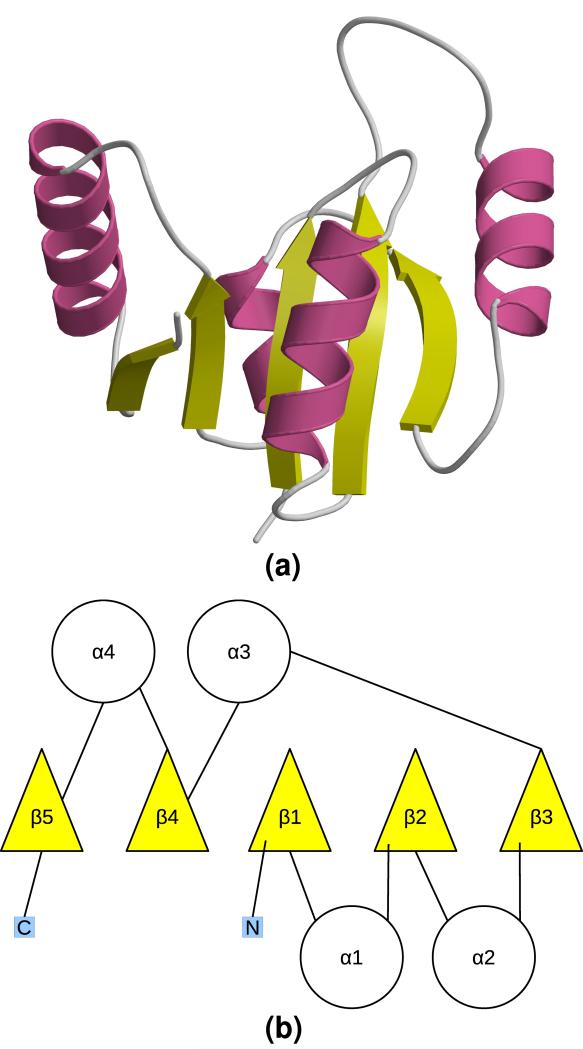
HUP domains adopt a Rossmann-like fold. As previously described, the HUP domain core is comprised of a five-stranded parallel β-sheet in a 5-4-1-2-3 configuration, surrounded on both sides by two α-helices (Aravind et al., 2002). Figure 1 illustrates the HUP domain core in three-dimensions and with a TOPS-like diagram (Westhead et al., 1999); core β-strands and α-helices are numbered sequentially along the sequence in the TOPS diagram, and this numbering scheme is used throughout the text; both the TOPS diagram and the three-dimensional structure of the core in Figure 1 illustrate the reference orientation referred to in the text. The core is defined as the set of elements of secondary structure that are common to all members of the HUP superfamily.
Figure 1.
HUP domain core: (a) three-dimensional representation of the HUP domain core, using CATH domain 2ielA00; (b) TOPS diagram (Westhead et al., 1999) illustrating the core secondary structure elements, which are numbered as referred to in the text. The N- and C-termini are represented as blue squares marked N and C, respectively. Both (a) and (b) illustrate the reference orientation referred to throughout the text. All three-dimensional molecular graphics were generated using Molscript (Kraulis, 1991) and rendered with Raster3D (Merritt et al., 1997).
Manual classification of functional sub-groups and analysis of the relationships between them
Nine Functional Sub-Groups (or FSGs) were defined in the HUP superfamily using information collected from the literature and several databases (see Methods), and all 85 non-redundant domains from that superfamily in CATH were assigned to them.
Here, FSGs were designed so as to group together proteins sharing obvious functional similarities that distinguish them from the rest of the superfamily (e.g. an identical catalytic mechanism for enzymes) but nevertheless allowing for some degree of functional diversity (e.g. different substrate specificities for enzymes). For the most part, these FSGs were defined on the basis of publications from experimental researchers working on members of this superfamily (Aravind et al., 2002), in order to ensure the final FSGs would match as closely as possible groupings suggested by experts in the available literature. Enzymatic FSGs group together enzymes with identical catalytic mechanisms but different substrate specificities, and non-enzymatic FSGs group together proteins participating in similar biological processes and sharing similar ligand-binding properties. Protein domains within FSGs were further classified according to their functional specificity (e.g. substrate specificity for enzymes).
Description of functional sub-groups (FSGs)
Table 1 shows the list of domains used in this study, sorted by FSGs and specificity, and provides information on their function and structure. Abbreviations given for each FSG in Table 1 are used throughout the text.
Table 1.
85 HUP domains from our dataset organised by FSGs. Column 1 lists the domain IDs from CATH, Column 2 gives the functional specificity, Column 3 gives the EC number, Column 4 shows the Multi-Domain Architecture (comma-separated list of CATH superfamilies to which domains belong, and simple graphical representation where different domains are represented by boxes separated by lines – HUP domains are always represented as white empty boxes), and Column 5 gives the quaternary structure. FSGs in which the HUP domain is catalytic are labelled with [E]. All these data were obtained from combined sources including articles describing the proteins and annotations from public databases such as UniProt, KEGG and Gene3D (see Methods). Domain IDs that have identical data in the other columns are together on the same line in the table.
