Abstract
The potential benefits of using population isolates in genetic mapping, such as reduced genetic, phenotypic and environmental heterogeneity, are offset by the challenges posed by the large amounts of direct and cryptic relatedness in these populations confounding basic assumptions of independence. We have evaluated four representative specialized methods for association testing in the presence of relatedness; (i) within-family (ii) within- and between-family and (iii) mixed-models methods, using simulated traits for 2906 subjects with known genome-wide genotype data from an extremely isolated population, the Island of Kosrae, Federated States of Micronesia. We report that mixed models optimally extract association information from such samples, demonstrating 88% power to rank the true variant as among the top 10 genome-wide with 56% achieving genome-wide significance, a >80% improvement over the other methods, and demonstrate that population isolates have similar power to non-isolate populations for observing variants of known effects. We then used the mixed-model method to reanalyze data for 17 published phenotypes relating to metabolic traits and electrocardiographic measures, along with another 8 previously unreported. We replicate nine genome-wide significant associations with known loci of plasma cholesterol, high-density lipoprotein, low-density lipoprotein, triglycerides, thyroid stimulating hormone, homocysteine, C-reactive protein and uric acid, with only one detected in the previous analysis of the same traits. Further, we leveraged shared identity-by-descent genetic segments in the region of the uric acid locus to fine-map the signal, refining the known locus by a factor of 4. Finally, we report a novel associations for height (rs17629022, P< 2.1 × 10−8).
INTRODUCTION
Isolated populations have a long history in genetic mapping studies of inherited disorders with advantages including reduced environmental, phenotypic and genotypic heterogeneity when compared with outbred populations (1–9). In particular, the reduction in genotypic heterogeneity observed in isolated populations due to founder effects, bottlenecks and genetic drift, may allow otherwise rare mutant alleles to rise to a higher frequency in these populations while at the same time narrowing the spectrum of candidate mutations (10,11). The increased chance for homozygosity has been a key factor in identifying mutations responsible for rare monogenic diseases in isolated populations (3,12,13). Here we investigate the advantages and limitations of exploiting homozygosity in isolated populations for the analysis of alleles effecting complex traits (14–16). Genome-wide association studies (GWAS) performed in outbred populations have thus far identified many common variation contributing to complex diseases (17). One limitation in these studies is that rare variants that are not in linkage disequilibrium with the common variants assayed are usually not detected (18). However, when the populations analyzed in a GWAS have a substantial number of individuals that share a recent common ancestor, not only common variants (19,20), but sometimes also rare variants affecting complex disorders can be identified (21–23).
Genetic analysis of isolated populations pose unique challenges for traditional GWAS methods that have been mainly focused on outbred populations (24). The crux of the difference is that in isolated populations the likelihood of any two individuals in the population to be related is not negligible. The resulting direct and cryptic relatedness confounds assumptions of independence between genotypes of different individuals, as well as between their heritable phenotypes. Isolated populations therefore contain a large amount of cross-individual correlations, which pose problems for most mapping methods. Further, consanguinity may be present so that two alleles in a random individual may also be correlated. The hidden correlation in the data can cause an overdispersion of the naïve test scores for association and consequently, false positive associations. Hence, the major challenge for performing GWAS in isolated populations is to account for this non-random intra- and inter-individual correlation.
Although standard association tests assume independence of genotypes and phenotypes across samples, several specialized approaches for association do account for underlying relatedness expected in isolated populations. In this work, we set out to select and evaluate different methods to map complex traits in an isolated, founder population. Many methods exist that rely on knowledge of the underlying family structure in the population. These approaches overcome the confounding effects of non-random correlation via deconstruction of the population into family units and the analysis of association independently within each unit (within family variance) (25–27). Some of these ‘family-based’ methods can be extended by adding the variance between families to the within family variance (28–32). A different type of ‘population-based’ approach does not utilize prior knowledge of family structure, but rather explicitly models the relatedness between all pairs of individuals based on their genotypes, and incorporates this variance into a mixed model for association (30,33–38). Such models have recently been extended to genome-scale human studies and have been shown to effectively control for population structure (37,38). Here our emphasis is on the evaluation of such methods in the context of extensive relatedness in study samples. We compared the performance of four representative methods that account differently for relatedness while testing for genome-wide association: (i) focusing on allele transmission to offspring within families (25) (ii) measuring association within as well as between families (28,29,32) and finally (iii) capturing the relatedness between all individuals in the population to construct a mixed model to test for association (37). Our simulations showed the mixed-model method to have increased statistical power to detect association, offering a 1.8–2-fold improvement over the family-based approaches.
As a proof of principle, we then used the mixed-model method for genome scans relating to metabolic traits and electrocardiographic measures in 2906 related individuals from the Island of Kosrae, Federated States of Micronesia, who were previously genotyped for over 350 000 SNPs. We reanalyzed data for 17 phenotypes previously studied in this cohort (19,39), along with 8 additional phenotypes. As positive controls, we observe nine genome-wide significant associations with known loci of measured levels of plasma cholesterol, high-density lipoprotein (HDL), low-density lipoprotein (LDL), triglycerides (TGs), thyroid stimulating hormone (TSH), homocysteine (HOMO), C-reactive protein (CRP) and uric acid, with only one detected in previous analysis of the same traits (19). In addition, we refined a broad signal peak for uric acid levels (chr11:59.1–65.3 Mb) by analysis of identity-by-descent (IBD) shared genetic segments that underlie the peak and dissected the full set of long-range shared haplotypes in this region. We identified a single 3% carrier frequency haplotype that accounted for all the signal in the region, and replicated one of the two previously known signals, refining that signal by a factor of 4. Finally, we show a region of novel association for height (HGT) (rs17629022, P< 2.1 × 10−8).
RESULTS
Computational performance
Five representative programs for performing association in the context of relatedness [family-based association test (FBAT), FBAT+Wald, a family-based test of association with quantitative phenotypes offered in the PLINK framework (PLINK/QFAM-total), efficient mixed model association expedited (EMMAX) and sequential oligogenic linkage analysis routines (SOLAR)] were assessed for speed and maximum memory allocation to determine their suitability for a genome-scale association analysis across thousands of individuals. The average computation time for association per marker for each program is given in Table 1. FBAT, FBAT+Wald, EMMAX and PLINK/QFAM-total ran at speeds <0.06 s/SNP, with FBAT performing >2-fold faster than the other three methods, compared with the SOLAR method which was >100-fold slower. The slow speed of the SOLAR method prohibited its further inclusion in the study. The maximum virtual memory allocation required by the remaining three methods did not exceed 1.7 GB of RAM during the runtime of each program.
Table 1.
Computation time (average computation time per second per SNP) and maximum memory allocation (megabytes of RAM) for each association methods (FBAT, FBAT+WALD, PLINK/QFAM-total, EMMAX and SOLAR)
| FBAT | FBAT+Wald | PLINK/QFAM-total | EMMAX | SOLAR | |
|---|---|---|---|---|---|
| Speed (sec/SNP) | 0.013 | 0.021 | 0.057 | 0.039 | 8.74 |
| Memory (MB RAM) | 1660 | 1660 | 760 | 790 | 3200 |
Comparison of method accuracy and efficacy on known-answer data
We evaluated the performance of different approaches to genome-wide association in the context of relatedness by comparing representative tools for each approach. The family-based association test (FBAT) is a method that considers allele transmission to offspring within families (25). The FBAT group has recently suggested an extension of their method that combines the within-family FBAT score with a rank-based, between-family score, derived from a Wald test of the whole cohort, that is robust to overdispersion, into one single test statistic (FBAT+Wald), which we also tested (32). Plink QFAM for quantitative traits, within- and between-mode (PLINK/QFAM-total), uses a linear regression-based between/within family approach similar to a model described previously (26,40). In the PLINK/QFAM-total framework, confounding effects of family structure are controlled by independent within- and between-family permutation strategies for estimating exact significance levels, where the empirical within- and between-family permuted P-values are combined to a single score. We corrected for any residual overinflation of the test statistic by standard genomic control adjustment (28,29). Finally, EMMAX is a method that first uses high-density genotypes to empirically estimate levels of relatedness between every pair of individuals, which are captured in a kinship matrix. The kinship matrix is then incorporated into a linear mixed model to adjust for correlation in the phenotypic distribution during association mapping (37). We used all four methods to perform genome-wide association analysis in a highly related, population-based cohort from the Island of Kosrae, Micronesia, that is comparable in size (n= 2906) to published single study case/control cohorts.
