Abstract
Complex structures are generated and maintained through energy flux. Structures embody information, and biological information is stored in nucleic acids. The progressive increase in biological complexity over geologic time is thus the consequence of the information-generating power of energy flow plus the information-accumulating capacity of DNA, winnowed by natural selection. Consequently, the most important component of the biological environment is energy flow: the availability of calories and their use for growth, survival, and reproduction. Animals can exploit and adapt to available energy resources at three levels. They can evolve different anatomical forms through nuclear DNA (nDNA) mutations permitting exploitation of alternative energy reservoirs, resulting in new species. They can evolve modified bioenergetic physiologies within a species, primarily through the high mutation rate of mitochondrial DNA (mtDNA)–encoded bioenergetic genes, permitting adjustment to regional energetic environments. They can alter the epigenomic regulation of the thousands of dispersed bioenergetic genes via mitochondrially generated high-energy intermediates permitting individual accommodation to short-term environmental energetic fluctuations. Because medicine pertains to a single species, Homo sapiens, functional human variation often involves sequence changes in bioenergetic genes, most commonly mtDNA mutations, plus changes in the expression of bioenergetic genes mediated by the epigenome. Consequently, common nDNA polymorphisms in anatomical genes may represent only a fraction of the genetic variation associated with the common “complex” diseases, and the ascent of man has been the product of 3.5 billion years of information generation by energy flow, accumulated and preserved in DNA and edited by natural selection.
Keywords: evolution, mitochondria, natural selection, human origins, common diseases
Charles Darwin and Albert Russel Wallace hypothesized that the environment acts on individual variation via natural selection to create new species (1, 2). However, nothing in the concept of natural selection requires that biological systems should evolve toward ever greater complexity. Yet, throughout the more than 3.5 billion years of biological evolution (3), life has generated ever more complex forms. What, then, drives increasing biological complexity, and what are its implications for the ascent of man?
Bioenergetics and the Origin of Biological Complexity
In a thermodynamically isolated system, complex structures decay toward randomness. However, in nonequilibrium systems, the flow of energy through the system generates and sustains structural complexity, and nonhomogeneous structures embody information (4, 5).
On Earth, the flux of energy through the biosphere is relatively constant. If the flow of energy were the only factor generating complexity, complexity would soon achieve a steady state between the production and decay of structure. Biology is not static, because the information embedded in biological structures can be encoded and duplicated by informational molecules, DNA and RNA. Therefore, biological complexity increases because a portion of the information generated by energy flow through each generation is added to the accumulated information stores from previous generations. The increasingly complex information can then be used to recreate the more complex structures, as long as there is sufficient energy flow (Mathematical Formulations).
The flow of energy through biological structures permits them to reproduce, thus duplicating their DNA. In the process of DNA copying, errors occur. The duplicated mutant DNA changes the physiology and structure of the progeny. These progeny must compete for the available energy resources within the environment. Those that are more effective at acquiring and/or expending the available energy will sustain their energy flux and thus survive and reproduce. This competition for limited energy resources is the basis of natural selection, which edits the duplicated information based on its efficiency of energy use. Hence, the origin of biological complexity is the interplay between the organizing principle of energy flow, the accumulation of information in nucleic acids, and the winnowing of that information to optimize the use of the available energy flux for information propagation (Mathematical Formulations).
During the origin of life, biomolecular systems interacted directly with energy flux, resulting in the formation and polymerization of ribonucleic acids and their subsequent conformational changes to form catalysts to facilitate biochemical reactions (6). Hence, energy flux was directly linked to the accrual of information within nucleic acids. Subsequently, systems evolved by which the nucleic acid information could be converted into the more flexible proteins permitting more complex structures.
Today, the primary energy source for terrestrial life is the flux of high-energy photons from the Sun through the biosphere. The high-energy photons are collected by plant chloroplasts, descendants of symbiotic cyanobacteria, and the energy used to split water to hydrogen and oxygen. The resulting hydrogen (reducing equivalents) is fixed to carbon to generate glucose. From plant glucose, solar energy flows in the form of reducing equivalents through the biosphere. Animals eat the plants, acquiring the carbohydrates and their stored reducing equivalents. Carbohydrate breakdown products then enter the mitochondria, descendants of symbiotic α-protobacteria, and the mitochondria strip the hydrogens off the hydrocarbons and react them with oxygen to generate water, releasing the stored energy (7).
Therefore, it is the information-generating power of energy flux plus the information storage capacity of nucleic acids, winnowed by natural selection, that continually drives biology to increased complexity. Dobzhansky argued that, “Nothing in biology makes sense except in the light of evolution,” but nothing in biology exists without energy flux. Therefore, to understand the origin of species and the ascent of man, we must understand how energy flows through the biosphere, creating the environment; how this energy flow increases biological information; and how the edited information results in complexity and thus man.
Energy and the Environment: Three Levels of Bioenergetics
From this analysis, it is clear that the central aspect of an organism’s “environment” is energy flow. The energy environment of a biological system is the balance between the energy available to a system and the demands made on the biological systems’ energy supply for survival and reproduction. As life is about the preservation and transmission of information, reproduction is the prime directive.
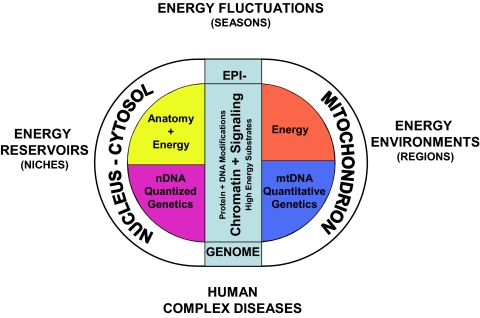
Organisms within the biosphere interface with energy flux at three levels: (i) the source of and requirement for the energy available to a species within its niche, the energy reservoir (ii); the regional differences in energy requirements and availability of subpopulations within a species, the energy environment; and (iii) the short-term fluctuations in energy availability and demands made on the individual during life by biological and environmental cycles, the energy fluctuations (Fig. 1).
Fig. 1.
