Abstract
M1 macrophages play a major role in worsening muscle injury in the mdx mouse model of Duchenne muscular dystrophy. However, mdx muscle also contains M2c macrophages that can promote tissue repair, indicating that factors regulating the balance between M1 and M2c phenotypes could influence the severity of the disease. Because interleukin-10 (IL-10) modulates macrophage activation in vitro and its expression is elevated in mdx muscles, we tested whether IL-10 influenced the macrophage phenotype in mdx muscle and whether changes in IL-10 expression affected the pathology of muscular dystrophy. Ablation of IL-10 expression in mdx mice increased muscle damage in vivo and reduced mouse strength. Treating mdx muscle macrophages with IL-10 reduced activation of the M1 phenotype, assessed by iNOS expression, and macrophages from IL-10 null mutant mice were more cytolytic than macrophages isolated from wild-type mice. Our data also showed that muscle cells in mdx muscle expressed the IL-10 receptor, suggesting that IL-10 could have direct effects on muscle cells. We assayed whether ablation of IL-10 in mdx mice affected satellite cell numbers, using Pax7 expression as an index, but found no effect. However, IL-10 mutation significantly increased myogenin expression in vivo during the acute and the regenerative phase of mdx pathology. Together, the results show that IL-10 plays a significant regulatory role in muscular dystrophy that may be caused by reducing M1 macrophage activation and cytotoxicity, increasing M2c macrophage activation and modulating muscle differentiation.
INTRODUCTION
Duchenne muscular dystrophy (DMD) is caused by genetic deletion of dystrophin, a subsarcolemmal protein that strengthens the muscle cell membrane (1,2). Loss of dystrophin renders myofibers susceptible to contraction-induced injury (2), leading to muscle degeneration and invasion by a heterogeneous infiltrate of immune cells (3). Although the immune cell infiltrate in dystrophin-deficient muscle is composed of macrophages, T-cells, neutrophils, eosinophils and mast cells, the number of macrophages vastly exceeds that of other inflammatory cell populations (4–9). Depletion of macrophages in the mdx mouse model of DMD at the early, acute peak of muscle pathology produced large reductions in lesions in the plasmalemma of muscle fibers (6), showing that muscle macrophages that are present during the acute, degenerative stage of mdx dystrophy are highly cytolytic, and that they play a central role in the pathogenesis of muscular dystrophy.
Inhibiting the immune response in dystrophic muscle can delay the progression of muscular dystrophy and prolong ambulation. For example, DMD patients treated with glucocorticoids, which are widely used for their immunosuppressant properties, have improved muscle strength and remain ambulatory longer than untreated patients (3,10,11). However, long-term use of glucocorticoids produces unwanted side effects, including weight gain, susceptibility to infection and cushingoid appearance (10,11), which has led to our interest in identifying better-tolerated, immune-based interventions to reduce inflammatory cell-mediated damage to dystrophic muscle. Interleukin-10 (IL-10) is an attractive candidate molecule because it is a pleiotropic cytokine that regulates lymphoid and myeloid cell function (12) and may thereby be able to modulate immune-cell-mediated damage of dystrophic muscle. IL-10 was initially discovered during screens for inhibitory factors produced by Th2 clones that prevented cytokine production by Th1 cells (13). In particular, IL-10 greatly inhibited the expression of interferon-gamma (IFN-γ) (14) and tumor necrosis factor-alpha (TNF-α) (14), both of which drive pro-inflammatory responses and activate cytotoxic, M1 macrophages (13). Once activated to the M1 phenotype, macrophages express cytokines such as TNF-α, IL-1β and IL-6 (15) that further promote inflammation and the M1 phenotype.
Although primary functions of IL-10 are suppressing the production of pro-inflammatory cytokines and deactivating cytolytic, M1 macrophages (16,17), IL-10 can also activate macrophages to the M2c phenotype (18,19). M2c macrophages express the common M2 markers, IL-4Rα, arginase-1 and CD206, but they also express CD163, which is not expressed by other M2 macrophage populations (18,20–23). CD163+ M2c macrophages are the predominant macrophage population during the regenerative stage of mdx muscular dystrophy, which suggests that they may influence muscle regeneration (24). Furthermore, addition of CD163+ macrophages to cultures of rat muscle cells increased myoblast proliferation (25), although data supporting a role for IL-10 in regulating muscle proliferation or differentiation in vivo have not yet been provided.