In order to assess the empirical power for each association method, we considered data for over 350 000 SNPs, with a minor allele frequency (MAF) >0.01, genotyped on an Affymetrix 500K platform in the Kosrae samples. We repeatedly analyzed association of these SNPs to a null, moderately heritable (h2 = 0.42 on Kosrae) phenotype, body mass index (BMI), which we modified afresh multiple times. Each modified phenotyped included a simulated genotype/phenotype association explaining 2% of the phenotypic variance to a different random SNP, additively combined to the true trait value. This effect size was chosen to echo milder effects detected and detectable in larger-sample studies that are now available. These data sets were constructed by selecting 1000 SNPs randomly across the genome (770 common SNPs after filtering, see Materials and Methods), where each phenotype was altered to reflect association with a different SNP in the random subset. In total, 770 genome-scale data sets containing modified SNP-phenotype association were generated and analyzed by all four methods.
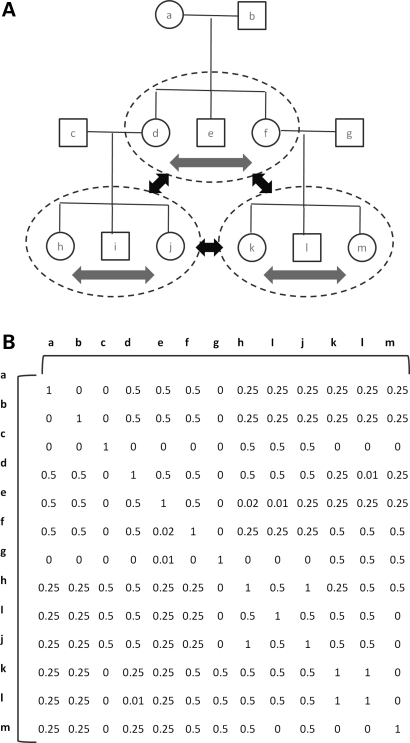
Following extensive pedigree construction based on genetic and oral information, >90% of the genotyped individuals on Kosrae form a single extended pedigree spanning 5+ generations and containing numerous consanguineous offspring and multiple marriage loops (19). To the best of our knowledge, large pedigrees with such complexity cannot be handled whole by methods that have a within-family component. Therefore, we broke our pedigree into smaller units of sibships-without-parents for the purposes of method comparison. This resulted in 586 sibships consisting of two or more individuals who share a mother and father (Fig. 1A). Any genotyped parents are considered only in the context of the parents’ sibship. Of the individuals not included in any sibship (n= 612), a subset was identified in which any two members of the subset were related to the degree of first cousins or less, as determined by genome-wide identity-by-descent sharing, resulting in 240 additional ‘sibships’ of size 1. To fairly compare FBAT and PLINK/QFAM-total to EMMAX and FBAT+Wald, we first analyzed only sibship individuals. However, to also quantify the benefits of using the entire data set, we repeated the analysis examining the entire cohort with EMMAX and the sibships for the FBAT and entire cohort for the Wald components of the FBAT+Wald method.
Figure 1.
Schema of the sibship and kinship matrix-based approaches to association studies in related cohorts (A) The extended Kosrae pedigree is broken into sibships without parents (indicated by the dotted gray circles). Parent-child or cousin relationships may exist between different sibships. Tests of association may be performed within sibships (gray arrows) and between sibships (black arrows). (B) A kinship matrix of all pairs of the same individual.
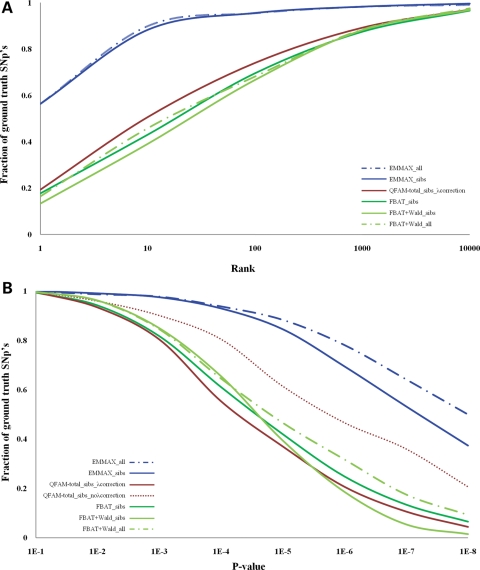
The empirical power for each method was recorded in two ways; the reported (i) genome-wide rank (Fig. 2A) and (ii) P-value (Fig. 2B) of the ground-truth SNP according to each method across repeated simulation iterations. While these criteria are equivalent when reported P-values are uniformly distributed (as required by the definition of a P-value), inclusion of both criteria facilitates evaluation of potential bias in such distributions. Comparison of the within-family only (FBAT) versus combined within- and between-family (PLINK/QFAM-total and FBAT+Wald) versus mixed model (EMMAX) tests demonstrated that the greatest power, as measured by rank order of the true effect, was obtained by the use of the mixed-model method. EMMAX had 88% power to rank the true SNP in the top 10 genome-wide, a 1.7-, 2- and 2.1-fold improvement over PLINK/QFAM-total, FBAT and FBAT+Wald, respectively (Fig. 2A). In addition, 56% of truly associated SNPs achieved genome-wide significance (gws) for the mixed-model method compared with <16% for the other methods (Fig. 2B—see Discussion). Although, the inclusion of the entire cohort (an additional ∼300 individuals that did not fall into sibships) in the FBAT+Wald method increased the number of truly associated SNPs that achieved gws to 21%, this was less than half of the gws SNP's EMMAX detected. The improvement in power was also observed in comparison of the PLINK/QFAM-total naive test scores (i.e. not adjusted for genomic control) with EMMAX, indicating that the dampening effect of the lambda correction does not account for the total power difference between the two methods (Fig. 2B). Finally, the EMMAX analysis of the entire cohort showed a further 1.2-fold increase in power for the top 10 ranked ground truth SNPs, with an additional 10% of SNP's surpassing gws compared with the EMMAX analysis of the sibships.
Figure 2.
Empirical estimation of power of association for four representative association methods for related cohorts (A) The aggregate rank of the ground truth SNP across 770 simulated data set for association performed with FBAT, FBAT+Wald, PLINK/QFAM-total+genomic control and EMMAX with sibship structured (n= 2007), simulated data sets and FBAT+Wald and EMMAX with all individuals (n= 2317), simulated data sets. (B) The aggregate P-values of the ground truth SNP for association performed with FBAT, FBAT+Wald, Plink/QFAM-total+genomic control, Plink/QFAM-total without genomic control and EMMAX with sibship structured, simulated data sets and FBAT+Wald and EMMAX with all individuals, simulated data sets.
Results from the genome-wide association analysis of 25 metabolic and electrocardiographic traits
On the basis of our observations from the simulated data, we selected the mixed-model method as the most powerful method for analyzing our study population. We first built a kinship matrix of pairwise identity-by-state metrics based on high-density markers for the entire pedigree, which was incorporated into the mixed model for association mapping (37). We analyzed 25 metabolic and electrocardiographic traits, using measurements from 3 clinical screenings carried out on Kosrae in 1994, 2001 and 2003 (19,39). The details of the phenotypes, including population means, trait heritability's, covariate and score inflation adjustments, are given in Supplementary Material, Tables S1 and S2 and (19,39). We had previously published the results of the genome-wide association analysis of 17 of these traits using the PLINK/QFAM-total framework (19,39). Here, we reanalyzed data for these 17 published phenotypes using the EMMAX framework, along with another 8 previously unreported phenotypes from the island.