Three hypothesized levels of eukaryotic animal cell adaptation to energy resources and demands. The primary contributor to the biological environment is the flux of energy through the biosphere. The dichotomy between structure and energy in eukaryotics results from the symbiotic origin of the eukaryotic cell involving the proto-mitochondrion and the proto-nucleus-cytosol. The mitochondrion became specialized in energy production and retained core genes for controlling energy production within the mtDNA. The nuclear-cytosol became specialized in structure with the accrual of the developmental genes in the nDNA. Because growth and reproduction must be coordinated with the availability of energy, the status of the energetic flux through the cellular bioenergetic systems, particularly the mitochondrion, came to be communicated to the nucleus-cytosol by alterations in the nDNA chromatin, the epigenome, and cytosol signal transduction systems, based on the production and availability of high energy intermediates, reducing equivalents, and ROS produced primarily by the mitochondrion. As a consequence, biological systems interface with the energy environment at three levels: the species level in which nDNA gene variation alters anatomical forms to exploit different environmental energy reservoirs, the species population level in which primarily mtDNA bioenergetic genetic variation permits adaptation to long-term regional differences in the niche energetic environment, and the individual level in which high-energy intermediates reflecting cyclic changes in environmental energetics drive the modification of the epigenome and the signal transduction pathways. [Reproduced with permission from ref. 44 (Copyright 2009, Cold Spring Harbor Laboratory Press).]
For a species, its energy reservoir is delineated by its food supply and its capacity to survive to reproduce within that niche. In most cases, this requires a specialized anatomy, and anatomical features frequently define species. The switch to new niches as they become available takes animals in the range of tens of thousands to hundreds of thousands of years. So mutations in anatomical genes must accumulate in that time frame to permit speciation. Once the anatomical structures to exploit the energy reservoir are in place, then purifying selection maintains those structures as long as the energy reservoir is stable and can support the species’ population. As anatomy is controlled by developmental genes, and as these genes are located on the chromosomes, nuclear DNA (nDNA) mutations are required to create a new species. The time that it takes for the necessary anatomical mutations to accumulate for speciation is determined by the nDNA mutation rate. For multicellular animals, the nDNA gene mutation rate is low, and hence the accumulation of adaptive nDNA mutations is slow.
For different subpopulations of a species, the energy environment can differ due to alternative climatic zones, differences in availability and type of calories, and differing demands on energy resources. These differences can result from migration, climatic change, changes in predation or parasitism, and so forth. Such environmental changes occur over hundreds to thousands of years, and because they are physiological, they require genetic changes in bioenergetic genes.
Bioenergetic genes are distributed throughout the genome and include hundreds to thousands of nDNA-encoded bioenergetic genes plus dozens of mtDNA-encoded bioenergetic genes. The mtDNA bioenergetic genes are the most functionally important because they are central to mitochondrial energy production. The mtDNA genes also have a much higher mutation rate than nDNA genes. Thus, adaptive bioenergetic mtDNA mutations arise in populations within hundreds to thousands of years and permit rapid physiological adaptation to changes in the regional energy environment. As regional subpopulations become established, additional nDNA mutations in bioenergetic genes arise to further solidify the physiological changes (8, 9). Over longer time periods, anatomical mutations can be added leading toward speciation.
An individual’s energy environment fluctuates in cycles throughout its life. These cycles can occur over the individual’s life span responding to intra- and intergenerational influences, recur annually in response to seasons, occur monthly relative to the reproductive cycle, or recur daily based on activity and feeding. All of these cyclic changes require reversible alterations in bioenergetic physiology, which cannot be achieved through static genetic changes.
Cyclic changes that occur over tens of years require moderate stability. These are achieved through epigenomic changes: modification of DNA by methylation or of histones through phosphorylation, acetylation, and methylation. Shorter-term reversible changes are accomplished through modulation of transcription factors and alterations in signal transduction pathways. All of these changes must be cued to changes in the energetic environment. Therefore, cyclic changes in the epigenome and signaling pathways are all mediated by changes in the intracellular concentrations of energetic intermediates including ATP for phosphorylation, acetyl-CoA for acetylation, NAD+ for Sirtuin-mediated deacetylation, S-adenosylmethionine (SAM) for methylation, oxidation-reduction (redox) state for thiol-disulfide regulation, and reactive oxygen species (ROS) for driving oxidative reactions (10).
Energy Reservoirs and Speciation
Since the publication of the Darwin–Wallace hypothesis of natural selection, numerous examples have been reported of anatomical changes associated with speciation. The earliest report of anatomical changes associated with exploitation of alternative energy resources was that of Darwin’s Galapagos finches, discussed by Darwin and Gould before the Geological Society of London in January 1839. More recently, the change in beak size of these finches has been attributed in part to changes in calmodulin expression (11). Comparable studies have continued for more than a hundred years, culiminating in the recent report that pelvic loss in stickleback fish is due to deletion of a tissue-specific enhancer of the Pituitary homeobox transcription factor 1 (Pitx1) gene (12). Although these studies confirm the importance of anatomical change in speciation, they belie the complexity of the physiological adaptations that are required for a species to occupy a new bioenergetic niche.
Energy Environments and Subpopulation Radiation
To understand the radiation of subpopulations of a species, it is necessary to study the intraspecific variation of a single species that occupies a wide range of regional energetic environments. The best studied species in this regard is Homo sapiens.
A striking feature of the radiation of mammalian and primate genomic elements is that mtDNA sequences show a much greater sequence evolution rate than do nDNA sequences (13–15). This rapid mtDNA radiation is reflected in the high degree of functional and regional variation of human mtDNAs (16). The mtDNA genes of all animals encode the core proteins of OXPHOS, so mtDNA mutations directly affect bioenergetic physiology and provide the ideal genetic system for adaptation to changes in regional energy environments.
Mitochondrial Bioenergetics and the mtDNA.
The unique capacity of the mtDNA to regulate bioenergetics has its roots in the symbiotic origin of the eukaryotic cell. Current theory postulates that a glycolytic motile microorganism, the proto-nucleus-cytosol, formed an association with an oxidative α-protobacterium, the proto-mitochondrion, about 2 billion years ago, probably in response to the rise in atmospheric oxygen generated by free-living cyanobacteria.