| Domain ID | Specificity | EC number | Multiple Domain Architecture (CATH superfamily codes) | PQS |
|---|---|---|---|---|
| FSG 1. Electron Transfer Flavoproteins (ETF) | ||||
| 2a1uA01 1efpA02 1o97D01 |
ETFα | Non-enzyme | 3.40.50.620,3.40.50.1220 |
heterodimer |
| 1efvB00 1efpB00 1o97C00 |
ETFβ | Non-enzyme | 3.40.50.620
|
Heterodimer |
| FSG 2. Aminoacyl-tRNA synthetases (AATRS) [E] | ||||
| 2cyaA01 2cybA01 1zh0A01 2cycA01 |
Tyr-tRNA synthetase | 6.1.1.1 | 3.40.50.620,1.10.240.10 |
Homodimer |
| 2pidA01 | Tyr-tRNA synthetase | 6.1.1.1 | 3.40.50.620,1.10.240.10,NA.NA.NA.NA |
Homodimer |
| 1n3lA01 | Tyr-tRNA synthetase | 6.1.1.1 | 3.40.50.620,1.10.240.10,2.40.50.140 |
Homodimer |
| 1h3fB01 1jilA01 1wq4A01 2ts1A01 |
Tyr-tRNA synthetase | 6.1.1.1 | 3.40.50.620,1.10.240.10,3.10.290.10 |
Homodimer |
| 1y42X01 | Tyr-tRNA synthetase | 6.1.1.1 | 3.40.50.620,1.10.240.10,3.10.290.10,NA.NA.NA.NA |
homodimer |
| 1i6kA01 2ip1A01 2yy5A01 |
Trp-tRNA synthetase | 6.1.1.2 | 3.40.50.620,1.10.240.10 |
homodimer |
| 1r6uB01 | Trp-tRNA synthetase | 6.1.1.2 | 1.10.287.10,3.40.50.620,1.10.240.10 |
homodimer |
| 1wkbA01 | Leu-tRNA synthetase | 6.1.1.4 | 3.40.50.620,3.90.740.10,1.10.730.10,NA.NA.NA.NA |
monomer |
| 1obhA01 | Leu-tRNA synthetase | 6.1.1.4 | 3.40.50.620,3.90.740.10,2.30.210.10,1.10.730.10 |
monomer |
| 1ileA01 | Ile-tRNA synthetase | 6.1.1.5 | 3.40.50.620,3.90.740.10,1.10.730.10 |
monomer |
| 1irxA01 | Lys-tRNA synthetase | 6.1.1.6 | 3.40.50.620,NA.NA.NA.NA,NA.NA.NA.NA,NA.NA.NA.NA,1.10.10.350 |
monomer? |
| 1gaxA01 | Val-tRNA synthetase | 6.1.1.9 | 1.10.730.10,3.40.50.620,3.90.740.10,3.30.1170.10,1.10.287.380 |
monomer |
| 2csxA01 | Met-tRNA synthetase | 6.1.1.10 | 3.40.50.620,2.170.220.10,1.10.730.10 |
monomer |
| 2d5bA01 | Met-tRNA synthetase | 6.1.1.10 | 3.40.50.620,2.170.220.10,1.10.730.10,2.40.50.140 |
homodimer |
| 1pg2A01 | Met-tRNA synthetase | 6.1.1.10 | 3.40.50.620,2.20.28.20,1.10.730.10 |
homodimer |
| 1li5A01 | Cys-tRNA synthetase | 6.1.1.16 | 3.40.50.620,NA.NA.NA.NA |
monomer |
| 1j09A01 | Glu-tRNA synthetase | 6.1.1.17 | 3.40.50.620,3.90.800.10,1.10.1160.10,1.10.8.70,1.10.10.350 |
monomer |
| 2o5rA01 | Glu-tRNA synthetase | 6.1.1.17 | 3.40.50.620,3.90.800.10,1.10.1160.10,1.10.8.70,1.10.10.350 |
monomer |
| 1qtqA05 | Gln-tRNA synthetase | 6.1.1.18 | 3.40.50.620,3.90.800.10,1.10.1160.10,2.40.240.10 |
monomer |
| 1f7uA01 | Arginyl-tRNA synthetase | 6.1.1.19 | 3.30.1360.70,3.40.50.620,1.10.730.10 |
monomer |
| 1iq0A01 | Arg-tRNA synthetase | 6.1.1.19 | 3.30.1360.70,3.40.50.620,1.10.730.10 |
monomer? |
| FSG 3. ATP-pyrophosphatases (ATP-PPASE) [E] | ||||
| 1kqpA00 1wxiA00 |
NAD synthetase | 6.3.1.5 | 3.40.50.620
|
homodimer |
| 1m1zA02 | beta-lactam-synthase | 6.3.3.4 | 3.60.20.10,3.40.50.620 |
homodimer |
| 1k92A01 1korB01 1vl2A01 1kh1C01 |
Argininosuccinate synthase |
6.3.4.5 | 3.40.50.620,3.90.1260.10,1.20.5.470 |
homotetramer |
| 2dplA01 | GMP synthase | 6.3.5.2 | 3.40.50.620,3.30.300.10 |
not_sure |
| 1gpmA02 2ywbA02 |
GMP synthase | 6.3.5.2 | 3.40.50.880,3.40.50.620,3.30.300.10 |
homotetramer |
| 1ct9B02 | Asparagine synthetase B | 6.3.5.4 | 3.60.20.10,3.40.50.620 |
homodimer |
| 1q15D02 | CarA | NA | 3.60.20.10,3.40.50.620 |
homotetramer |
| FSG 4. Cryptochromes/DNA photolyases (C-DNAP) | ||||
| 1tezA01 | Deoxyribodipyrimidine photo-lyase [8-HDF] |
4.1.99.3 | 3.40.50.620,1.25.40.80,1.10.579.10 |
monomer |
| 2j07A01 | Deoxyribodipyrimidine photo-lyase [FMN] |
4.1.99.3 | 3.40.50.620,1.25.40.80,1.10.579.10 |
monomer |
| 1dnpA01 | Deoxyribodipyrimidine photo-lyase [MTHF] |
4.1.99.3 | 3.40.50.620,1.25.40.80,1.10.579.10 |
monomer |
| 1np7B01 | Cryptochrome DASH | Non-enzyme | 3.40.50.620,1.25.40.80,1.10.579.10 |
monomer |
| FSG 5. Nucleotidyltransferases (NT) [E] | ||||
| 1ej2A00 1f9aA00 |
Nicotinamide-nucleotide adenylyltransferase |
2.7.7.1 | 3.40.50.620
|
homohexamer |
| 1kqnB00 | Nicotinamide-nucleotide adenylyltransferase 1 |
2.7.7.1 | 3.40.50.620
|
homohexamer |
| 1nupB00 | Nicotinamide-nucleotide adenylyltransferase 3 |
2.7.7.1 | 3.40.50.620
|
homotetramer |
| 1lw7A01 | Nicotinamide-nucleotide adenylyltransferase [bi] |
2.7.7.1 | 3.40.50.620,3.40.50.300 |
homotetramer |
| 1mrzA01 | Riboflavin kinase/FMN adenylyltransferase |
2.7.7.2 | 3.40.50.620,2.40.30.30 |
monomer? |
| 1qjcA00 1tfuA00 1o6bA00 1vlhB00 1od6A00 |
Phosphopantetheine adenylyltransferase |
2.7.7.3 | 3.40.50.620
|
homohexamer |
| 1v47B02 | Sulfate adenylyltransferase |
2.7.7.4 | 3.10.400.10,3.40.50.620 |
Homodimer? |
| 1jhdA01 | Sulfate adenylyltransferase |
2.7.7.4 | 3.10.400.10,3.40.50.620 |
Homodimer |
| 1r6xA02 | Sulfate adenylyltransferase |
2.7.7.4 | 3.10.400.10,3.40.50.620,3.40.50.300 |
Homohexamer |
| 1yumA00 1k4kB00 |
Nicotinate-nucleotide adenylyltransferase |
2.7.7.18 | 3.40.50.620
|
Monomer |
| 1kamA00 | Nicotinate-nucleotide adenylyltransferase |
2.7.7.18 | 3.40.50.620
|
Homodimer |
| 1cozA00 | Glycerol-3-phosphate cytidylyltransferase |
2.7.7.39 | 3.40.50.620
|
homodimer |
| FSG 6. Pantothenate Synthetases (PS) [E] | ||||
| 1mopA01 1ihoA01 |
Pantothenate synthetase | 6.3.2.1 | 3.40.50.620,3.30.1300.10 |
homodimer |
| 1v8fA01 2ejcA01 |
Pantothenate synthetase | 6.3.2.1 | 3.40.50.620,3.30.1300.10 |
homodimer? |
| FSG 7. tRNA specific 2-thiouridylases (TRMU) [E] | ||||
| 2derB01 2hmaA01 |
tRNA-specific 2- thiouridylase |
2.1.1.61 | 3.40.50.620,2.30.30.280,2.40.30.10 |
monomer? |
| FSG 8. Phosphoadenylyl-sulfate reductases (PAPSR) [E] | ||||
| 1surA00 | Phosphoadenylyl-sulfate reductase |
1.8.4.8 | 3.40.50.620
|
homodimer |
| FSG 9. Universal Stress Proteins A (USPA) | ||||
| 1mjhB00 | MJ0577 | NA | 3.40.50.620
|
homodimer |
| 1jmvA00 | Universal Stress Protein A |
Non-enzyme | 3.40.50.620
|
homodimer |
| Poorly characterised proteins | ||||
| 1tq8A00 | Uncharacterised protein | NA | 3.40.50.620
|
homotetramer? |
| 1q77A00 | Uncharacterised protein | NA | 3.40.50.620
|
homotetramer? |
| 1ru8A01 | Uncharacterised protein | NA | 3.40.50.620,3.90.1490.10 |