We observe minimal score inflation using the EMMAX method where the inflation factor λ is estimated to be in the range from 1.03 to 0.96 for all 25 traits (Supplementary Material, Table S1). This contrasts with previously reported λ values that are significantly >1 for a subset of the traits analyzed using PLINK/QFAM-total (λ range: 2.05–1.10) (19). We note that genome-wide analysis with λ = 1.1 inflation of P-values results in a 4.8-fold increase in the expected number of false-positive results attaining gws, with practical implication for the feasibility of follow-up.
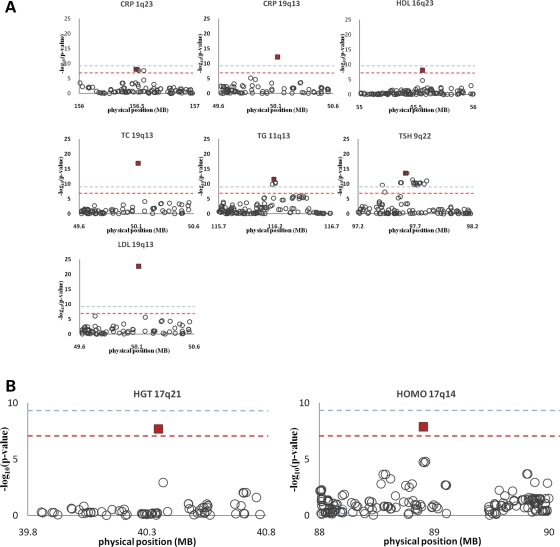
Table 2 provides the top SNP per region emerging genome- and study-wide significant from the EMMAX analysis of all 25 traits. We determined a gws threshold based on ∼310 k effective independent tests (∼350−320 k actual tests) to be 1.6 × 10−7 (see Materials and Methods). As positive controls, we observed associations to eight genomic regions for seven traits exceeding our gws threshold. Top SNPs are in or near to known loci for plasma cholesterol (APOE; rs4420638, P ≤ 1.47 × 10−17), HDL (CETP; rs1800775, P ≤ 7.03 × 10−9), LDL (APOE; rs4420638, P ≤ 1.47 × 10−23), TGs (APOC3/A5; rs7396835, P ≤ 2.42 × 10−12), TSH (FOXE1; rs1877431, P ≤ 6.32 × 10−14), HOMO (NOX4; rs1836883, P≤ 1.3 × 10−8) and CRP (APOE; rs4420638, P ≤ 6.16 × 10−13 and CRP; rs3093077, P ≤ 7.10 × 10−9) (41–47). These eight regions are shown in detail in Figure 3A. Five of these signal peaks also surpass our study-wide significance threshold of 6.4 × 10−9 (see Materials and Methods). Only one of these positive controls had been detected at gws in the previous analysis of the same traits via PLINK/QFAM method; TGs (APOC3/A5; rs7396835, P ≤ 1.2 × 10−9) (19). In addition we observe two other known positive controls significant at FDR <0.3 (Supplementary Material, Table S5); LDLs (HMGCR; rs3846663, P ≤ 8.77 × 10−7) and HDL (APOE; rs4420638, P ≤ 6.33 × 10−7) (41,48).
Table 2.
Genome- and study-wide significant SNPs from the analysis of 25 traits
| Trait | na | CHR | POS (MB)b | SNP | A1c | A2d | MAF | effect size |
% varg | EMMAX (P-value) | FBAT (P-value) | FBAT+WALD (P-value) | PLINK/QFAM (P-value) | Gene regionh | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| βe | s.e.f | ||||||||||||||
| TC | 2753 | ||||||||||||||
| 19 | 45.4 | rs4420638 | G | A | 0.182 | 0.274 | 0.033 | 2.42 | 1.47 × 10−17 | 3.43 × 10−5 | 2.31 × 10−5 | 3.43 × 10−7 | APOC1/APOE/TOMM40 | ||
| LDL | 2775 | ||||||||||||||
| 19 | 45.4 | rs4420638 | G | A | 0.182 | 0.316 | 0.033 | 3.24 | 2.57 × 10−23 | 7.54 × 10−5 | 1.72 × 10−4 | 1.91 × 10−7 | APOC1/APOE/TOMM40 | ||
| HDL | 2774 | ||||||||||||||
| 16 | 57.0 | rs1800775 | A | C | 0.394 | 0.183 | 0.026 | 1.82 | 7.03 × 10−9 | 4.32 × 10−5 | 3.21 × 10−4 | 5.82 × 10−5 | CETP | ||
| TG | 2754 | ||||||||||||||
| 11 | 116.7 | rs7396835 | T | C | 0.372 | 0.214 | 0.026 | 2.36 | 2.42 × 10−12 | 4.33 × 10−7 | 2.31 × 10−8 | 1.22× 10−9 | APOA5/APOCIII | ||
| CRP | 1872 | ||||||||||||||
| 19 | 45.4 | rs4420638 | G | A | 0.182 | −0.318 | 0.041 | 3.05 | 6.16 × 10−13 | 8.56 × 10−6 | 1.02 × 10−6 | 1.60 × 10−6 | APOC1/APOE/TOMM40 | ||
| 1 | 159.7 | rs3093077 | C | A | 0.247 | 0.245 | 0.037 | 2.25 | 7.10 × 10−9 | 5.41 × 10−2 | 2.11 × 10−3 | 2.31 × 10−3 | CRP | ||
| HOMO | 1870 | ||||||||||||||
| 11 | 89.3 | rs1836883 | T | C | 0.321 | 0.210 | 0.036 | 1.75 | 1.29 × 10−8 | 6.99 × 10−4 | 5.67 × 10−4 | 1.17 × 10−8 | NOX4 | ||
| TSH | 1858 | ||||||||||||||
| 9 | 100.5 | rs1877431 | A | G | 0.224 | −0.299 | 0.037 | 3.46 | 6.32 × 10−14 | 1.10 × 10−2 | 3.21 × 10−3 | 3.5 × 10−6 | FOXE1 | ||
| HGT | 2364 | ||||||||||||||
| 17 | 43.0 | rs17629022 | C | T | 0.064 | 0.481 | 0.058 | 2.83 | 2.00 × 10−8 | 8.36 × 10−3 | 7.56 × 10−3 | 1.72 × 10−3 | GFAP | ||
| URT | 1861 | ||||||||||||||
| 11 | 63.9 | rs2186571 | A | G | 0.028 | −1.433 | 0.101 | 9.79 | 1.77 × 10−34 | 4.20× 10−20 | 2.19× 10−18 | 2.11× 10−27 | URAT1i | ||
This table lists the top SNP in a genetic region for the association using the EMMAX method of 25 traits pertaining to metabolic syndrome and electrocardiographic conductance that surpasses genome-wide significance (P≤ 1.6 × 10−7). SNPs that also surpassed study-wide significance (P≤ 6.4 × 10−9) are indicated in italics. P-values for association for the same SNPs using the other three methods tested in this study are also given.
an, the number of phenotyped individuals for the trait.
bPOS(MB): chromosomal physical position in megabases; Genome Reference Consortium build 37 (GRCh37).
cA1, the minor allele.
dA2, the major allele.
eβ, the effect size (linear mixed-model regression coefficient).
fs.e., the standard error of the effect size.
g%var, the percent variance explained (oneway ANOVA).
hCandidate genes in the region of the top SNP.
irs2186571 is >400 kb upstream of URAT1 (see Results for explanation).
Figure 3.