IL-10 can also have direct effects on muscle cells. Treatment of C2C12 myoblasts with IL-10 prior to treating with pro-inflammatory cytokines can influence the induction of signaling pathways that are activated by cytokine exposure. For example, pre-treatment of myoblasts with IL-10 prevented the induction of JNK phosphorylation by TNF-α (26). Furthermore, IL-10 can block the induction of IL-6 expression by TNF-α in myoblasts and antagonize the inhibitory effects of TNF-α and IL-1β on IGF-1 signaling in myoblasts (26,27). Those findings indicate that modulation of IL-10 levels in inflamed muscle may affect the response of muscle to the inflammatory process, as well as influencing the inflammatory process itself.
The present investigation tests the hypothesis that IL-10 regulates macrophage function during muscular dystrophy and thereby affects muscle pathology. IL-10 null mdx mice (IL-10−/−/mdx mice) were generated to test whether IL-10 reduces muscle fiber injury by inhibiting the cytotoxic activities of M1 macrophages and to test whether manipulation of IL-10 expression in vivo affects the strength or endurance of mdx mice. In vitro assays were also used to test whether IL-10 stimulation of macrophages isolated from mdx muscle reduces their cytotoxicity and assay whether macrophage lysis of muscle cells is influenced by ablation of IL-10 expression. We then tested whether IL-10 influences M2c activation of macrophages in dystrophic muscle by assaying the expression levels of genes associated with M2c macrophages during the acute phase of mdx pathology and during the regenerative phase. Finally, we tested whether IL-10 can regulate muscle regeneration through direct effects on muscle cell proliferation and differentiation in vitro and in vivo. Collectively, our findings provide insight into the diverse processes that are regulated by IL-10 during muscular dystrophy and suggest IL-10 may have value as a therapeutic agent for the treatment of DMD.
RESULTS
Expression of IL-10 and its receptor are elevated in dystrophic muscle during the acute onset of pathology and during muscle regeneration
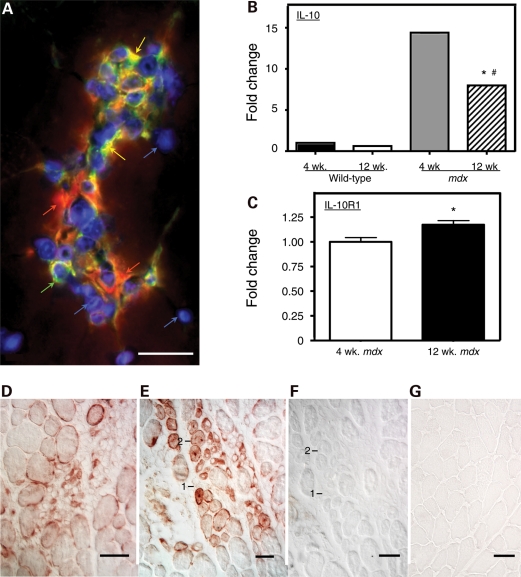
The distribution of satellite cells, M1 macrophages and M2 macrophages in inflammatory lesions in mdx muscle was assayed by double-immunolabeling for F4/80 antigen and CD206. F4/80 is a macrophage specific marker and CD206 is expressed by M2 macrophages (28) and by satellite cells (29). Our observations showed that F4/80+/CD206− M1 macrophages and F4/80+/CD206+ M2 macrophages are co-distributed in inflammatory lesions in mdx muscle (Fig. 1A). Real-time PCR analysis for IL-10 and IL-10R1 expression in mdx muscle showed that IL-10 expression was elevated between 8- and 15-fold in mdx quadriceps muscle, compared with wild-type muscle, with a significant decline in IL-10 expression in mdx muscle between 4-week-old and 12-week-old mdx mice (Fig. 1B). However, real-time PCR data show a small but significant increase in IL-10R1 expression between 4 weeks and 12 weeks of age in mdx quadriceps (Fig. 1C). Immunohistochemical localization of IL-10R1 in mdx muscles from 4-week and 12-week-old mice showed that IL-10R1 was present on the surface of mononucleated cells in inflammatory lesions and on myotubes and regenerative muscle fibers (Fig. 1D–G).