homodimer? |
| 2dumC00 | Uncharacterised protein | NA | 3.40.50.620
|
homodimer? |
| 2pg3A00 | Uncharacterised protein | NA | 3.40.50.620
|
homodimer? |
| 2ielA00 | Uncharacterised protein | NA | 3.40.50.620
|
homooctamer? |
| 2pfsA01 | Uncharacterised protein | NA | 3.40.50.620
|
homodimer? |
Three FSGs consist of non-enzymatic HUP domains, whereas six consist exclusively of enzymes and display remote similarities in their catalytic mechanism. For example, one of these enzymatic FSGs is labelled Class I Aminoacyl-tRNA synthetases (AATRS), and consists of enzymes that all catalyse the ATP-dependent attachment of amino acids to the acceptor end of their cognate tRNAs. Enzymes with different amino acid specificities can however be found within this FSG. More details on FSGs are given in Supp Mat S1.
Mapping of functional sub-groups onto HUP domain sequence space
Figure 2 illustrates the functional space for the HUP superfamily when considering all predicted HUP domains in known sequences and not only the HUP domains of known structure used to define the FSGs. There are 34568 predicted HUP domains in Gene3D version 6.0 (Yeats et al., 2008), and most have poorly understood or unknown functions. Figure 2 was generated by considering non-redundant representatives of these predicted HUP domains, and further details on the procedure to generate it are given in Supp Mat S2. Each dot in Figure 2 represents a domain sequence. Any pair of domains is linked if they share detectable sequence similarity. Dots are coloured according to the FSG to which they are likely to belong based on their functional annotation in UniProt. It must be noted that a significant proportion (around 50%) of predicted HUP domains cannot be assigned to any of the nine FSGs on the basis of their functional annotations alone (because no functional annotations are available for them), and that yet unknown novel FSGs will most likely be defined as more members of the HUP superfamily become functionally and structurally characterised. Figure 2 shows that most FSGs are subdivided into smaller clusters that correspond roughly to function specificities, and the different FSGs are not easily discriminated by sequence similarity measures. Further analysis of this plot is provided in Supp Mat S2.
Figure 2.

Sequence space in the HUP superfamily. Each dot represents a domain and domains are coloured according to the FSG to which they are likely to belong (ETFs: red, AAtRSs: green, ATP-PPases: greenblue, C-DNAPs: violet, NTs: yellow, PSs: cyan, PAPSs: brown, tRMUs: light-blue, USPAs and unclassified: white). Captions in the Figure represent different functional specificities within these FSGs.
Functional links between FSGs
In spite of their obvious diversity, some aspects of function seem to be rather well conserved between the different HUP domains. For example, all HUP domains except ETFα and members of the cryptochrome/DNA photolyase family, bind adenine-nucleotides in the main active site cavity, at the C-terminal end of the core β-strands. Moreover, several HUP enzymatic sub-groups do present similarities in the types of reactions they catalyse. These and other functional links between FSGs are discussed further in Supp Mat S3.
Substrate/cofactor distribution in FSGs
The majority of HUP domains bind to ATP or AMP. However, the other ligands are very diverse, ranging from large bulky molecules such as tRNAs in AATRSs to small ions such as sulfate in sulfate adenylyltransferase. Ligands bound by all HUP domains in our dataset are shown in Table S16.
Conservation of Functionally Important Positions in HUP Superfamily
A structural alignment of all representative structures at the 60% sequence identity level shows that very few residues are significantly conserved across the whole superfamily. Those that are conserved are all hydrophobic residues which are buried in the domain core. Within FSGs, functional residues tend to be better conserved even though some key residues can vary because features like substrate-specificity, which determine the nature of catalytic residues to a certain extent, are not conserved. Detailed results from our residue conservation analysis, including multiple sequence alignments, conservation scores and functional residue annotations are provided at http://www.cathdb.info/wiki/docu.php?id=hupfam.
Comparative Structural Analysis of Functional Sub-Groups
Multiple Domain Architectures (MDAs) in the different FSGs
HUP domains are found in a wide variety of multi-domain contexts. MDAs for all proteins in the dataset are shown in Table 1. The majority of extra domains that combine with HUPs are inserted at either termini of the HUP domain (see Supp Mat S4). It is clear that recombination with other domains can affect a domain’s function (Bashton et al., 2007). Here, we have identified seven different ways by which combination with extra domains affects the function of the proteins containing HUP domains (see Supp Mat S5 and Table S17). An important point is that many different MDAs can be found within an FSG, but that domains with different specificities within an FSG can possess the same MDAs. Therefore, MDAs do not constitute a sufficient criterion to assign a relative to a particular FSG, or to a particular functional specificity.
Global Domain Structural Similarity as a Marker of Protein Function
We examined the extent to which global structure similarity between HUP domains distinguishes between the FSGs. Our results, presented in more detail in Supp Mat S6 indicate that structural data contains a signal for functional classification of HUP domains, but that this signal is weakened by the prominence of the common superfamily core when comparing whole domain structures. This points to the importance of relying on structural embellishments to the core to get insights on function, as explored in the next section.