As the symbiosis matured, the two organisms consolidated their metabolic pathways and exchanged genes, natural selection enriching for more efficient forms. During the ensuing intersymbiont reorganization, most of the genes of the mitochondrial genome were transferred to the nDNA to become interspersed among the existing nuclear-cytosol bioenergetic genes. Ultimately, one genetic and metabolic combination was sufficiently energetically efficient to permit the advent of multicellularity. In this proto-multicellular eukaryote, 98% of the protein coding genes of the mitochondrial genome had been transferred to the nucleus, encompassing all of the polypeptide genes for mitochondrial growth, reproduction, and metabolism plus ≈80 polypeptide genes for OXPHOS (7, 17, 18).
The mtDNAs of multicellular animals all retained roughly the same 13 OXPHOS polypeptide genes. These include seven (ND1, 2, 3, 4L, 4, 5, and 6) of the 45 subunits of OXPHOS complex I, one [cytochrome b (cytb)] of the 11 subunits of complex III, three (COI, II, and III) of the 13 subunits of complex IV, and two (ATP 6 and 8) of the ≈16 subunits of complex V. Animal mtDNAs also retain the rRNAs and tRNAs genes for mitochondrial protein synthesis and a control region for regulating mtDNA replication and transcription (7, 17, 18).
Carbohydrates and fats are metabolized through the mitochondrial intermediate acetyl-CoA, by the tricarboxylic acid (TCA) cycle and β-oxidation pathways. These pathways strip the reducing equivalents off of the hydrocarbons and transfer them to mitochondrial NAD+ and FAD. The resulting reducing equivalents (electrons) are transferred from NADH + H+ and FADH2 to complexes I and II, respectively, initiating the electron transport chain (ETC). From complexes I and II, the electrons are transferred to coenzyme Q (CoQ) and then through complex III, cytochrome c, and complex IV to reduce 1/2 O2 into H2O. The energy that is released as the electrons pass through complexes I, III, and IV is used to transport protons out across the mitochondrial inner membrane to generate a transinner membrane electrochemical potential (ΔP = Δψ + ΔμH+). The energy stored in this capacitance, ΔP, can then be used by the ATP synthase, complex V, to condense ADP + Pi to ATP, the ATP being exported to the cytosol by the adenine nucleotide translocators (ANTs). ΔP can also be used to drive many other functions including the import of cytosolic Ca2+ into the mitochondrial matrix (7, 17, 18).
If excess electrons accumulate in complexes I and III and CoQ, they can be donated directly to O2 to give superoxide anion (O2.−), a potent oxidizing agent. Mitochondrial O2.− can be converted to hydrogen peroxide (H2O2) by the matrix Mn superoxide dismutase (MnSOD) or the intermembrane space Cu/ZnSOD. The H2O2 can acquire an additional electron, producing the highly reactive hydroxyl radical (·OH), or can be reduced to water by glutathione peroxidase. Consequently, the core ROS species (O2. −, H2O2, and ·OH) are primarily of mitochondrial origin (7, 17, 18).
The mitochondrion also incorporates a self-destruct system, the mitochondrial permeability transition pore (mtPTP). The mtPTP can be activated by a decline in either ΔP or high-energy phosphates or an increase in mitochondrial matrix Ca2+ level or ROS toxicity.
The efficiency by which OXPHOS generates ATP is called the coupling efficiency. This is determined by the efficiency with which complexes I, III, and IV convert the oxidation of reducing equivalents into ΔP and the efficiency by which complex V converts ΔP into ATP. A tightly coupled OXPHOS system maximizes ATP generation per calorie burned. A less coupled system must burn more calories for the same amount of ATP, resulting in a higher caloric intake and greater heat production (7).
All of the proton translocating complexes of OXPHOS (complexes I, III, IV, and V) must be balanced to ensure that one complex is not disproportionately permeable to protons and thus shorts ΔP. This is achieved by having the core electron and proton transport genes retained on a single piece of nonrecombining DNA, the exclusively maternally inherited mtDNA. This requires that each new mutation be tested by natural selection in the context of the previously existing variants encoded by that mtDNA (7).
mtDNA Variation in Adaptation and Disease.
Because each cell has hundreds of mitochondria and thousands mtDNAs, new mtDNA mutations generate an intracellular mixture of mutant and normal mtDNAs, heteroplasmy. The percentage of mutant and normal mtDNAs can be unequally distributed at cytokinesis, such that the percentage of mutant mtDNAs can drift during successive mitotic and meiotic cell divisions, replicative segregation.
As the percentage of deleterious mtDNA mutations increases, the energy output of the cell declines until it drops below the minimum energy output required for that cell type to function and symptoms ensue, the bioenergetic threshold. To date, more than 200 pathogenic mtDNA mutations have been identified, and these cause all of the symptoms seen in the common metabolic and degenerative disease including diabetes and metabolic syndrome, forms of blindness, deafness, neurodegenerative disease, myopathy, cardiomyopathy, renal dysfunction, and hepatic failure. Mutations in the mtDNA also contribute to cancer and aging (7, 17).
The high mtDNA mutation rate means that deleterious mtDNA mutations are very common. The frequency of recognized mitochondrial diseases is already estimated at 1/4,000–1/5,000 (19), and the de novo mtDNA mutation rate observed in cord blood, as assessed through 15 known pathogenic mutations, has been reported as 1 in 200 (20). Given the high mtDNA mutation rate and the great importance and conservation of the mtDNA genes, the cumulative mtDNA genetic load should drive animal species to extinction. This paradox is resolved because the mammalian ovary encompasses a selective system that systematically eliminates those proto-oocyes that harbor the most severely deleterious mtDNA mutations (21, 22). Consequently, only oocytes with mildly deleterious, neutral, or beneficial mtDNA variants are ovulated and can be transmitted into the next generation. New mtDNA variants are constantly being introduced into animal populations, thus modifying individual energy metabolism. These variants provide the physiological variability required for subpopulations to adjust to new regional energetic environments.
As an mtDNA harboring an adaptive mutation becomes enriched in a new energy environment, additional neutral or advantageous mtDNA mutations accumulate sequentially along that regional maternal lineage. This creates distinctive branches of the mtDNA tree, each a cluster of related mtDNA haplotypes known as a haplogroup. Because the number of possible adaptive mtDNA mutations is finite, the same adaptive mutations have been observed repeatedly on different mtDNA backgrounds around the world. This convergent evolution confirms that these mtDNA mutations are adaptive.