Ten regions of genome-wide significant associations. (A) We observe eight genome-wide significant associations to known loci for plasma cholesterol (TC), high-density lipoprotein (HDL), low-density lipoprotein (LDL), triglycerides (TGs), thyroid stimulating hormone (TSH), homocysteine (HOMO) and C-reactive protein (CRP), with only one detected in the previous analysis of the same traits. (B) Region of novel association for height (HGT) (rs17629022, P< 2.1E-8). The trait and chromosomal location are given for each plot. The y-axis show's –log10 P-value (LOD) and the x-axis shows the chromosomal physical position in megabases. The maroon square represents the location of the top scoring SNP. The red and pale blue dashed lines indicate the genome- and study-wide significance thresholds, respectively.
To examine the genetics of common traits on Kosrae when compared with outbred populations, we assessed the effect sizes of known loci observed in our study. Specifically, we identified 53 established associations in large, mainly Caucasian studies across seven traits (BMI, CRP, HDL, HGT, LDL, TG and URT), where the associations had sufficiently high effect size and/or allele frequency to be detectable at nominal significance in the ∼3000-strong Kosrae study (Supplementary Material, Table S4) (41,47–55). For SNPs not directly typed on the Affymetrix array, association results are reported for a proxy on the Affymetrix chip with strong correlation (r2> 0.95) to the original SNP in both HapMap Caucasian (CEU) and Asian [Han Chinese (CHB), Chinese in Denver (CHD), Japanese (JPT) and Gujarati Indians (GIH)] panels. Of the 53 SNPs, only 21 had signals that were detectable on Kosrae at nominal significance, indicating that more than half the associated SNPs were tagging causal variants that were either too rare to be detected or not present on the Island. Sixteen of the 21 detectable signals on Kosrae had effects in the same direction as observed for those makers in the Caucasian cohorts (P< 0.01), and 5 were in the opposite direction. Of the former set of 16 SNPs, 12 showed stronger effect sizes on Kosrae, a greater number than expected by chance (P< 0.027). However, in this comparison, we cannot rule out the possibility that effect sizes may be inflated upward due to winners curse, confounding this estimation.
We observed two interesting genome-wide significant signals, one strong signal for uric acid levels that resides >500 kb upstream of two independent known associations (rs2186571, P< 1.8 × 10−34), and one signal for HGT which represented a novel association in the Kosraen cohort (rs17629022, P< 2.1 × 10−8) (Supplementary Material, Fig. S3A and Fig. 3B, respectively). The novel SNP rs17629022 for HGT has a MAF of 0.06 and a strong effect size of 0.48 standard deviations per allele (for normalize z-scores, μ = 0, σ = ± 1). This corresponds to an average 1.28′′ (95% confidence intervals: 0.97′′–1.66′′) and 4.23′′ (95% confidence interval: 2.48′′–5.97′′) average increase in HGT in carriers (n= 271) and minor allele homozygotes (n= 13), respectively, compared with major allele homozygotes. The SNP resides on chromosome 17q21 in a gene-rich region containing GAP, CCDC103 and FAM187A. In all, of the 10 hit SNPs associated via the EMMAX method, two were also found to be associated with gws level by PLINK/QFAM and FBAT+Wald; TGs (APOC3/A5; rs7396835, P ≤ 1.2 × 10−9 and P ≤ 2.3 × 10−8, respectively) and urate (rs2186571, P< 2.1 × 10−27 and P ≤ 2.2 × 10−18, respectively), and one by FBAT; urate (rs2186571, P< 4.2 × 10−20), see Table 2.
Analysis of the uric acid trait revealed a broad peak in the genome scan on chromosome 11q13.1 (59.2–65.3 Mb) containing 54 SNPs that surpassed the study-wide significance threshold (Supplementary Material, Table S5 and Fig. S3A). The top SNP, rs2186571, represents a novel association with uric acid. rs2186571/A has a MAF of 0.028 and a strong effect size of −1.14 standard deviations per allele, accounting for ∼10% of uric acid level variance on Kosrae, where carriers of rs2186571/A have a mean uric acid level of 3.6 mg/dl compared with the major allele homozygotes level of 5.5 mg/dl. However, rs2186571 resides ∼500 kb upstream of two variants on chromosome 11q13.1 recently found to be associated with uric acid levels (56). To determine whether the strong uric acid signal represented a novel observation or might tag either of the previously observed signals, we performed a haplotype-based fine-mapping of the signal region and conditional analysis.
Fine mapping the interval of uric acid association
Common shared long-range haplotypes were detected via pairwise IBD genetic segment matching between all individuals in the pedigree using the GERMLINE software (56). The IBD segments were at least 3 cM in length and allowed for up to 1% mismatching due to genotyping error. IBD segments were then clustered to groups of similar haplotypes, with >500 kb overlapping sequence and sharing >99% sequence identity, as described in detail previously (57). Clusters of shared haplotypes were then independently mapped to uric acid levels, which identified a single strong mapping cluster, where the specific sharing boundaries of the haplotype mapped to a ∼2 Mb sub-region (chr11:63.7–65.5) of the uric acid GWAS signal peak (Supplementary Material, Fig. S3A). This ‘uric acid haplotype’ (rs4980499 to rs1074156) is unique to a group of 96 individuals (carrier frequency 0.03), is strongly associated with uric acid (P< 9.07 × 10−46) and has an effect size of −2.12 standard deviations per allele (accounting for ∼20% of uric acid level variance on Kosrae; 95% confidence intervals 16.8–21.6%). Conditioning the uric acid levels for the uric acid haplotype effect abolishes signal at the rs218571 SNP (P< 0.83), whereas a significant signal remains (P< 2.1 × 10−5) at the uric acid haplotype when the levels are adjusted for the effect of rs218571 (Supplementary Material, Table S4). Genome-wide scan of haplotype conditioned uric acid levels reveals no remaining genome-wide significant signals (Supplementary Material, Fig. S3B). Therefore, the uric acid haplotype likely captures the full signal tagged by the top GWAS SNP.
We next determined whether the uric acid haplotype represented a novel variant or a refinement of either of two local previously shown uric acid variants, rs17300741 and rs505802. The two known variants reside in/near organic ion transporter 4 (OAT4) and urate transporter 1 (URAT1), respectively, which are known candidate genes for renal urate anion exchange and are thought to regulate blood uric acid levels (58,59). Neither variant was directly assayed in the Kosrae genotype panel. However, two assayed variants on the 500 k Affymetrix chip, rs12362644 and rs10897518 that flank rs17300741 and rs505802, respectively, were observed to be in complete linkage disequilibrium (r2= 1) with each variant in all HapMap Asian panels (CHB, CHD, JPT, GIH). We therefore concluded that rs12362644 and rs10897518 genotyped on Kosrae would be strong tags for the unobserved rs17300741 and rs505802 variants, respectively. Indeed, signals for association were observed for rs12362644 (P < 8.16 × 10−5) and rs10897518 (P < 3.41 × 10−17), the latter being study-wide significant. Analysis of the uric acid haplotype conditioned on rs12362644 and rs10897518 abolished the signal at these two variants (P< 0.41 and P< 0.93, respectively). However, the strong signal at the uric acid haplotype persisted (P< 8.48 × 10−23). Conversely, when uric acid levels are conditioned on the uric acid haplotype, a moderate signal for rs12362644 (tagging rs17300741) remained 2.66 × 10−3; however, the signal at rs1087518 (tagging rs505802) was abolished (P< 0.94) (Supplementary Material, Table S4), indicating that, while there is some correlation between rs12362644 and the uric acid haplotype, the haplotype effect is not explained by the effect of rs12362644, but that the variant tagged by rs10897518 is carried on the uric acid haplotype background (Supplementary Material, Table S4). Moreover, the comparing the 0.06 frequency of rs1087518 on Kosrae to the lower 0.015 frequency of the uric acid haplotype indicates a refinement of the signal by a factor of 4, dramatically reducing the sequence and sample search space for pinpointing the underlying causal variant.