Secondary structure embellishments in different FSGs
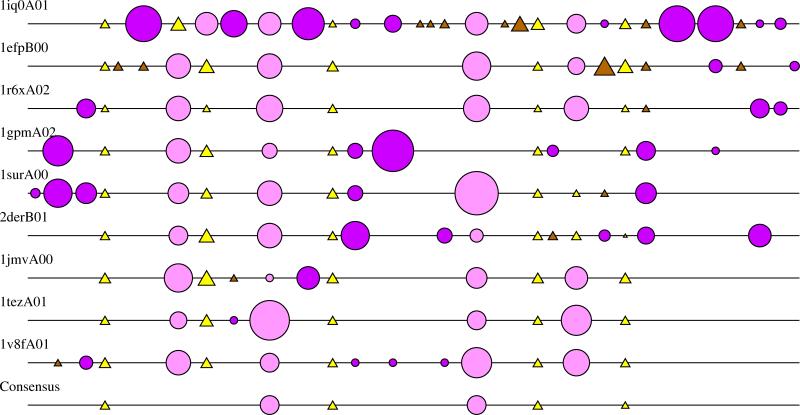
Based on the previously mentioned definition of the HUP domain core, we have identified so-called secondary structure embellishments in each FSG. We define these embellishments as insertions relative to the core, which contain at least three residues and one distinct element of secondary structure. Figure 3 provides an alignment of the secondary structures of representatives from the nine different FSGs, and thereby illustrates the core secondary structures which are common to all members of the superfamily, as well as the embellishments causing differences in secondary structure contents between the different FSGs.
Figure 3.
2DSEC diagram (Reeves et al., 2006) showing an alignment of the secondary structure elements of representatives from the nine FSGs in the HUP superfamily. Each line in the plot represents a protein. Elements of secondary structure are represented by circles for α-helices and triangles for β-sheets. Conserved and non-conserved α-helices are coloured pink and magenta, respectively. Conserved and non-conserved β-sheets are coloured yellow and brown, respectively. The size of circles and triangles reflects the size of the corresponding secondary structures. The conserved elements constitute the common core of the superfamily.
For each FSG, we analysed (a) the topological insertion points of embellishments, (b) the three-dimensional location of the embellishments relative to the HUP domain core, and (c) whether the embellishments are directly involved in molecular function.
Topological Insertion Points of Embellishments
Manual observation of all embellishments shows that these are common at both termini, and after core β strands 2, 3 and 4, but that they almost never occur after core α helices (see Figure S11b). This suggests a bias for embellishments to be inserted after core β strands rather than core α helices. Embellishments are also very rare in the loop that follows core β strand 1, possibly due to the crucial role of that loop for binding nucleotides in most HUP domains (Aravind et al., 2002).
Three-dimensional location of embellishments relative to the core
Here, all three-dimensional locations are given relative to the reference orientation shown in Figure 1a, where the top and bottom of the fold are towards the C-terminus and the N-terminus of core β strands, respectively.
As a result of them being preferentially inserted after core β strands, most embellishments are located at the top of the core. On the other hand, embellishments are very rarely observed at the bottom of the core. The few of those that do are N-terminal embellishments that are involved in connecting the HUP domains to previous domains in the same polypeptide. Supplementary Material Table S19 provides a more detailed description of the three-dimensional location of all embellishments relative to the core.
The clear-cut preference for embellishments to be located towards the top of the Rossmann-fold seems consistent with the fact that such embellishments are generally directly involved in function, and thus participate in functional differentiation between related proteins. Indeed, the main active site of most HUP domains is located at the top of the core β-sheet, in the region where most embellishments are observed. These insertions often modify the shape of the active site, and experimental results have demonstrated their importance for substrate- or cofactor-binding in several HUP domain-containing proteins. The absence of embellishments at the bottom of the core between core α helices and core β strands, on the other hand, may suggest strong selection pressure against insertions in these regions, possibly because such insertions would be detrimental to the stability of the three-dimensional structures of HUP domains.
Function of Embellishments
The importance of HUP embellishments in different aspects of molecular function is summarized in Table 2 and illustrated in Figure 4. We have identified five distinct categories of molecular function and checked whether individual embellishments were involved in any of these (see Table 2).
Table 2.
Insertion points (columns) and functional role (rows) of embellishments observed in the different FSGs of the HUP domain superfamily. FSGs are identified by their abbreviations introduced in Table 1. The first line “Ligand” covers all embellishments involved in catalysis, or in binding ligands, substrates and cofactors. See text for a complete description of the other categories.
| N-β1 | β1-α1 | α1-β2 | β2-α2 | α2-β3 | β3-α3 | α3-β4 | β4-α4 | α4-β5 | β5-C | |
|---|---|---|---|---|---|---|---|---|---|---|
| LIGAND | AAtRSs | C-DNAPs;PSs | AAtRSs;TRMUs | PAPSs | ATP-PPases;NTs;TRMUs;PAPSs | |||||
| DOMAIN | ATP-PPases | C-DNAPs | AAtRSs;ATP-PPases;TRMUs | ATP-PPases;TRMUs | ATP-PPases | |||||
| SUBUNIT | NTs | AAtRSs;ATP-PPases;PSs;PAPSs | ETFs | ETFs;ATP-PPases | ||||||
| PROTEIN | ETFs | ETFs;PAPSs | ||||||||
| FLEXIBLE | AAtRSs |
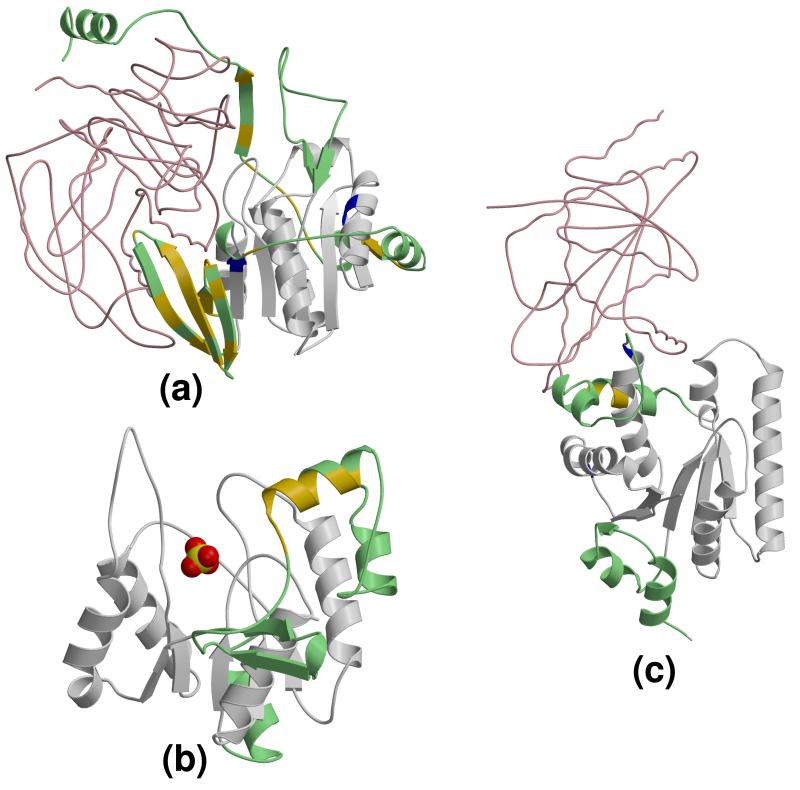
Figure 4.