Figure 1.
IL-10-mediated signaling between macrophages and muscle cells is enhanced in muscular dystrophy. (A) Inflamed lesions in 4-week-old mdx muscle contain M1 macrophages, M2 macrophages and satellite cells. Cross-section of quadriceps muscle was labeled with anti-F4/80 (red), which binds all macrophage phenotypes, anti-CD206 (green), which binds M2 macrophages and satellite cells, and DAPI reagent (blue), which binds DNA to show the position of nuclei. M1 macrophages are CD206−/F4/80+ (red). M2 macrophages are CD206+/F4/80+ (orange). Satellite cells are CD206+/F4/80− (green). Bar = 50 µm. (B) Quantitative, real-time PCR results for relative mRNA levels for IL-10 in quadriceps muscles of 4-week-old or 12-week-old wild-type or mdx mice. Expression levels are relative to 4-week-old wild-type muscles, for which the expression level is set at one unit. Asterisks indicate significantly different from 4-week-old wild-type quadriceps at P< 0.001. Hash symbol indicates significantly different from 4-week-old quadriceps from mice that are the same genotype at P< 0.001. Each experimental group included quadriceps from five mice. Error bars are too small to appear in the figure. (C) Quantitative, real-time PCR results for relative mRNA levels for IL-10R1 in quadriceps muscles of 4-week-old or 12-week-old mdx mice. Expression levels are relative to 4-week-old muscles, for which the expression level is set at one unit. Asterisks indicate significantly different from 4-week-old quadriceps at P< 0.05. Each experimental group included quadriceps from five mice. Bars represent sem. (D–G) Cross-sections of quadriceps muscle immunolabeled for IL-10R1 or antibody control preparations. Bars = 50 µm. (D) Section of 4-week-old mdx muscle showing an inflamed lesion containing cells expressing IL-10R1 (red). (E) Section of 4-week-old mdx muscle showing regenerative muscle fibers (labeled 1 or 2) that express IL-10R1 (red). (F) Section adjacent to the section shown in (E). The section was incubated with anti-IL-10R1 from which immunoglobins specific for IL-10R1 were depleted from the antibody solution by incubation with mouse IL-10R1 prior to labeling the section. The fibers labeled 1 or 2 are the same regenerative fibers as those labeled 1 or 2 in (E). (G) Section from 4-week-old wild-type muscle immunolabeled for IL-10R1 using treatment conditions that were identical to those used to label the mdx muscle section shown in (E).
IL-10 reduces activation and cytotoxicity of mdx muscle M1 macrophages in vitro and in vivo
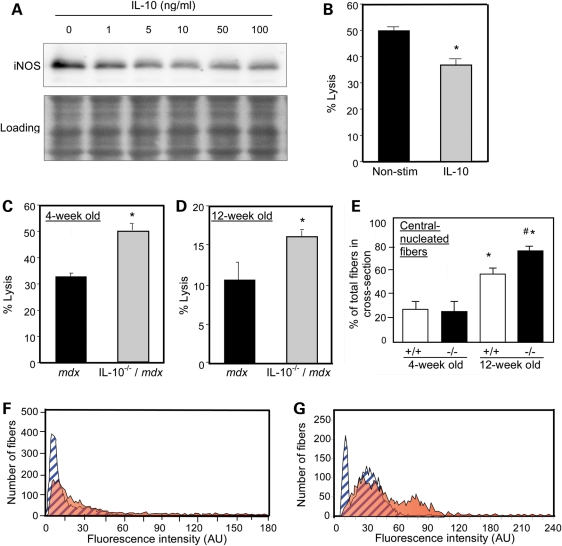
Previous work has shown that M1 macrophages lyze mdx muscle fibers via iNOS-dependent mechanisms during the early, acute phase of mdx pathology (24) and that IL-10 can inhibit iNOS expression (30). Findings presented here show that treatment of mdx muscle macrophages with IL-10 ex vivo significantly reduced iNOS expression and attenuated macrophage cytotoxicity (Fig. 2A and B). Macrophages that were isolated from muscles of either 4-week-old or 12-week-old mdx mice that were null mutants for IL-10 were significantly more cytotoxic than macrophages isolated from mdx mice that expressed IL-10 (Fig. 2C and D). Finally, we tested whether IL-10-mediated signaling could influence mdx muscle fiber damage in vivo by assaying muscle membrane damage and fiber regeneration in mdx mice that expressed IL-10 or in which the IL-10 gene had been ablated (Fig. 2E–G). Influx of the fluorescent, extracellular marker dye Procion orange was used as an indicator of muscle membrane damage, which showed that loss of IL-10 significantly increased muscle membrane damage in both 4-week-old and 12-week-old mdx mice (Fig. 2F and G). Loss of IL-10 in mdx muscle also caused elevations in central-nucleated, regenerative fibers in 12-week-old mice (Fig. 2E), reflecting increased regeneration following increased muscle damage in 4- and 12-week-old IL-10−/−/mdx mice.