Each regional indigenous human population has its own distinctive mtDNAs. African mtDNAs belong to macrohaplogroup L, which encompasses the greatest mtDNA sequence diversity implying an African origin for the mtDNA tree (23–25). Of all of the African mtDNA variants, only two mtDNAs successfully left Africa and colonized Eurasia, founding macrohaplogroups M and N. Only macrohaplogroup N radiated into Europe, generating the European-specific lineages H, I, J, Uk, T, U, V, W, and X. Both macrohaplogroups M and N radiated into Asia, generating a plethora of mtDNA lineages. Of these, only A, C, and D became enriched in northeastern Siberia and were in a position to cross the Bering land bridge to colonize the Americas (7, 16).
The regional specificity of mtDNA lineages suggests that mtDNA variation permitted humans to live in different climatic zones, perhaps through regulation of OXPHOS coupling efficiency and thus thermal regulation (7). Accordingly, mtDNA variation but not nDNA variation correlates with regional temperature extremes (26). In mtDNAs harboring the two founder macrohaplogroup N missense mutations, ND3 nucleotide 10398 (amino acid change A114T) and ATP6 nucleotide 8701 (amino acid change A59T), several mitochondrial physiological parameters have been shown to be altered (27). Furthermore, a mtDNA control region variant has been found to change mitochondrial transcription and copy number (28).
Although mild mtDNA mutations may be adaptive in one local energy environment, the same mutation might be maladaptive in another energy environment. Consistent with this conjecture, mtDNA haplogroups have been found to be important risk factors for a wide range of common metabolic and degenerative diseases and to influence various cancers and longevity (17, 29).
Once a subpopulation has become established in a region through adaptive mtDNA mutations, additional mutations can arise in nDNA bioenergetic genes to further enhance physiological adaptation and contribute to speciation (8). Examples of such variants in human populations include polymorphisms in the nDNA-encoded mitochondrial uncoupling protein genes (30–33) and in the bioenergetic transcription factor genes for the peroxisome-proliferator-activated receptor γ (PPARγ) (34) and PPARγ-coactivator 1α (PCG-1α) (35, 36). These polymorphic genes have also been found to be risk factors for obesity and diabetes in certain populations. In large-scale population studies that cut across regional energy environments, the associations with PPARγ and PGC-1α are lost (37–40). This paradox may result from the mixing of populations from different energy environments which harbor alternative region-specific adaptive genetic variants, such that the impact of each individual regional variant is averaged out.
Energy Fluctuation and Cyclic Adaptation
Individual adjustments to cyclic changes in the energy environment must be reversible. Therefore, cyclic changes cannot be due to DNA sequence changes, but must be due to changes in bioenergetic gene expression. Relevant cyclic bioenergetic changes encompass a wide temporal range from intergenerational effects to daily fluctuations. The more long-term cyclic modulations occur as epigenomic changes at the chromatin level, whereas the shorter-term changes involve alterations of transcription factors, signal transduction pathways, and protein activation.
Epigenomic Regulation of Bioenergetics.
Because the primary environmental variable is energetics, and because the bioenergetic genes are dispersed across the chromosomes and mtDNA, responses to environmental fluctuation must involve pan genomic regulation of bioenergetic genes. The modulation of the epigenome by intracellular concentrations of high-energy intermediates provides the necessary link between the energetic state of the environment and the modulation of cellular gene expression. When calories were abundant, the organism must grow and reproduce, which requires the up-regulation of gene expression. When calories were limiting, the organism must become quiescent, requiring the shutdown of gene expression (10).
Epigenomic regulation occurs at the chromatin level. The nDNA is packaged in nucleosomes encompassing 146–147 base pairs of DNA wrapped around a complex of two copies each of histones H2A, H2B, H3, and H4. The amino-terminal tails of the histones are positively charged, such that they bind electrostatically to the phosphate backbone of the DNA and inhibit transcription. However, when the histone tails become phosphorylated by kinases using ATP, or acetylated by histone acetyltransferases (HAT) using acetyl-CoA, the positive charges are neutralized, the affinity of the histone tails to DNA is reduced, and the chromatin opens to permit transcription. Methylation of DNA and of histone tails by methyltransferases using SAM can also modulate the affinity of proteins for DNA.
ATP is generated by both glycolysis and OXPHOS when caloric reducing equivalents are prevalent. Mammalian cell acetyl-CoA is generated primarily in the mitochondrion during pryruvate or fatty acid oxidation. Within the mitochondrion, the acetyl-CoA is converted to citrate by condensation with oxaloacetate (OAA) via citrate synthetase. Citrate can be exported into the cytosol, where it is cleaved back to acetyl-CoA and OAA by ATP-citrate lyase (10). Mitochondrial acetyl-CoA can also be exported out of the mitochondrion as acetylcarnitine by the carnitine/acylcarnitine acetyltranslocase. In the cytosol, acetylcarnitine reverts back into acetyl-CoA for use in histone acetylation (41).
SAM is produced in the cytosol by the reaction L-methionine + ATP. ATP is generated by the mitochondrion and glycolysis, whereas the methyl groups to convert homocystine to methionine come from the mitochondrion. Therefore, all of the primary substrates for chromatin modification are produced by the bioenergetic pathways, which in turn are fueled by the availability of calories in the environment (10).
Evidence that the epigenome regulates bioenergetics comes from the facts that pathogenic mtDNA mutations result in symptoms similar to those attributed to the epigenomic disease and that several epigenomic diseases have been associated with mitochondrial dysfunction. Epigenomic diseases affect imprinting, methylation, and chromatin organization (42). The epigenome can regulate dispersed bioenergetic genes in either the cis configuration for adjacent genes or in the trans configuration for dispersed genes. Current knowledge about chromatin organization suggests that the cis regulation occurs with chromatin loop domains and that trans regulation occurs by diffusible trans acting factors or by bringing together dispersed genes into transcriptional islands, in part through shared enhancer sequences (10).