Examples of HUP embellishment being involved in different aspects of molecular function. HUP domains are displayed in cartoons. HUP domain cores are coloured in grey and presented in the reference orientation (see Figure 1) in all subfigures. Embellishments to the HUP domain cores are coloured in different shades of green and blue. (a) tRNA 2-thiouridylase (PDB 2deu) in complex with a tRNA fragment (displayed in wireframe and coloured orange) and an AMP molecule (displayed and coloured in CPK). The light blue and dark green embellishments are important for binding the tRNA. (b) Carbapenam synthetase (PDB 1q15) contains two domains. The N-terminal domain is coloured orange, and the C-terminal domain is the HUP domain. All four embellishments to the HUP domain (coloured in light blue, light green, dark green and dark blue, respectively) are involved in contacts with the extra N-terminal domain. (c) Tyrosyl-tRNA synthetase (PDB 1h3f) is a homodimeric enzyme. One subunit is represented by a light-pink α-trace, whereas the HUP domain of the other subunit is displayed in cartoons. A major HUP domain embellishment (coloured light blue) is important for contacts between the two subunits. For clarity reasons, not all residues are shown in this figure. (d) Electron transfer flavoproteins (PDB 1efv) consist of two different chains, designated α (coloured light gold and displayed in cartoons) and β. Embellishments to the HUP domain in subunit β (coloured light blue and light green) are very important for stabilising the complex with subunit α.
As mentioned above, a large number of embellishments observed in the HUP domains are located in the vicinity of the main active site, at the top of the core β-sheet. Such embellishments are often involved in modifying the shape of the active site, dynamically protecting it from the solvent, or also directly contacting the substrates and cofactors thus affecting specificities. In particular, embellishments inserted after core strands 2 and 3, or at the C-terminus, are often observed to be important for catalysis (see Figure 4a). Embellishments from 7 out of 9 FSGs participate in ligand-binding or catalysis in the main active site region, the two exceptions being non-enzymatic FSGs of ETFs and USPAs (see Table 2).
Several embellishments from different FSGs are also involved in contacts with other domains along the polypeptide chain, or with other protein subunits. Embellishments involved in inter-domain contacts are often inserted at the top of the HUP domain core as well (see Figure 4b). In four FSGs, embellishments inserted after core strand 3 are involved in contacts with other subunits, and such embellishments are often located towards the back of the HUP domain core (see Figure 4c).
Finally, in spite of the limited amount of structural data available for examining this type of interface in this superfamily, two examples could be identified where HUP embellishments participate in contacts with other proteins, as in electron-transfer flavoproteins, where subunits α and β have embellishments that are extensively involved in the complex interface (see Figure 4d).
Automated Approaches to Recover Manual Function Classification
The functional classification of HUP domains presented above was obtained manually through a cumbersome and lengthy process. Such a manual approach would be difficult to implement on a larger-scale, for example in an attempt to classify functions in all large and diverse superfamilies. Therefore, it is important to explore automated approaches that would at least guide classification in other superfamilies. In this section we examine the ability of automated methods to recover the different levels of our manual classification.
Recovering Functional Specificity Groups using Sequence Signals
First, we considered the possibility of recovering our functional specificity groups by exploiting sequence profiles derived from predicted HUP domains in all known sequences. GeMMA is an automated method that subdivides related sequences into fine-grained clusters (or GeMMA families) according to function (Lee et al., 2010).
When checking their contents in terms of the 85 HUP domains from our dataset, we find that GeMMA families match fairly well to groups of HUP domains with similar functional specificities, with only one case where domains with different specificities are found in the same GeMMA family, and four cases where domains with the same specificity are found in different GeMMA families (See Table S21). In all these cases but one, functional reasons can be found to explain the discrepancies. The most notable is that Ile-, Leu- and Val-tRNA synthetases end up in the same GeMMA family in spite of their different specificities. However, it has long been recognized that these three enzymes have extremely similar mechanisms, to the extent that they often catalyse each other’s reaction by mistake, and that these common errors necessitate the existence of specific correction mechanisms (Nureki et al., 1998). Interestingly, an archaeal Leu-tRNA synthetase representative, which features a unique structural extension and lacks another, bacterial-specific insertion, ends up in its own GeMMA family (Fukunaga et al., 2005). The other mismatches are explained further in Supp Mat S7.
As expected, catalytic residues show a great degree of conservation within the functionally specific GeMMA families. Either catalytic or ligand-binding residue data is available for a total of 24 GeMMA families, but only 11 have both. We observe that in all but two of these 11 families, catalytic residues are more conserved than ligand-binding residues, which are themselves better conserved than residues not known to be functional. The exceptions are the GeMMA families corresponding to EC numbers (i.e. functional specificities) 6.3.5.4 and 6.3.4.5, both belonging to the FSG of ATP-PPases, and in which catalytic residues are less conserved than ligand-binding residues. However, not enough functional residue data is available in this superfamily for safely drawing general trends in terms of their variability. The different patterns of residue conservation in different GeMMA families result in very recognisable sequence signatures that characterise them (see Figure S13). See also http://www.cathdb.info/wiki/docu.php?id=hupfam for more detailed data regarding our analysis of residue conservation and functional residues in this superfamily.
Recovering FSGs using Local Structural Motifs
We have observed that different secondary structure decorations to the core are associated with variations in functionally important molecular interactions. Therefore we examined whether such local structural differences could be exploited to classify relatives into their FSGs.
FLORA is an automated method which identifies distinctive structural motifs in predefined sets of functionally related homologous proteins and can use these motifs to predict the function of novel structures (Redfern et al., 2009). Here, we examined whether FLORA could be used to recover the manually curated FSGs.
To this aim, FLORA was used to identify distinctive motifs (or “templates”) for each FSG and we tested its ability to recognise other members of the same FSG using these motifs (see Methods for details).
The performance of FLORA was measured by re-classifying each of the 73 HUP domains from FSGs with at least 3 members (see Methods), using a leave-one-out approach. Each HUP domain was removed sequentially and FLORA templates were built for each FSG using the remaining 72 domains. The left-out domain was then re-assigned to the template it matched best. Using this approach, all 73 HUP domains were assigned to their correct FSG. Figure 5 illustrates a few examples where the motifs detected by FLORA clearly relate to the functions of HUP domains. All FLORA motif residues are shown in Table S20. These results suggest that a method capturing structurally conserved local motifs can assist in classifying structures to their appropriate FSGs in this superfamily.
Figure 5.