Figure 2.
IL-10 deactivates M1 macrophages and reduces muscle damage in mdx mice. (A) Macrophages isolated from 4-week-old mdx mice were stimulated with a range of IL-10 concentrations (1–100 ng/ml) and lyzed after 24 h of stimulation. Macrophage lysates were separated by SDS–PAGE and relative levels of iNOS were assayed by western blotting. The membrane used for immunoblotting was stained with Ponceau red (‘Loading’) before application of the antibody to verify equal protein loading in each lane. (B) Macrophages purified from 4-week-old mdx muscles were stimulated with IL-10 (10 ng/ml) for 24 h prior to co-culturing with C2C12 myotubes. After 24 h of co-culture, cell lysis was decreased 26% by IL-10. Asterisks indicate P< 0.05 compared with non-stimulated control. (C and D) Macrophages isolated from 4-week-old (C) or 12-week-old (D) IL-10−/−/mdx mice displayed increased cytotoxic potential compared with IL-10+/+/mdx controls (30–50% increase in IL-10−/−/mdx versus IL-10+/+/mdx). Asterisks indicates significant difference at P< 0.05 compared with IL-10+/+/mdx. (E) Quantification of the number of central-nucleated fibers in cross-sections of quadriceps muscles from 4-week-old or 12-week-old mdx mice that expressed IL-10 (+/+) or were IL-10 null mutants (−/−). Asterisk indicates significantly different from 4-week-old IL-10+/+/mdx quadriceps at P< 0.05. Hash symbol indicates significantly different from age-matched IL-10+/+/mdx mice quadriceps. Each experimental group included quadriceps from six mice. Bars represent sem. (F and G) Measurements of fluorescence intensity in muscle fibers of 4-week-old (F) and 12-week-old mice (G) indicated increases in myofiber injury in IL-10−/−/mdx mice compared with IL-10+/+/mdx controls. Blue, striped peaks represent data from IL-10+/+/mdx. Orange peaks represent data from IL-10−/−/mdx mice.
IL-10 null mutation reduces mdx mouse strength
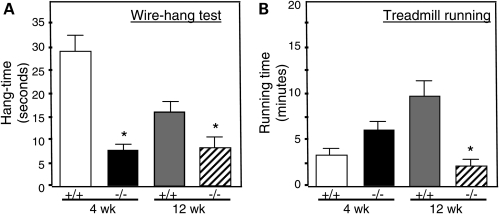
We tested whether the increase in muscle membrane damage caused by ablation of IL-10 signaling in mdx mice was associated with changes in strength or endurance. Null mutation of IL-10 caused large, significant reductions of strength in 4-week-old and 12-week-old mice that were assayed in the wire-hang test (Fig. 3A). Similarly, treadmill running time was greatly reduced in 12-week-old mdx mice in which IL-10 was mutated, although there was no significant effect on the running time of 4-week-old mice (Fig. 3B).
Figure 3.
Null mutation of IL-10 reduces strength and endurance of mdx mice. Mouse strength was assessed in the wire-hang test (A) and endurance was measured by time-to-fatigue in treadmill running (B). Asterisk indicates significant difference at P< 0.05 compared with IL-10+/+/mdx at same age. Bars indicate sem.