DISCUSSION
To determine which association method would be best powered to analyze a highly related isolate population, we compared four tests utilizing different approaches for capturing underlying population structure. One of the methods measured variance within family units and three of the methods added the variance between families using different strategies to control for the overinflation of the naïve score that resulted from the sample structure. Two of the approaches uniformly rescaled the markers by adding a rank-based P-value to a score or adjusting the score for overinflation by genomic control. These approaches reduce false positives, but also lead to decreased power for true positives. The mixed-model approach modified the test statistic in a marker specific way, which in turn changed the ranks of associated results, so that false positives are reduced, but true positives did not change. In our evaluation of four representative methods for mapping disease alleles in highly related isolated populations, we found that the mixed-model method significantly outperformed the other three methods. The power difference was indeed in part due to the over-conservative uniform adjustments, but they did not account for all the power difference between the two methods. The kinship matrix incorporated in the mixed model likely captured more between-family variance than the other methods. Further, as the mixed-model method was not restricted to families, an analysis across the whole sample increased the power.
Handling hidden and direct relatedness in the context of a mixed-model has both theoretical and practical advantages over other approaches for association testing in closely knit, isolated cohorts. Theoretically, it handles relatives distant and close, demographic trends and complex pedigrees; variation both within and between families is utilized; and the test statistic produced is uniformly distributed, with no artificial overdispersion of the P-value. Practically, the ability to include all samples in the analysis gave a >2-fold boost in power and our empirical results support the use of mixed models for association in populations such as our samples. Finally, the proof of the pudding is the analysis and reanalysis of 25 biomedical traits from the island using the more powerful mixed-model approach yielding 10 significant hits, only one of which had been detected by other methods. More important were the discovery of a putative novel association for HGT, and a novel long shared haplotype that accounts for ∼20% variance in uric acid levels on the Island.
These two novel associations exemplify both the limitations and opportunities for performing genetic mapping in isolated populations. We demonstrate association for HGT in a region not previously observed, despite multiple large-scale genetic studies in Caucasians (59,60). It is possible that the underlying mutation is of stronger effect or higher frequency on the island, making it easier to detect, alternatively the causal variant may also be Asian specific, or even private to the island. If the latter case is true, then the signal is unlikely to be confirmed by replication. It should also be noted that the signal for the HGT SNP did not reach study-wide significance and could represent a false positive. On the other hand, we observe an extremely strong signal for uric acid level, also present in Caucasian populations, which we could refine by a IBD-based haplotype fine-mapping facilitated by the abundance of long shared haplotypes in isolated populations (54,61). Further, as the uric acid haplotype tags only one of the known variants, our finding supports independence between the two reported signals in the region.
In conclusion, it is worthwhile reviewing our work in the context of recent ideas and results in the association analysis. Specifically, the emerging picture is that despite comprehensive scans of association with common SNPs of multiple complex phenotypes, only a limited fraction of their heritable component has been identified (17,62). Expanding the search for associated alleles to variants that are rare in the general population provides another piece of this heritable component, with multiple gene sequencing studies implicating such variants with moderate to high penetrance (63–67). Our experiences in association testing in an isolated population can serve as a model for other studies of similar cohorts. Methodologically, mixed models optimally extract association information from such samples. More importantly, in terms of study design and choice of cohort, we observe some of the previously theorized advantages and limitations of isolated populations. We showed that some variants that are associated with traits in other populations do not replicate on Kosrae, due to bottleneck effects rather than lack of power. On the other hand, observed associations show strong effects and statistical support for a cohort of this size, potentially due to increased genetic and environmental homogeneity. Finally, as GWAS cohorts increase in size to the six-digit range, and inclusion of somewhat related individuals becomes a practical modus operandi, results in this work are expected to become relevant to a wide range of populations.
MATERIALS AND METHODS
Study population and phenotypes
A full description of the screening and genotyping of the Kosraen cohort was described elsewhere (19). Briefly, we surveyed 3148 highly related individuals from the Pacific Island of Kosrae in three separate screenings carried out in 1994, 2001 and 2003, who represent >75% of the adult population on the Island. Informed consent was obtained from each individual screened and so were self-reported family histories and lifestyle information. Fasting blood was collected and centrifuged. Plasma and buffy coats were frozen and shipped to Rockefeller University, NY, for serological assays and DNA extraction. Phenotypes were measured for 25 metabolic and electrocardiographic traits. Seventeen traits which were previously described [total cholesterol, LDL, HDL, TG, CRP, TSH, HGT, weight (WGT), waist circumference, BMI, percent body fat, leptin hormone levels, fasting blood sugar, systolic blood pressure, diastolic blood pressure, PR interval and QRS interval] (19,32) and an additional eight traits [insulin sensitivity, glomerular filtration rate, homocysteine (HOMO), folic acid (FLT), uric acid (URT), Sokolow-Lyon voltage, Cornell voltage and RR (inverse heart rate) interval (RRD)] [Supplementary Material, Table S2 and (19,32) for details]. IRB approval was obtained from all participating institutions. A first pass reconstruction of the extended pedigree of the 3000-strong cohort included denser sampling of a further ∼1000 related non-genotyped individuals to fill out the pedigree and careful cross-referencing of patient records. More than 90% of the cohort formed a single extended 5+ generation pedigree. Genome-wide SNP data were subjected to identity-by-state analyses and identity-by-descent estimation performed in PLINK (29) to correct and validate the pedigree structure. Full details of the pedigree construction are described elsewhere (19).
Genotypes and quality control
A total of 2906 study participants were successfully genotyped on the Affymetrix 500K platform; data were generated at Affymetrix, South San Francisco, CA, USA. Genotypes were called with the BRLMM algorithm and a minimum call rate of 95% were achieved. A total of 446 802 SNPs passed quality control filters and between ∼122K-91K SNPs that were monomorphic or very rare (MAF <0.01) for each phenotype were also excluded. The final data set yielded between 354 901 and 323 902 SNPs per phenotype with MAF >0.01 for the analysis (Supplementary Material, Table S1).
Data simulation
The performance for each association method was evaluated by analyzing simulated data sets constructed from real BMI phenotype data in which no genome-wide significant associations were found (19). For simulation, 1000 SNPs were randomly selected from Kosraen 500 k Affymetrix genotypes and 770 remained after filtering MAF >0.01. Using these SNPs, we generated a total of 770 genome-scale (∼350 k SNPs each) simulated data sets, in which each phenotype (BMI) was ‘spiked’ to represent a dependency with one of 770 SNPs, which contains an effect explaining an additional 2% of phenotypic variance. Each simulated data set, comprising 2906 Kosraen individuals, was deconstructed into a pedigree of 586 sibships of size ≥2 plus 240 individuals who were less than first cousins (a total of 2007 individuals) that could be handled by all four methods. The four methods (FBAT, FBAT+Wald, Plink/QFAM-total and EMMAX) were used to perform association for each of the 770 sibship-structured, simulated data sets. Finally, in order to assess the full power of the both the FBAT+Wald and EMMAX methods, the simulated data sets were also reanalyzed, this time including an extra 310 individuals who did not fit into the sibship pedigrees (n = 2317).