Functional role of motifs detected by FLORA and embellishments in HUP domains. In all three subfigures, the HUP domain is displayed in cartoons, with the core of the domain coloured grey, the embellishments coloured green, the motifs identified by FLORA coloured blue, and the residues that are identified by FLORA and that are also in embellishments are coloured in yellow. For clarity, extra domains are not shown in this figure. (a) Human Electron Transfer Flavoprotein subunit β (PDB structure 1efv). FLORA detects motifs in two embellishments that are very important for mediating the interaction of subunit β with the other subunit in the complex (displayed as a pink coil). (b) Yeast ATP sulfurylase (PDB 1r6x); a sulfate ion displayed as CPK indicates the location of the active site. One single motif is detected by FLORA in ATP sulfurylase; it is centred on a helix that is part of a large C-terminal embellishment that is specific to the FSG of nucleotidyltransferases. In addition, this helix is located on the top of the main active site, and several of its residues have been shown to be important for substrate binding in other members of this FSG such as phosphopantetheine adenylyltransferase (Izard, 2002). (c) Human tyrosyl-tRNA synthetase (PDB 1n3l). Here, only two very small motifs are detected by FLORA. One lies within a large embellishment that is common to all aminoacyl-tRNA synthetases, and both are located in the interface with the other subunit (displayed as a pink coil) of the tyrosyl-tRNA synthetase homodimer.
Discussion
Here, we describe extensive functional diversity in the HUP superfamily. Differences in structural embellishments are a common structural cause for functional change between homologues in this superfamily, and structural embellishments are shown to be often directly involved in function for roles such as catalysis and molecular binding. The major functional subdivisions (i.e. FSGs) in this superfamily are generally characterised by embellishments that are inserted in specific points relative to the common superfamily core (even though embellishments can vary in shape and length within the FSGs, depending on the specificity of the domains in which they are found). Embellishments are therefore promising for function classification and prediction. For example, a set of extra β-strands in ETF subunits constitute a prominent structural feature of these proteins and could in theory be used to predict whether an uncharacterised HUP domain sequence is an ETF. Embellishments have been the subject of extensive studies for their structural (Jiang et al., 2007), functional (Reeves et al., 2006;Sandhya et al., 2008;Sandhya et al., 2009) and evolutionary properties (Wolf et al., 2007). An increasing body of work suggests that protein fragments of the size of most embellishments in this study may be very useful for predicting protein function from structural data (Kolodny et al., 2006;Manikandan et al., 2008), and detailed structural and functional studies of embellishments such as the one presented here may provide valuable data for developing approaches for doing so.
In addition to providing a very detailed benchmark for functional classification in the important superfamily of HUP domains, results from this study shed new light on issues at stake when attempting to classify domains according to function. This is important if we are ever to be able to do so automatically. Functional classifications of different levels of complexity have been derived for a small number of superfamilies but these were all obtained manually (Nagano et al., 2002;Leipe et al., 2002), and no automated approaches to apply on a large-scale have been proposed so far. Standard annotation schemes such as EC numbers can be helpful in this context but they should be used with caution as all of them suffer from significant drawbacks (Rison et al., 2000;Babbitt, 2003). Two useful resources which provide classification of related proteins according to function are the SFLD for mechanistically diverse superfamilies (Pegg et al., 2006) and PANTHER which classifies proteins in families and subfamilies indexed by function (Thomas et al., 2003). However both of these resources depend on significant levels of manual curation and therefore do not solve the issue.
Whilst it is relatively straightforward for us to classify protein domains with identical functions into functional specificity groups, classification at coarser levels of functional similarity (i.e. FSG-level in this study) is highly challenging. This is mainly because functional links between members of different FSGs are such that FSGs may differ depending on the functional criterion being used to define them. This is in part the result of the complex evolutionary processes that have given rise to novel functions within a superfamily (Todd et al., 2001;Gerlt et al., 2001).
Therefore, defining FSGs was the most difficult issue in this work. Our analysis soon revealed that no existing approaches gave reasonable groupings of proteins, at this level of functional similarity, and that the only possible way to identify FSGs was by manual curation, using the available literature. Yet, identifying such FSGs and the common properties of the proteins in them can be very useful notably for predictive purposes. For example, it can be very helpful to know whether a protein is an aminoacyl-tRNA synthetase or not (i.e. FSG-level definition), even if the amino acid on which it acts is still unknown (i.e. functional specificity-level definition). The manual curation necessary for defining FSGs is a time-consuming process, and it is not practical in the long term for classifying proteins from all superfamilies.
Therefore, guidance is needed from automated classification protocols which exploit specific features distinguishing functionally diverse relatives within superfamilies. A number of methods have been developed for automated functional classification (Brown et al., 2007;Redfern et al., 2008;Lee et al., 2010), most of which rely on the identification of specificity-determining residues which are conserved within functional families but not conserved across them (Reva et al., 2007;Capra et al., 2008). The patterns of residue conservation that we observed in the HUP superfamily suggest that such methods might be helpful for classification of some of the specificity groups but not for the FSGs as residue positions are generally not well conserved within those. Indeed, the automated sequence-based tools we explored to help us in the classification did not work as the sequence signal was not useful for classifying at the FSG level (see Figure 2 where FSGs are thoroughly mixed).
In this study, we illustrate that sequence data is useful for classifying functions at the specificity level but that using it for FSG-level classification remains difficult, and that structural data is important for this level (see above). Following this logic, we propose an automated classification protocol (see Supp Mat S9 and Figure S15 for more details) whereby functional specificity groups are first identified using approaches such as GeMMA. Once specificity groups are identified, they can in some cases be clustered on the basis of existing annotations of characterised members of these groups. For example, if all GeMMA families with the same EC number (down to the 3rd digit) are merged together, we find that merged GeMMA clusters (hereby referred to as “predicted FSGs”) correspond to the manually curated enzymatic FSGs (i.e. contain the same domain members of known structure). Exceptions include two domain structures (1np7B01 and 1q15D02) that we were able to classify manually in enzymatic FSGs using information from the literature (see Table 1), but that are missing in the predicted FSGs because these domains, and all other members of their respective GeMMA families, are not annotated with EC numbers in public databases. The 3D structures in each predicted FSGs can then be used as input for a method such as FLORA to derive structural templates, which can then be used to classify the unannotated members of the predicted FSGs. For example, applying FLORA to derive templates from the structures in the predicted FSGs and using these templates to scan the unclassified domains (1np7B01 and 1q15D02, see above) allowed us to classify them correctly into their predicted FSGs, thereby improving the match between the predicted and the manually curated FSGs.
Thus, a multi-step approach that combines GeMMA and FLORA, or any other analogous methods, can recover a complex functional classification of domains in a superfamily with reasonable accuracy and can be used to maintain and extend the classification automatically. Importantly, this protocol allowed us to classify (and thereby functionally annotate) 78% of the 34568 sequences in the HUP superfamily into FSGs (a total of 27138 sequences – see Figure S15). Accuracy remains an issue but automated guidance would at least be very valuable to ease the highly cumbersome process of manual curation. This is very promising in the context of annotating the numerous functionally uncharacterised sequences that belong to large and diverse superfamilies.