Imprinting diseases generally involve cis-acting epigenetic defects. In Angelman and Prader-Willi syndromes, the perturbation of cell function involves genetic inactivation of the active allele on chromosome 15q11–13 in the context of an inactive imprinted allele on the opposite chrosomome. The pathophysiology of Angelman syndrome apprears to be mitochondrial, as analysis of an Angelman murine model has revealed that the hippocampal neurons have a reduced synaptic vesicle density and shrunken mitochondria and the brain has a partial defect in OXPHOS complexes II + III (10).
The pathophysiology of Beckwith-Wiedemann syndrome and Wilm’s tumor may also involve bioenergetic dysfunction. Both of these diseases are associated with loss of imprinting (LOI) on chromosome 11q15.5 within a chromatin loop domain encompassing the insulin-like growth factor 2 (IGF2) gene. IGF2 may act through the PI3K-Akt-FOXO pathway to modulate energy metabolism (10, 42).
Rett syndrome and the laminopathies may be epigenomic diseases that act in trans to affect mitochondrial function. Rett syndrome is caused by mutations in the methyl-CpG binding protein 2 (MeCP2), which binds to meCpG islands throughout the chromosomes (43). As abnormal mitochondria and mitochondrial function have been reported in several Rett patient studies, loss of MeCP2 might disrupt the coordinate regulation of nDNA energy gene expression (10). The laminopathies are caused by mutations in the laminin A/C gene (LMNA), which disrupt the nuclear architecture and potentially transcriptional islands. Mutations in the LMNA gene have been found to produce similar phenotypes to those found in mtDNA mutations and a study of cells harboring LMNA mutations revealed mitochondrial defects. Therefore, various epigenomic defects may affect mitochondrial function implying that an important function of the epigenome is to coordinate the expression of the dispersed bioenergetic genes (10).
Bioenergetic Regulation of Signal Transduction and Metabolism.
To respond to more rapid energy environment fluctuations, animal cells modify transcription factors and signal transduction systems via high energy intermediates. High and low blood sugar results in the secretion of insulin by the pancreatic α cells and glucagon by the pancreatic β cells, respectively. Insulin binds to the insulin receptor, which signals, through phosphatidyl-inositol 3 kinase (PI3K) and Akt/PKB, to phosphorylate and inactivate the FOXO transcription factor. When not phosphorylated, FOXO binds to the PCG-1α promoter and increases PGC-1α expression, which up-regulates mitochondrial biogenesis and OXPHOS. Thus, in the presence of glucose, FOXO is inactivated, OXPHOS is down-regulated, and glycolysis is favored. In the absence of glucose FOXO is active and OXPHOS is up-regulated to burn fat. Similarly, glucacon binds to the glucagon receptor to activate adenylylcyclase, and the resulting cAMP activates protein kinase A (PKA) to phosphorylate CREB. Activated CREB also binds to the PGC-1α promoter and up-regulates OXPHOS. Low glucose thus doubly induces OXPHOS by inhibiting insulin signaling and enhancing glucagon signaling (7).
The PI3K pathway is also linked via the tuberous sclerosis protein complex (TSC) to the mTORC1 regulation of nutrient metabolism. mTORC1 is also modulated by AMP kinase, which is activated by reductions in high-energy phosphates. Virtually every signal transduction pathway is modulated by ATP-mediated phosphorylation, so almost all cellular processes are regulated by the availability of high-energy intermediates (10, 18).
Changes in cellular redox state are also important in regulating transcription factors and metabolic pathways. The redox state of the cell reflects the flux of reducing equivalents from the mitochondrion, through the nucleus-cytosol, and on to the other cellular compartments. Reducing equivalents enter the mitochondrion as NADH + H+ at −250 mV and flow through the ETC and other cellular pathways down to oxygen at +600 mV. The importance of the subcellular redox status is illustrated by the class III histone deacetylase Sirt1. Sirt1 removes acetyl groups from proteins in the presence of NAD+ via the reaction: acetyl-lysine + NAD+ → lysine + nicotinamide + 2′-O-acetyl-ADP ribose. Although the oxidized NAD+ is a required coreactant, the reduced form of NAD+, NADH + H+, cannot be used by Sirt1. Therefore, deacetylation is coupled to the cellular redox state. The FOXO and PGC-1α transcription factors are inactivated by acetyl-CoA–mediated acetylation. They can be reactivated by deacetylation by Sirt1 + NAD+. When glucose is abundant, glycolysis reduces cytosolic NAD+ to NADH + H+ in the process of generating pyruvate. The pyruvate is converted to acetyl-CoA in the mitochondrion, and the acetyl-CoA is exported back into the cytosol, and is used to acetylate and inactivate PGC-1α and FOXO. Because the cytosolic NAD+ is reduced to NADH + H+, Sirt1 cannot deacetylation FOXO and PGC-1α and OXPHOS is inhibited whereas glycolysis is favored. By contrast, when fatty acids and ketone bodies (acetoacetate and β-hydroxybutyrate) are metabolized, they are burned entirely within the mitochondrion, and the cytosolic NAD+ remains oxidized. The combination of Sirt1 + NAD+ then deacetylates and activates the FOXO and PGC-1α transcription factors, up-regulating OXPHOS to oxidize fats and ketones (44).
The redox regulation of cellular metabolism goes far beyond its effects in Sirt1 activity. In the mitochondrion, a substantial portion of the NADH is oxidized via the ETC using O2 to generate ΔP, but the redox state of a portion of the NADH + H+ is increased by the nicotinamide nucleoside transydrogenase (Nnt), using energy from ΔP to drive the transfer of reducing equivalents from NADH + H+ to NADPH + H+ with a redox potential of −405 mV. Mitochondrial NADPH + H+ can then drive the reduction of oxidized glutathione (GS-SG) to reduced glutathione (2GSH), and GSH can act through the glutathione peroxidases to detoxify mitochondrial ROS and other radicals. NADPH + H+ also provides reducing equivalents for mitochondrial thioredoxin-2(SH)2/SS [Trx2(SH)2/SS], to drive peroxidoxins to reduce radical species and to mediate the modulation of the redox status of thiol-disulfides of an array of mitochondrial enzymes directly regulating their activity (18, 45).