Association methods
We performed comparative analysis for three different association approaches: a within-family test versus two within- and between-family tests versus a mixed-model test. We tested each representative tool under additive models and with moderate parameters to optimize the behavior of association mapping. FBAT (version 2.0.2c) (25) was chosen to represent a within-family test. We set minsize to 3, which is the required parameter of minimum size of family to include for analysis, without loss of any data and biallelic test under an additive model was performed with the default settings. We selected two approaches that combine a within-family test with a between-family score. The first was the QFAM-total procedure implemented in the PLINK framework (plink version 1.05) (29). We ran the within- and between-family test (–qfam-total), combined with a 1 M permutations (–aperm 1 000 000) to calculate the within- and between-family permutation-based combined empirical P-value. We also calculated any overdispersion of the test statistic using the –adjust flag, which adjusts the empirical P-value by the genomic control inflation factor. The second within- and between-family test was an extension of the FBAT test. For this we calculated a ‘screening statistic’ by first deriving a rank-based P-value from a Wald test performed in Plink (–assoc) and we then combined the FBAT and rank-based Wald score with equal weights as outlined (32) to produce a FBAT+Wald score. Finally, we selected EMMAX (pre-release beta version) for representing a mixed-model method which theoretically can handle all the relatedness of the cohort (34,37). We first calculated an identity-by-state kinship matrix for individuals using the Affymetrix genotypes in EMMAX (emmax-kin –v –h –s –d 10), and then added the kinship matrix to the mixed model for association testing [emmax-kin –v –h –s –d 10 –k (kinship-matrix)] (37). As both FBAT and PLINK/QFAM-total do not support association mapping in complex pedigree structures, the pedigree was broken down into sibships (siblings-without-parents) as described previously (19) and these sibships were used for testing for the FBAT and PLINK/QFAM-total association methods. For a fair comparison, the Wald score of the FBAT+Wald test was derived using just sibships (FBAT+Wald_sibs) and the EMMAX analysis was performed by using just the sibships both for the construction of the kinship matrix and the mixed-model association testing (EMMAX_sibs). For power quantification, the FBAT+Wald and EMMAX analyses were repeated, this time including an extra 310 individuals who did not fit into the sibship pedigrees (n = 2317). In the case of FBAT+Wald, these extra individuals were added to the ‘screening statistic’ (FBAT+Wald_all), and for EMMAX the extra individuals were used both to construct the kinship matrix and included in the mixed model (EMMAX_all).
Association performance
We evaluated the performance of each method by two metrics. First, a rank-based score measured the P-value rank of the ground truth SNP in its own data set. We compared the aggregate of rank of ground truth SNP for the four different association methods and considered the one that assigns more high ranks to ground truth SNPs to be the more powerful method. The second metric was a P-value-based score, which measures the P-value of the ground truth SNP in its respective data set. With the sum of ground truth SNPs exceeding a particular P-value threshold, we alternatively compared the power of each method.
Association mapping on real data
EMMAX was selected for association study of real Kosraen data. A total of 25 quantitative traits were adjusted for age and gender effects, converted to z-scores and outliers (>3 standard deviations from the mean) were removed (Supplementary Material, Tables S1 and S2). Low frequency genotypes (MAF >0.01) were removed. A kinship matrix which captured the relatedness between all pairs of genotyped individuals (n= 2906) was incorporated into the EMMAX mixed model to test for association with each trait. Minimal score inflation was seen in the nominal P-values for all traits (Supplementary Material, Table S1 and Fig. 2). The effect size (β) is given as the difference in standard deviations from the mean per allele and was calculated as the regression coefficient of the linear mixed model. The percent variance explained was calculated by oneway ANOVA. As there is a high degree of linkage disequilibrium in our data, we determined a gws threshold by first extrapolating an approximate testing burden from the trait data. The median minima P-value from 14 null traits was 1.6 × 10−6, indicating an estimated testing burden of ∼310 000 (actual number of tests was ∼350–320 K). The gws threshold to account for multiple testing for each trait was determined by Bonferroni correction based on the estimated testing burden, i.e. 0.05/310 000 = P≤ 1.6 × 10−7. A study-wide significance threshold that also accounts for testing multiple traits in the same study was determined conservatively using the Bonferroni correction: 1.6 × 10−7/25 = P≤ 6.4 × 10−9.
Identifying haplotypes
Genotypes were phased using the BEAGLE-trio algorithm in one run according to the recommended parameters: 1 sample, 10 iterations (68,69). Phased genotypes were then analyzed for pairwise IBD matching by the GERMLINE algorithm (56) using the default parameters: minimum match length of 3 cM, maximum 2 homozygote errors, 0 heterozygote errors and window size of 128 markers. In other words, haplotypes were registered whenever pairwise comparison revealed a window of allele-call identity at least 3 cM in length with up to 1% mismatch allowed for genotyping error.
Clustering haplotypes and haplotype-based association
Clustering GERMLINE-derived haplotypes has been previously described in detail (57). In brief, a set of individuals that share any size overlap of genetic segments were first identified, then the specific sharing boundary positions were mapped between individuals in that set, and each set was dubbed ‘a haplotype cluster’. A single individual can be a member of maximum two haplotype clusters at any given marker and multiple haplotype clusters across all markers in the genome. A total of 93 629 distinct haplotype clusters were identified after filtering for haplotype clusters of ≥0.01 frequency ≥500 kb in length. The haplotype clusters were associated with uric acid levels using EMMAX (see above) and a 13 genome-wide significant haplotypes spanning chr11:36.2–94.2 Mb emerged (P< 8.52 × 10−8), with a chr11: 63.4–65.2 sub-region being most significantly associated (P< 9.06 × 10−46).
Computation
All analysis of the simulated data sets and real data were performed on a 3.0 GHz Intel Xeon dual core 64-bit cluster containing 100 nodes (with four processors each), where each node had 8–16 GB of RAM.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by grants from the Starr Foundation and Howard Hughes Medical Institute for all measurement of metabolic traits and genotypes. Measurements of electrocardiographic traits in Kosrae were supported by a K23 from the National Heart, Lung and Blood Institute (grant number 080025), a Doris Duke Charitable Foundation Clinical Scientist Development Award and a Burroughs Wellcome Fund Career Award for Medical Scientists to CNC. Support from a Women & Science fellowship was provided to EEK. Support from the National Science Foundation (grant numbers CAREER 0845677, EMT 0829882) was provided to IP.