Finally, the detailed study of HUP domains presented here provides explanatory mechanisms for understanding how functional diversity can emerge within large and diverse superfamilies, i.e. via a combination of large structural insertions that cause divergence of coarser groups with generally different functional mechanisms, followed by more fine-tuned divergence within these groups via specific residue mutations.
Experimental Procedures
Dataset
All structural analyses presented in this paper were performed using a set of 85 domains selected from CATH v3.2 (Cuff et al., 2009b) superfamily 3.40.50.620 at the 60% sequence identity level (this cut-off was chosen as it has been shown that proteins sharing more than 60% sequence identity generally have the same function (Addou et al., 2009)), apart from the FLORA analysis which was performed using a subset of this dataset (see below). This set can be obtained from the CathDomainList file, which is available for download on the CATH website (http://www.cathdb.info/). In a few cases, domain boundaries had to be re-defined for structural comparisons to make sense from an evolutionary point of view. To ensure we were considering all functions with a known structure in the HUP superfamily, Cys-tRNA synthetase domain structure 1li5A01, which was not yet classified in CATH v3.2 was manually added to the initial list of domains in the dataset.
Definition of Functional Sub-Groups
FSGs were initially defined based on an extensive analysis of the available literature on the superfamily. Known functional information and expert opinions on the division of the HUP superfamily into relevant functional groupings were considered, together with annotations gathered from several databases including PDBsum (Laskowski, 2009), UniProt (UniProt Consortium, 2009), KEGG (Kanehisa et al., 2008) and Gene3D (Yeats et al., 2008). GO terms (The Gene Ontology Consortium, 2000), Multiple Domain Architectures and EC numbers (for enzymes) were also taken into account. CATH domains were mapped to these annotations using our in-house database Gene3D (Yeats et al., 2008). Multi-domain architectures for the 85 proteins with HUP domains in our dataset were obtained by considering all their CATH domain assignments in Gene3D.
Embellishment analysis
Embellishments were defined as insertions relative to the common superfamily core that consisted of at least 3 residues and contained at least one element of secondary structure. Embellishments were identified manually by looking at structures of HUP domains and subtracting from them the structural core (see Figure 1). The structural core itself was manually defined in a previous step by analysing structures from all members of the superfamily in CATH (Cuff et al., 2009b), and identifying those elements of secondary structures that were conserved in at least 80% of domains. The program 2DSEC (Reeves et al., 2006) was used to produce two-dimensional projections of a multiple structure alignment of representatives from the nine FSGs generated with CORA (Orengo, 1999).
Functional roles of embellishments were identified from the relevant literature, from PDB structures of the HUP domains in complex with their ligands, and from databases detailing functional roles of individual residues such as the Catalytic Site Atlas (Porter et al., 2004) for catalytic residues and LigASite (Dessailly et al., 2008) and PDBsum (Laskowski, 2009) for ligand-binding residues.
FLORA analysis
FLORA (Redfern et al., 2009) is a program that aims to detect structural features that distinguish protein domains that share the same function from other related domains with different functions. Details of the algorithm as applied in this work are given in Supp Mat S8. Here, we have adopted a leave-one-out approach to test the ability of FLORA to re-assign HUP domains to their correct FSG. The dataset used for the FLORA analysis consists of the 73 domains from our original dataset that belong to FSGs with at least 3 structures (see Table 1), as necessitated by the leave-one-out procedure. FLORA can be obtained from the authors upon request.
GeMMA analysis
GeMMA is a program which follows an agglomerative clustering protocol in order to cluster together a set of input sequences into sequence families (Lee et al., 2010). GeMMA performs iterative all-against-all profile-profile comparison of a set of sequence clusters followed by merging of the most similar clusters and then re-alignment of the merged clusters, and uses existing software for multiple sequence alignment and profile-profile comparison. Clustering stops when profile-profile comparisons reach an e-value cut-off of 1e-30 that was derived by maximising the overlap between GeMMA families and subfamilies of sequences having the same function. GeMMA was run using all 34568 HUP sequences in the Gene3D v6.0 database (Yeats et al., 2008) as input, resulting in a total of 2436 GeMMA families. Both GeMMA and the input set of sequences in FASTA format can be obtained from the authors upon request.
Analysis of residue conservation
At the level of the whole superfamily, residue conservation was obtained by producing a multiple structure alignment of representative HUP domains at the 60% sequence identity level using CORA (Orengo, 1999), and computing conservation scores from this multiple structure alignment using Scorecons (Valdar, 2002).
Within FSGs, residue conservation scores were computed as follows: first, all GeMMA families containing one of the 85 domains in our structure dataset were mapped to their corresponding FSG, by automatically checking the UniProt (UniProt Consortium, 2009) description lines of sequences in the GeMMA families and comparing them with the functions describing the FSGs. The program cd-hit (Li et al., 2006) was then run on the set of GeMMA families that map to a given FSG, so as to remove redundancy at 40% sequence identity cut-off. T-Coffee (Notredame et al., 2000) was then run on the final set of non-redundant sequences to re-align all the FSG sequences from the different corresponding GeMMA families. Finally, Scorecons (Valdar, 2002) was used to compute conservation scores from these T-Coffee alignments.
Scorecons was also used to identify conserved positions within GeMMA families. Catalytic residues were obtained from the CSA (Porter et al., 2004) and binding residues from LigASite (Dessailly et al., 2008).