To a limited extent, the reducing equivalents of mitochondrial NADH + H+ and NADPH + H+ can also be transferred to the cytosol. Reducing equivalents from NADH + H+ can be exported via the mitochondrial inner membrane aspartate–malate shuttle. Mitochondrial NADPH + H+ can be exported to the cytosol via citrate, which is converted to malate. Malate is then oxidized by the cytosolic malic enzyme to pyruvate in association with the reduction of NADP+ to NADPH + H+. Cytosolic NADPH + H+ can also be generated by glucose 6-phosphate dehydrogenase (18).
The cytosolic NADPH + H+ redox state is approximately −393 mV. This can drive cytosolic glutathione reductase and associated glutathione peroxidases to buffer cytosolic ROS and the glutaredoxins to regulate the redox status of proteins. Cytosolic NADPH + H+ also determines the redox status of the cytosolic and nuclear thioredoxin-1(SH)2/SS [Trx1(SH)2/SS]. Trx1(SH)2/SS donates reducing equivalents to cytosolic peroxidoxins to control radicals, and to the thiol/disulfides of enzymes and transcription factors to regulate their activity. Trx1(SH)2/SS directly regulates proteins such as Oct-4, but also regulates the redox status of the bifunctional apurinic/apyrimidinic endonuclease/redox factor-1(APE/ Ref1Red/Ox). The redox state of APE/ Ref1Red/Ox, in turn, modulates the activity of a variety of transcription factors including activator protein–1 (AP1, c-Jun), NF-E2–related factor–2 (Nrf2), NF-κB, p53, glucocorticoid receptor (GR), estrogen receptor (ER), and hypoxia-inducible factor–1α (HIF-1α) (18, 45).
Mitochondrially modulated ROS production also regulates the activity of a wide spectrum of enzymes, including tyrosine and serine/threonine kinases, multiple phosphatases, and NFκB-mediated cytokine and inflammatory responses (18). ROS levels as well as oxygen tension directly regulate the activation of the HIF-1α transcription factor. HIF-1α is constitutively synthesized but is inactivated in the presence of high O2 by hydroxylation via prolylhydroxylase domain protein 2 (PHD2). Reduced O2 and mitochondrially generated ROS production can inhibit PHD2 activity, stabilizing HIF-1α. HIF-1α together with HIF-1β then act as a transcription factor to induce the expression of glycolytic enzymes and vascularization and hematopoietic factors, alter the oxygen affinity of OXPHOS complex IV by inducing subunit COX4-2 and the mitochondrial LON protease to degrade subunit COX4-1, induce pyruvate dehydrogenase (PDH) kinase 1 to inhibit PDH, and thus block the conversion of pyruvate to acetyl-CoA, induce MXI-1 to inhibit Myc thus reducing expression of PGC-1α, and induce BNIP3 to initiate the autophagic degradation of the mitochondria (18, 46). Hence, energy flux through the animal cell regulates virtually every aspect of cellular growth, differentiation, quiescence, and death.
Bioenergetics and the Ascent of Man
This energetic-information hypothesis on the origin of biological complexity has fundamental implications for the ascent of man. Since Vesalius’ anatomical catalog published over 450 years ago, Western medicine has taken a predominantly anatomical perspective of medicine, the anatomical paradigm of disease. Similarly, since the discovery of the Mendelian laws of inheritance about 150 years ago, it has been assumed that all genes are inherited in a Mendelian fashion, the Mendelian paradigm of genetics. Since all anatomical genes are chromosomal and thus also Mendelian, these two paradigms provided an internally consistent perspective on biology and medicine for 100 years. The anatomical and Mendelian paradigms of medicine have produced many advances, such as understanding the molecular basis of diseases resulting from severely deleterious nDNA mutations in structural genes. Because of these successes, it has been assumed that if a disease Mendelizes it is genetic and if it does not it is “complex,” the later implying an interaction between multiple Mendelian genes plus the environment.
Medicine pertains exclusively to humans, a single species. Consequently, the most important variables in intraspecific adaptation to local environmental changes, which are primarily energetic, should be alterations in bioenergetic genes, either genetic or epigenetic. Mutations in the mtDNA are more common than nDNA mutations and epigenomic changes can rapidly change the expression of bioenergetic genes. Efforts to explain common “complex” diseases like diabetes based exclusively on the analysis of nDNA variation in tissue-specific genes would then be expected to be relatively unproductive, as has been the case. In reality, common diseases may not be particularly “complex,” they may simply be energetic and non-Mendelian.
The discovery that the mammalian ovary harbors a selective system to eliminate the most deleterious mtDNA mutations explains why the high mtDNA mutation rate does not drive mammalian populations to extinction from overwhelming mtDNA genetic load. Since the mtDNA only encodes OXPHOS genes and OXPHOS genes are expressed in every cell of the body, intraovarian selection can monitor the physiological consequences of mitochondrial OXPHOS defects within proto-oocytes and eliminate those with the most severe bioenergetic aberrations.
By contrast, most nDNA-encoded developmental genes are not expressed in the gametes, so gametes harboring severely deleterious developmental mutations cannot be phenotypically identified and eliminated within the gonads. Purifying selection of deleterious nDNA mutations must occur postconception, at the individual organism level. This greatly increases the genetic load and energy wastage caused by deleterious nDNA mutations. To avoid introduction of too many deleterious nDNA mutations into the population, the nDNA mutation rate must be kept low. Still, the nDNA mutation rate cannot be zero, as maintaining this level of fidelity would be too energy expensive and would also eliminate the capacity of organisms to adapt to new energy reservoirs. Genetic load then places an upper limit on the combination of the nDNA mutation rate and the genetic target size, the amount of protein coding information in the organism’s genome. In animal species, a steady state may have been achieved between nDNA mutation rate and gene target size when the genome complexity reached that of the invertebrates. This may explain why the number of protein coding genes is similar between Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, and Homo sapiens, and that most of the increased structural complexity in vertebrates has been achieved by alternative splicing and elaboration of complex temporal and spatial gene regulation. Because of these nDNA constraints and the direct interface between organismal bioenergetics and changes in the energetic environment, the dominant mechanism for intraspecific adaptation to environmental change occurs through bioenergetics.