ACKNOWLEDGEMENTS
We are sincerely grateful to the Government and Department of Health of Kosrae for their partnership and especially to the people of Kosrae for making this study possible.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Ober C., Abney M., McPeek M.S. The genetic dissection of complex traits in a founder population. Am. J. Hum. Genet. 2001;69:1068–1079. doi: 10.1086/324025. doi:10.1086/324025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angius A., Bebbere D., Petretto E., Falchi M., Forabosco P., Maestrale B., Casu G., Persico I., Melis P.M., Pirastu M. Not all isolates are equal: linkage disequilibrium analysis on Xq13.3 reveals different patterns in Sardinian sub-populations. Hum. Genet. 2002;111:9–15. doi: 10.1007/s00439-002-0753-z. doi:10.1007/s00439-002-0753-z. [DOI] [PubMed] [Google Scholar]
- 3.Peltonen L., Jalanko A., Varilo T. Molecular genetics of the Finnish disease heritage. Hum. Mol. Genet. 1999;8:1913–1923. doi: 10.1093/hmg/8.10.1913. doi:10.1093/hmg/8.10.1913. [DOI] [PubMed] [Google Scholar]
- 4.Gulcher J., Kong A., Stefansson K. The genealogic approach to human genetics of disease. Cancer J. 2001;7:61–68. [PubMed] [Google Scholar]
- 5.Freimer N.B., Reus V.I., Escamilla M., Spesny M., Smith L., Service S., Gallegos A., Meza L., Batki S., Vinogradov S., et al. An approach to investigating linkage for bipolar disorder using large Costa Rican pedigrees. Am. J. Med. Genet. 1996;67:254–263. doi: 10.1002/(SICI)1096-8628(19960531)67:3<254::AID-AJMG3>3.0.CO;2-N. doi:10.1002/(SICI)1096-8628(19960531)67:3<254::AID-AJMG3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 6.Zamel N., McClean P.A., Sandell P.R., Siminovitch K.A., Slutsky A.S. Asthma on Tristan da Cunha: looking for the genetic link. The University of Toronto Genetics of Asthma Research Group. Am. J. Respir. Crit Care Med. 1996;153:1902–1906. doi: 10.1164/ajrccm.153.6.8665053. [DOI] [PubMed] [Google Scholar]
- 7.Abney M., McPeek M.S., Ober C. Estimation of variance components of quantitative traits in inbred populations. Am. J. Hum. Genet. 2000;66:629–650. doi: 10.1086/302759. doi:10.1086/302759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aulchenko Y.S., Heutink P., Mackay I., Bertoli-Avella A.M., Pullen J., Vaessen N., Rademaker T.A., Sandkuijl L.A., Cardon L., Oostra B., van Duijn C.M. Linkage disequilibrium in young genetically isolated Dutch population. Eur. J. Hum. Genet. 2004;12:527–534. doi: 10.1038/sj.ejhg.5201188. doi:10.1038/sj.ejhg.5201188. [DOI] [PubMed] [Google Scholar]
- 9.van der Walt J.M., Scott W.K., Slifer S., Gaskell P.C., Martin E.R., Welsh-Bohmer K., Creason M., Crunk A., Fuzzell D., McFarland L., et al. Maternal lineages and Alzheimer disease risk in the Old Order Amish. Hum. Genet. 2005;118:115–122. doi: 10.1007/s00439-005-0032-x. doi:10.1007/s00439-005-0032-x. [DOI] [PubMed] [Google Scholar]
- 10.Newman D.L., Hoffjan S., Bourgain C., Abney M., Nicolae R.I., Profits E.T., Grow M.A., Walker K., Steiner L., Parry R., et al. Are common disease susceptibility alleles the same in outbred and founder populations? Eur. J. Hum. Genet. 2004;12:584–590. doi: 10.1038/sj.ejhg.5201191. doi:10.1038/sj.ejhg.5201191. [DOI] [PubMed] [Google Scholar]
- 11.Arcos-Burgos M., Muenke M. Genetics of population isolates. Clin. Genet. 2002;61:233–247. doi: 10.1034/j.1399-0004.2002.610401.x. doi:10.1034/j.1399-0004.2002.610401.x. [DOI] [PubMed] [Google Scholar]
- 12.Gudmundsson G., Matthiasson S.E., Arason H., Johannsson H., Runarsson F., Bjarnason H., Helgadottir K., Thorisdottir S., Ingadottir G., Lindpaintner K., et al. Localization of a gene for peripheral arterial occlusive disease to chromosome 1p31. Am. J. Hum. Genet. 2002;70:586–592. doi: 10.1086/339251. doi:10.1086/339251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindqvist A.K., Steinsson K., Johanneson B., Kristjansdottir H., Arnasson A., Grondal G., Jonasson I., Magnusson V., Sturfelt G., Truedsson L., et al. A susceptibility locus for human systemic lupus erythematosus (hSLE1) on chromosome 2q. J. Autoimmun. 2000;14:169–178. doi: 10.1006/jaut.1999.0357. doi:10.1006/jaut.1999.0357. [DOI] [PubMed] [Google Scholar]
- 14.Peltonen L., Palotie A., Lange K. Use of population isolates for mapping complex traits. Nat. Rev. Genet. 2000;1:182–190. doi: 10.1038/35042049. doi:10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 15.Heutink P., Oostra B.A. Gene finding in genetically isolated populations. Hum. Mol. Genet. 2002;11:2507–2515. doi: 10.1093/hmg/11.20.2507. doi:10.1093/hmg/11.20.2507. [DOI] [PubMed] [Google Scholar]
- 16.Gulcher J.R., Kong A., Stefansson K. The role of linkage studies for common diseases. Curr. Opin. Genet. Dev. 2001;11:264–267. doi: 10.1016/s0959-437x(00)00188-x. doi:10.1016/S0959-437X(00)00188-X. [DOI] [PubMed] [Google Scholar]
- 17.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. doi:10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodmer W., Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. doi:10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe J.K., Maller J.B., Pe'er I., Neale B.M., Salit J., Kenny E.E., Shea J.L., Burkhardt R., Smith J.G., Ji W., et al. Genome-wide association studies in an isolated founder population from the Pacific Island of Kosrae. PLoS. Genet. 2009;5:e1000365. doi: 10.1371/journal.pgen.1000365. doi:10.1371/journal.pgen.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., O'Connell J.R., McArdle P.F., Wade J.B., Dorff S.E., Shah S.J., Shi X., Pan L., Rampersaud E., Shen H., et al. From the cover: whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc. Natl. Acad. Sci. U. S. A. 2009;106:226–231. doi: 10.1073/pnas.0808358106. doi:10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabatti C., Service S.K., Hartikainen A.L., Pouta A., Ripatti S., Brodsky J., Jones C.G., Zaitlen N.A., Varilo T., Kaakinen M., et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 2009;41:35–46. doi: 10.1038/ng.271. doi:10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollin T.I., Damcott C.M., Shen H., Ott S.H., Shelton J., Horenstein R.B., Post W., McLenithan J.C., Bielak L.F., Peyser P.A., et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322:1702–1705. doi: 10.1126/science.1161524. doi:10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kallio S.P., Jakkula E., Purcell S., Suvela M., Koivisto K., Tienari P.J., Elovaara I., Pirttila T., Reunanen M., Bronnikov D., et al. Use of a genetic isolate to identify rare disease variants: C7 on 5p associated with MS. Hum. Mol. Genet. 2009;18:1670–1683. doi: 10.1093/hmg/ddp073. doi:10.1093/hmg/ddp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourgain C., Genin E. Complex trait mapping in isolated populations: are specific statistical methods required? Eur. J. Hum. Genet. 2005;13:698–706. doi: 10.1038/sj.ejhg.5201400. doi:10.1038/sj.ejhg.5201400. [DOI] [PubMed] [Google Scholar]
- 25.Lange C., van Steen K., Andrew T., Lyon H., DeMeo D.L., Raby B., Murphy A., Silverman E.K., MacGregor A., Weiss S.T., et al. A family-based association test for repeatedly measured quantitative traits adjusting for unknown environmental and/or polygenic effects. Stat. Appl. Genet. Mol. Biol. 2004;3:Article17. doi: 10.2202/1544-6115.1067. doi: 10.2202/1544-6115.1067. [DOI] [PubMed] [Google Scholar]
- 26.Abecasis G.R., Cardon L.R., Cookson W.O. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 2000;66:279–292. doi: 10.1086/302698. doi:10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almasy L., Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998;62:1198–1211. doi: 10.1086/301844. doi:10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. doi:10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 29.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abecasis G.R., Cardon L.R., Cookson W.O., Sham P.C., Cherny S.S. Association analysis in a variance components framework. Genet. Epidemiol. 2001;21(Suppl. 1):S341–S346. doi: 10.1002/gepi.2001.21.s1.s341. [DOI] [PubMed] [Google Scholar]