Supplementary Material
Acknowledgments
The authors wish to acknowledge the European Union Framework Program 7 Impact grant and the Protein Structure Initiative of the National Institute for General Medicine at the National Institutes of Health, for financial support. We also thank Marie Manandise for helpful comments in writing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Addou S, Rentzsch R, Lee D, Orengo CA. Domain-based and family-specific sequence identity thresholds increase the levels of reliable protein function transfer. J. Mol. Biol. 2009;387:416–430. doi: 10.1016/j.jmb.2008.12.045. [DOI] [PubMed] [Google Scholar]
- Agarwal G, Rajavel M, Gopal B, Srinivasan N. Structure-based phylogeny as a diagnostic for functional characterization of proteins with a cupin fold. PLoS. One. 2009;4:e5736. doi: 10.1371/journal.pone.0005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Koonin EV. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA. Proteins. 2002;48:1–14. doi: 10.1002/prot.10064. [DOI] [PubMed] [Google Scholar]
- Babbitt PC. Definitions of enzyme function for the structural genomics era. Curr. Opin. Chem. Biol. 2003;7:230–237. doi: 10.1016/s1367-5931(03)00028-0. [DOI] [PubMed] [Google Scholar]
- Bashton M, Chothia C. The generation of new protein functions by the combination of domains. Structure. 2007;15:85–99. doi: 10.1016/j.str.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Brown DP, Krishnamurthy N, Sjolander K. Automated protein subfamily identification and classification. PLoS. Comput. Biol. 2007;3:e160. doi: 10.1371/journal.pcbi.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 2006;361:1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- Capra JA, Singh M. Characterization and prediction of residues determining protein functional specificity. Bioinformatics. 2008;24:1473–1480. doi: 10.1093/bioinformatics/btn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C, Gough J. Genomic and structural aspects of protein evolution. Biochem. J. 2009;419:15–28. doi: 10.1042/BJ20090122. [DOI] [PubMed] [Google Scholar]
- Cuff A, Redfern OC, Greene L, Sillitoe I, Lewis T, Dibley M, Reid A, Pearl F, Dallman T, Todd A, Garratt R, Thornton J, Orengo C. The CATH hierarchy revisited-structural divergence in domain superfamilies and the continuity of fold space. Structure. 2009a;17:1051–1062. doi: 10.1016/j.str.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff AL, Sillitoe I, Lewis T, Redfern OC, Garratt R, Thornton J, Orengo CA. The CATH classification revisited--architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009b;37:D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessailly BH, Lensink MF, Orengo CA, Wodak SJ. LigASite--a database of biologically relevant binding sites in proteins with known apo-structures. Nucleic Acids Res. 2008;36:D667–D673. doi: 10.1093/nar/gkm839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessailly BH, Redfern OC, Cuff A, Orengo CA. Exploiting structural classifications for function prediction: towards a domain grammar for protein function. Curr. Opin. Struct. Biol. 2009;19:349–356. doi: 10.1016/j.sbi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Yokoyama S. Crystal structure of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii reveals a novel editing domain orientation. J. Mol. Biol. 2005;346:57–71. doi: 10.1016/j.jmb.2004.11.060. [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu. Rev. Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- Glasner ME, Gerlt JA, Babbitt PC. Evolution of enzyme superfamilies. Curr. Opin. Chem. Biol. 2006;10:492–497. doi: 10.1016/j.cbpa.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Godzik A, Jambon M, Friedberg I. Computational protein function prediction: are we making progress? Cell Mol. Life Sci. 2007;64:2505–2511. doi: 10.1007/s00018-007-7211-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein RA. The structure of protein evolution and the evolution of protein structure. Curr. Opin. Struct. Biol. 2008;18:170–177. doi: 10.1016/j.sbi.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Izard T. The crystal structures of phosphopantetheine adenylyltransferase with bound substrates reveal the enzyme’s catalytic mechanism. J. Mol. Biol. 2002;315:487–495. doi: 10.1006/jmbi.2001.5272. [DOI] [PubMed] [Google Scholar]
- Jiang H, Blouin C. Insertions and the emergence of novel protein structure: a structure-based phylogenetic study of insertions. BMC. Bioinformatics. 2007;8:444. doi: 10.1186/1471-2105-8-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodny R, Petrey D, Honig B. Protein structure comparison: implications for the nature of ‘fold space’, and structure and function prediction. Curr. Opin. Struct. Biol. 2006;16:393–398. doi: 10.1016/j.sbi.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript - A Program to Produce Both Detailed and Schematic Plots of Protein Structures. Journal of Applied Crystallography. 1991;24:946–950. [Google Scholar]
- Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Redfern O, Orengo C. Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol. 2007;8:995–1005. doi: 10.1038/nrm2281. [DOI] [PubMed] [Google Scholar]
- Lee DA, Rentzsch R, Orengo C. GeMMA: functional subfamily classification within superfamilies of predicted protein structural domains. Nucleic Acids Res. 2010;38:720–737. doi: 10.1093/nar/gkp1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Manikandan K, Pal D, Ramakumar S, Brener NE, Iyengar SS, Seetharaman G. Functionally important segments in proteins dissected using Gene Ontology and geometric clustering of peptide fragments. Genome Biol. 2008;9:R52. doi: 10.1186/gb-2008-9-3-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. Raster3d version 2: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Nureki O, Vassylyev DG, Tateno M, Shimada A, Nakama T, Fukai S, Konno M, Hendrickson TL, Schimmel P, Yokoyama S. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- Orengo CA. CORA--topological fingerprints for protein structural families. Protein Sci. 1999;8:699–715. doi: 10.1110/ps.8.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg SC, Brown SD, Ojha S, Seffernick J, Meng EC, Morris JH, Chang PJ, Huang CC, Ferrin TE, Babbitt PC. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry. 2006;45:2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern OC, Dessailly B, Orengo CA. Exploring the structure and function paradigm. Curr. Opin. Struct. Biol. 2008;18:394–402. doi: 10.1016/j.sbi.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern OC, Dessailly BH, Dallman TJ, Sillitoe I, Orengo CA. FLORA: a novel method to predict protein function from structure in diverse superfamilies. PLoS. Comput. Biol. 2009;5:e1000485. doi: 10.1371/journal.pcbi.1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves GA, Dallman TJ, Redfern OC, Akpor A, Orengo CA. Structural diversity of domain superfamilies in the CATH database. J. Mol. Biol. 2006;360:725–741. doi: 10.1016/j.jmb.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 2007;8:R232. doi: 10.1186/gb-2007-8-11-r232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rison SC, Hodgman TC, Thornton JM. Comparison of functional annotation schemes for genomes. Funct. Integr. Genomics. 2000;1:56–69. doi: 10.1007/s101420000005. [DOI] [PubMed] [Google Scholar]
- Sandhya S, Pankaj B, Govind MK, Offmann B, Srinivasan N, Sowdhamini R. CUSP: an algorithm to distinguish structurally conserved and unconserved regions in protein domain alignments and its application in the study of large length variations. BMC. Struct. Biol. 2008;8:28. doi: 10.1186/1472-6807-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhya S, Rani SS, Pankaj B, Govind MK, Offmann B, Srinivasan N, Sowdhamini R. Length variations amongst protein domain superfamilies and consequences on structure and function. PLoS. One. 2009;4:e4981. doi: 10.1371/journal.pone.0004981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AE, Orengo CA, Thornton JM. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar WS. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr. Opin. Struct. Biol. 2004;14:208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Westhead DR, Slidel TW, Flores TP, Thornton JM. Protein structural topology: Automated analysis and diagrammatic representation. Protein Sci. 1999;8:897–904. doi: 10.1110/ps.8.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Y, Madej T, Babenko V, Shoemaker B, Panchenko AR. Long-term trends in evolution of indels in protein sequences. BMC. Evol. Biol. 2007;7:19. doi: 10.1186/1471-2148-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats C, Lees J, Reid A, Kellam P, Martin N, Liu X, Orengo C. Gene3D: comprehensive structural and functional annotation of genomes. Nucleic Acids Res. 2008;36:D414–D418. doi: 10.1093/nar/gkm1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.