Although there may be an upper limit on the amount of structural gene information that can be added to the nuclear genomes of higher animals, the flux of energy through the biosphere is continually adding information to the environment. Much of this physical and biological information is too transient to be of value to future animal generations and thus is not stored in DNA. The present position of food resources would be an example of such information. Still, this information is of benefit for the survival and reproduction of the individual. As a result, the continued evolution of information storage and retrieval systems needed to shift from the use of DNA to store the information to the use of DNA to build structures that could store transient information. These short-term information storage and retrieval systems ultimately became the brain.
The brain’s information is lost when the individual dies. Yet, some of this information may be beneficial to the individual’s relatives and descendants, requiring that this information be transmitted between related individuals. This provided the impetus for the evolution of language and learning, leading to culture, libraries, and computers.
Toward the end of their lives, Darwin and Wallace became estranged. Darwin argued that natural selection was sufficient to explain the origin of the existing biological world. Wallace believed that natural selection alone was insufficient to explain the existence of complex structures such as the human brain. From the bioenergetic perspective, Wallace’s reservations were justified, as complexity can be generated only through the information-generating power of energy flow and the cumulative information storage capacity of nucleic acids. It took more than 3.5 billion years for these systems to amass sufficient information to generate the human brain. Thus the missing concept that Wallace sought to explain the ascent of man is the interaction between energtics and information.
Mathematical Formulations
According to the second law of thermodynamics, an energetically isolated system will move toward equilibrium in association with increased disorder or entropy (S), ΔA = ΔU – TΔS, where A is Helmholtz free energy (energy for useful work), U is the total energy of the system, and T is the absolute temperature. In a system in which T is constant, disorder increases (ΔS is +) as ΔA declines, provided ΔU is constant. However, in a system where energy flows through the system, such that an equal quantity of energy enters and leaves the system, the total instantaneous energy (U) remains constant but the energy to perform work (ΔA) and thus produce change is increased. If ΔA becomes greater than U, then ΔS becomes –, disorder decreases, and the system becomes more ordered, which is the case for biological systems.
With sufficient information (I), any system can be described. The greater the disorder of a system (larger the S) the greater the information that is necessary for its description, S = kiI, were I is information and ki is a constant. However, energy flow generates structure, the nature of which is determined by the inherent properties of the system. To describe a system requires the information to describe the physical properties of the system, I(p), and the information inherent in the system that permits the creation of the ordered aspects of the system, I(o). By analogy, to describe a glass of ice water requires information about the physical properties of the ice and water but also information about the inherent properties of H2O that cause dipole interactions and crystalline lattice formation.
The information to describe the entire system is related to the energy of the entire system, U, and encompasses both the physical [I(p)u] and ordering [I(o)u] components of the system's information. Therefore, U = k1I(p)u + k2I(o)u and the information embodied in U is I(u) = I(p)u + I(o)u. By analogy, A = k3I(p)a + k4I(o)a and the information embodied in A is I(a) = I(p)a + I(o)a. In a physically isolated system, the usable information in A, I(a), is less than that in U, I(u), and the information content of the system decays. To maintain a steady state structure, additional energy must be added to A, through energy flux (Ef). This flow of energy will generate information through interaction with the components of the system, Ef = k5I(ef). This information is then added to the usable information of the system, I(a) = I(p)a + I(o)a + I(ef). Therefore, S = ([k1I(p)u + k2I(o)u] – [k3I(p)a + k4I(o)a + k5I(ef)])/T.
If the system is to avoid decay, Ef must generate information, I(ef), that is equal to or greater than the difference between I(u) and I(a). In a complex system that does not decay (ΔS is zero or negative), A = k3I(p)a + k4I(o)a + k5I(ef) ≥ k1I(p)u + k2I(o)u = U. Consequently, if k5I(ef) > [k1I(p)u + k2I(o)u] – [k3I(p)a + k4I(o)a], then information and order increase within the system.
Because the flux of energy across the Earth’s surface, Ef, is roughly constant, the amount of organizing information from Ef must also be constant. Hence, if this were the only factor, the biosphere would quickly come to stasis. The reason that this does not occur is because information pertaining to the ordering of the system, I(o), can be accumulated, using an appropriate information storage and retrieval system. In biology, this information storage system is nucleic acids, I(na). Therefore, in the biosphere a portion of I(ef), I(o)u, and I(o)a are retained as I(na): I(ef)na, I(o) na(u), and I(o) na(a). Hence, I(ef)na + I(o) na(a) must be > I(o) na(u) for the biosphere to continually increase in complexity.
The nucleic acid information present in the biosphere today, I(na), is the sum of the total nucleic acid information that has formed over 3.5 billion years of terrestrial biology, I(na)t, minus that portion of the total information that has been removed by natural selection or cataclysm, I(na)e. I(na) = I(na)t – I(na)e. The nucleic acid information in today's biosphere, I(na), divided by the sum of the energy flux through the biosphere over the past 3.5 billion years, represents the average efficiency by which energy flux has been converted into conserved biological information on Earth.
Acknowledgments
This work was supported by National Institutes of Health Grants NS21328, AG24373, DK73691, AG13154, and AG16573; California Institute for Regenerative Medicine Comprehensive Grant RC1-00353-1; and a Doris Duke Clinical Interfaces Award 2005.
Footnotes
The author declares no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution IV: The Human Condition,” held December 10–12, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/SACKLER_Human_Condition.
This article is a PNAS Direct Submission.