- 31.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. doi:10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 32.Won S., Wilk J.B., Mathias R.A., O'Donnell C.J., Silverman E.K., Barnes K., O'Connor G.T., Weiss S.T., Lange C. On the analysis of genome-wide association studies in family-based designs: a universal, robust analysis approach and an application to four genome-wide association studies. PLoS. Genet. 2009;5:e1000741. doi: 10.1371/journal.pgen.1000741. doi:10.1371/journal.pgen.1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J., Pressoir G., Briggs W.H., Vroh Bi I., Yamasaki M., Doebley J.F., McMullen M.D., Gaut B.S., Nielsen D.M., Holland J.B., et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006;38:203–208. doi: 10.1038/ng1702. doi:10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 34.Kang H.M., Zaitlen N.A., Wade C.M., Kirby A., Heckerman D., Daly M.J., Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. doi:10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aulchenko Y.S., de Koning D.J., Haley C. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177:577–585. doi: 10.1534/genetics.107.075614. doi:10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amin N., van Duijn C.M., Aulchenko Y.S. A genomic background based method for association analysis in related individuals. PLoS. One. 2007;2:e1274. doi: 10.1371/journal.pone.0001274. doi:10.1371/journal.pone.0001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. doi:10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Ersoz E., Lai C.Q., Todhunter R.J., Tiwari H.K., Gore M.A., Bradbury P.J., Yu J., Arnett D.K., Ordovas J.M., Buckler E.S. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010;42:355–360. doi: 10.1038/ng.546. doi:10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith J.G., Lowe J.K., Kovvali S., Maller J.B., Salit J., Daly M.J., Stoffel M., Altshuler D.M., Friedman J.M., Breslow J.L., Newton-Cheh C. Genome-wide association study of electrocardiographic conduction measures in an isolated founder population: Kosrae. Heart Rhythm. 2009;6:634–641. doi: 10.1016/j.hrthm.2009.02.022. doi:10.1016/j.hrthm.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fulker D.W., Cherny S.S., Sham P.C., Hewitt J.K. Combined linkage and association sib-pair analysis for quantitative traits. Am. J. Hum. Genet. 1999;64:259–267. doi: 10.1086/302193. doi:10.1086/302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N.P., Rieder M.J., Cooper G.M., Roos C., Voight B.F., Havulinna A.S., et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008;40:189–197. doi: 10.1038/ng.75. doi:10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elliott P., Chambers J.C., Zhang W., Clarke R., Hopewell J.C., Peden J.F., Erdmann J., Braund P., Engert J.C., Bennett D., et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA. 2009;302:37–48. doi: 10.1001/jama.2009.954. doi:10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M., et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. doi:10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandhu M.S., Waterworth D.M., Debenham S.L., Wheeler E., Papadakis K., Zhao J.H., Song K., Yuan X., Johnson T., Ashford S., et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. doi:10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gudmundsson J., Sulem P., Gudbjartsson D.F., Jonasson J.G., Sigurdsson A., Bergthorsson J.T., He H., Blondal T., Geller F., Jakobsdottir M., et al. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat. Genet. 2009;41:460–464. doi: 10.1038/ng.339. doi:10.1038/ng.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pare G., Chasman D.I., Parker A.N., Zee R.R., Malarstig A., Seedorf U., Collins R., Watkins H., Hamsten A., Miletich J.P., Ridker P.M. Novel associations of CPS1, MUT, NOX4, and DPEP1 with plasma homocysteine in a healthy population: a genome-wide evaluation of 13 974 participants in the Women's Genome Health Study. Circ. Cardiovasc. Genet. 2009;2:142–150. doi: 10.1161/CIRCGENETICS.108.829804. doi:10.1161/CIRCGENETICS.108.829804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aulchenko Y.S., Ripatti S., Lindqvist I., Boomsma D., Heid I.M., Pramstaller P.P., Penninx B.W., Janssens A.C., Wilson J.F., Spector T., et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009;41:47–55. doi: 10.1038/ng.269. doi:10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lettre G., Jackson A.U., Gieger C., Schumacher F.R., Berndt S.I., Sanna S., Eyheramendy S., Voight B.F., Butler J.L., Guiducci C., et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. doi:10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R., Elliott K.S., Lango H., Rayner N.W., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. doi:10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loos R.J., Lindgren C.M., Li S., Wheeler E., Zhao J.H., Prokopenko I., Inouye M., Freathy R.M., Attwood A.P., Beckmann J.S., et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. doi:10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ridker P.M., Pare G., Parker A., Zee R.Y., Danik J.S., Buring J.E., Kwiatkowski D., Cook N.R., Miletich J.P., Chasman D.I. Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women's Genome Health Study. Am. J. Hum. Genet. 2008;82:1185–1192. doi: 10.1016/j.ajhg.2008.03.015. doi:10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. doi:10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 53.Kim J.J., Lee H.I., Park T., Kim K., Lee J.E., Cho N.H., Shin C., Cho Y.S., Lee J.Y., Han B.G., et al. Identification of 15 loci influencing height in a Korean population. J. Hum. Genet. 2010;55:27–31. doi: 10.1038/jhg.2009.116. doi:10.1038/jhg.2009.116. [DOI] [PubMed] [Google Scholar]
- 54.Kolz M., Johnson T., Sanna S., Teumer A., Vitart V., Perola M., Mangino M., Albrecht E., Wallace C., Farrall M., et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS. Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. doi:10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dehghan A., Kottgen A., Yang Q., Hwang S.J., Kao W.L., Rivadeneira F., Boerwinkle E., Levy D., Hofman A., Astor B.C., et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–1961. doi: 10.1016/S0140-6736(08)61343-4. doi:10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gusev A., Lowe J.K., Stoffel M., Daly M.J., Altshuler D., Breslow J.L., Friedman J.M., Pe'er I. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 2009;19:318–326. doi: 10.1101/gr.081398.108. doi:10.1101/gr.081398.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kenny E.E., Gusev A., Riegel K., Lutjohann D., Lowe J.K., Salit J., Maller J.B., Stoffel M., Daly M.J., Altshuler D.M., et al. Systematic haplotype analysis resolves a complex plasma plant sterol locus on the Micronesian Island of Kosrae. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13886–13891. doi: 10.1073/pnas.0907336106. doi:10.1073/pnas.0907336106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ekaratanawong S., Anzai N., Jutabha P., Miyazaki H., Noshiro R., Takeda M., Kanai Y., Sophasan S., Endou H. Human organic anion transporter 4 is a renal apical organic anion/dicarboxylate exchanger in the proximal tubules. J. Pharmacol. Sci. 2004;94:297–304. doi: 10.1254/jphs.94.297. doi:10.1254/jphs.94.297. [DOI] [PubMed] [Google Scholar]
- 59.Hirschhorn J.N., Lettre G. Progress in genome-wide association studies of human height. Horm. Res. 2009;71(Suppl. 2):5–13. doi: 10.1159/000192430. doi:10.1159/000192430. [DOI] [PubMed] [Google Scholar]
- 60.Weedon M.N., Lango H., Lindgren C.M., Wallace C., Evans D.M., Mangino M., Freathy R.M., Perry J.R., Stevens S., Hall A.S., et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. doi:10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonnen P.E., Pe'er I., Plenge R.M., Salit J., Lowe J.K., Shapero M.H., Lifton R.P., Breslow J.L., Daly M.J., Reich D.E., et al. Evaluating potential for whole-genome studies in Kosrae, an isolated population in Micronesia. Nat. Genet. 2006;38:214–217. doi: 10.1038/ng1712. doi:10.1038/ng1712. [DOI] [PubMed] [Google Scholar]
- 62.Frazer K.A., Murray S.S., Schork N.J., Topol E.J. Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 2009;10:241–251. doi: 10.1038/nrg2554. doi:10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 63.Fearnhead N.S., Wilding J.L., Winney B., Tonks S., Bartlett S., Bicknell D.C., Tomlinson I.P., Mortensen N.J., Bodmer W.F. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. doi:10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cohen J.C., Kiss R.S., Pertsemlidis A., Marcel Y.L., McPherson R., Hobbs H.H. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. doi:10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 65.Kotowski I.K., Pertsemlidis A., Luke A., Cooper R.S., Vega G.L., Cohen J.C., Hobbs H.H. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am. J. Hum. Genet. 2006;78:410–422. doi: 10.1086/500615. doi:10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Romeo S., Pennacchio L.A., Fu Y., Boerwinkle E., Tybjaerg-Hansen A., Hobbs H.H., Cohen J.C. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat. Genet. 2007;39:513–516. doi: 10.1038/ng1984. doi:10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marini N.J., Gin J., Ziegle J., Keho K.H., Ginzinger D., Gilbert D.A., Rine J. The prevalence of folate-remedial MTHFR enzyme variants in humans. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8055–8060. doi: 10.1073/pnas.0802813105. doi:10.1073/pnas.0802813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Browning B.L., Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am. J. Hum. Genet. 2009;85:847–861. doi: 10.1016/j.ajhg.2009.11.004. doi:10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Browning B.L., Browning S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. doi:10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.