References
- 1.Darwin CR, Wallace AR. On the tendency of species to form varieties; and on the perpetuation of varieties and species by natural means of selection. J Proc Linnean Soc Lond Zool. 1858;3:46–50. [Google Scholar]
- 2.Darwin C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray of Albemarle Street; 1859. [Google Scholar]
- 3.Simpson S. Questioning the oldest signs of life. Sci Am. 2003;288:70–77. doi: 10.1038/scientificamerican0403-70. [DOI] [PubMed] [Google Scholar]
- 4.Morowitz HJ. Energy Flow in Biology, Biological Organization as a Problem in Thermal Physics. New York: Academic; 1968. [Google Scholar]
- 5.Rubí JM. The long arm of the second law. Sci Am. 2008;299:62–67. doi: 10.1038/scientificamerican1108-62. [DOI] [PubMed] [Google Scholar]
- 6.Ricardo A, Szostak JW. Origin of life on earth. Sci Am. 2009;301:54–61. doi: 10.1038/scientificamerican0909-54. [DOI] [PubMed] [Google Scholar]
- 7.Wallace DC. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- 8.Mishmar D, et al. Adaptive selection of mitochondrial complex I subunits during primate radiation. Gene. 2006;378:11–18. doi: 10.1016/j.gene.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 9.Lane N. Biodiversity: On the origin of bar codes. Nature. 2009;462:272–274. doi: 10.1038/462272a. [DOI] [PubMed] [Google Scholar]
- 10.Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abzhanov A, et al. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 12.Chan YF, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown WM, Prager EM, Wang A, Wilson AC. Mitochondrial DNA sequences of primates: Tempo and mode of evolution. J Mol Evol. 1982;18:225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- 14.Neckelmann N, Li K, Wade RP, Shuster R, Wallace DC. cDNA sequence of a human skeletal muscle ADP/ATP translocator: Lack of a leader peptide, divergence from a fibroblast translocator cDNA, and coevolution with mitochondrial DNA genes. Proc Natl Acad Sci USA. 1987;84:7580–7584. doi: 10.1073/pnas.84.21.7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallace DC, et al. Sequence analysis of cDNAs for the human and bovine ATP synthase beta subunit: Mitochondrial DNA genes sustain seventeen times more mutations. Curr Genet. 1987;12:81–90. doi: 10.1007/BF00434661. [DOI] [PubMed] [Google Scholar]
- 16.Wallace DC, Lott MT, Procaccio V. In: Emery and Rimoin’s Principles and Practice of Medical Genetics. 5th Ed. Rimoin DL, Connor JM, Pyeritz RE, Korf BR, editors. Philadelphia: Churchill Livingstone Elsevier; 2007. pp. 194–298. [Google Scholar]
- 17.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer AM, et al. Prevalence of mitochondrial DNA disease in adults. Ann Neurol. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 20.Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet. 2008;83:254–260. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan W, et al. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart JB, et al. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6:0063–0071. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson MJ, Wallace DC, Ferris SD, Rattazzi MC, Cavalli-Sforza LL. Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns. J Mol Evol. 1983;19:255–271. doi: 10.1007/BF02099973. [DOI] [PubMed] [Google Scholar]
- 24.Cann RL, Stoneking M, Wilson AC. Mitochondrial DNA and human evolution. Nature. 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 25.Merriwether DA, et al. The structure of human mitochondrial DNA variation. J Mol Evol. 1991;33:543–555. doi: 10.1007/BF02102807. [DOI] [PubMed] [Google Scholar]
- 26.Balloux F, Handley LJ, Jombart T, Liu H, Manica A. Climate shaped the worldwide distribution of human mitochondrial DNA sequence variation. Proc Biol Sci. 2009;276:3447–3455. doi: 10.1098/rspb.2009.0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazuno AA, et al. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2006;2:1167–1177. doi: 10.1371/journal.pgen.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suissa S, et al. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet. 2009;5:e1000474. doi: 10.1371/journal.pgen.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villarroya F, Iglesias R, Giralt M. PPARs in the control of uncoupling proteins gene expression. PPAR Res. 2006;2007:1–12. doi: 10.1155/2007/74364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano T, et al. A/G heterozygote of the A-3826G polymorphism in the UCP-1 gene has higher BMI than A/A and G/G homozygote in young Japanese males. J Med Invest. 2006;53:218–222. doi: 10.2152/jmi.53.218. [DOI] [PubMed] [Google Scholar]
- 32.Bulotta A, et al. The common -866G/A polymorphism in the promoter region of the UCP-2 gene is associated with reduced risk of type 2 diabetes in Caucasians from Italy. J Clin Endocrinol Metab. 2005;90:1176–1180. doi: 10.1210/jc.2004-1072. [DOI] [PubMed] [Google Scholar]
- 33.Cha MH, Shin HD, Kim KS, Lee BH, Yoon Y. The effects of uncoupling protein 3 haplotypes on obesity phenotypes and very low-energy diet-induced changes among overweight Korean female subjects. Metabolism. 2006;55:578–586. doi: 10.1016/j.metabol.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 34.Altshuler D, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 35.Ek J, et al. Mutation analysis of peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to type II diabetes mellitus. Diabetologia. 2001;44:2220–2226. doi: 10.1007/s001250100032. [DOI] [PubMed] [Google Scholar]
- 36.Muller YL, Bogardus C, Pedersen O, Baier L. A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor gamma coactivator-1 is associated with altered lipid oxidation and early insulin secretion in Pima Indians. Diabetes. 2003;52:895–898. doi: 10.2337/diabetes.52.3.895. [DOI] [PubMed] [Google Scholar]
- 37.Saxena R, et al. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 38.Zeggini E, et al. Wellcome Trust Case Control Consortium (WTCCC) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 41.Madiraju P, Pande SV, Prentki M, Madiraju SR. Mitochondrial acetylcarnitine provides acetyl groups for nuclear histone acetylation. Epigenetics. 2009;4:399–403. doi: 10.4161/epi.4.6.9767. [DOI] [PubMed] [Google Scholar]
- 42.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 43.Loat CS, et al. Methyl-CpG-binding protein 2 polymorphisms and vulnerability to autism. Genes Brain Behav. 2008;7:754–760. doi: 10.1111/j.1601-183X.2008.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallace DC. Mitochondria, bioenergetics, and the epigenome in eukaryotic and human evolution. Cold Spring Harb Symp Quant Biol. 2009. ePub ahead of print, http://symposium.cshlp.org/content/early/2009/11/25/sqb.2009.74.031. [DOI] [PMC free article] [PubMed]
- 45.Kemp M, Go YM, Jones DP. Nonequilibrium thermodynamics of thiol/disulfide redox systems: A perspective on redox systems biology. Free Radic Biol Med. 2008;44:921–937. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Semenza GL. Mitochondrial autophagy: Life and breath of the cell. Autophagy. 2008;4:534–536. doi: 10.4161/auto.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]