IL-10 promotes the M2c phenotype of mdx muscle macrophages in vitro and in vivo
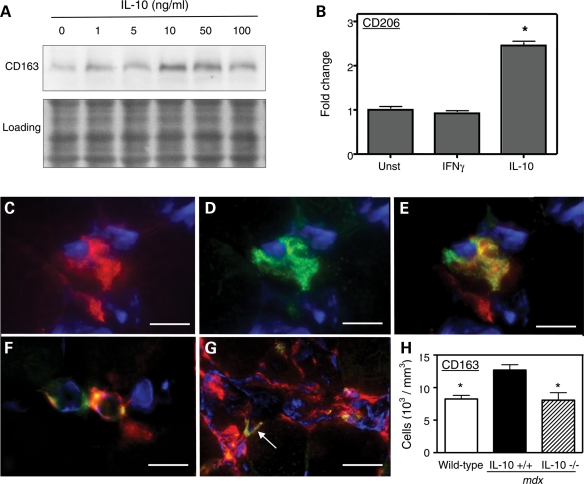
We also tested whether IL-10 could influence the pathology of muscular dystrophy by affecting the phenotype of M2 macrophages. Because previous investigations demonstrated that M2c macrophages are associated with regeneration of mdx muscle and comprise a larger proportion of the inflammatory infiltrate in regenerative mdx muscle (24), we tested whether IL-10 could promote the M2c phenotype in dystrophic muscle. Expression of CD163 and CD206 were used as indicators of the M2c phenotype. IL-10 stimulation of macrophages that were isolated from mdx muscle increased the expression of CD163 and CD206 in vitro, which confirmed that mdx muscle macrophages expressed IL-10R and showed that their stimulation with IL-10 skews them toward an M2c phenotype (Fig. 4A and B). Immunohistochemical observations confirmed that F4/80+/CD163+ M2c macrophages are present in inflammatory lesions in mdx muscles (Fig. 4C–E). However, resident macrophages that are present in much lower numbers in the muscles of healthy, wild-type mice also express CD163 (Fig. 4F), reflecting a role for M2c macrophages in normal physiological processes in muscle. Although the numbers of M2c macrophages increase greatly in mdx muscle, they remain a minor fraction of the total inflammatory infiltrate in inflammatory lesions (Fig. 4G).
Figure 4.
IL-10 induces M2c activation of macrophages. (A) Mdx muscle macrophages were stimulated with a range of concentrations of IL-10 for 24 h. Western analysis showed that maximal induction of CD163 occurred in response to 10 ng/ml of IL-10. The membrane used for immunoblotting was stained with Ponceau red (‘Loading') to verify equal protein loading in each lane. (B) CD206 expression was measured by quantitative, real-time PCR in peritonial macrophages cultured for 24 h in presence of 10 ng/ml of IFN-γ or IL-10. Unstimulated macrophages were used as controls. Asterisk indicates significantly different from unstimulated cells, P< 0.001. Each experimental group included five plates containing 6×106 cells. (C–G) M2c macrophages are present in dystrophic muscle. Cross-sections of 4-week-old mdx quadriceps were labeled with antibodies against CD163 (green) and F4/80 (red) to determine the presence of M2c macrophages in dystrophic muscle. Nuclei were labeled with DAPI reagent (blue). (C) F4/80+ macrophage. Bar = 12 µm. (D) Macrophage shown in (C) double-labeled with anti-CD163. Bar = 12 µm. (E) Merged image of (C) and (D). Bar = 12 µm. (F) C57 muscle contains resident macrophages that express CD163. Bar = 15 µm. (G) M2c macrophages represent a minor population of macrophages present in inflamed lesions (arrow). Bar = 20 µm. (H) Quantitative immunohistochemistry shows that null mutation of IL-10 in mdx mice reduces CD163+ macrophages in 12-week-old IL-10−/−/mdx mice quadriceps muscles (striped bar; 10 mice analyzed) compared with IL-10+/+/mdx mice quadriceps (black bar; 11 mice analyzed), to levels that do not differ from wild-type mice (white bar; nine mice analyzed). Asterisks indicate significant difference from IL-10+/+/mdx at P< 0.05. Bars represent sem.
Quantitative immunohistochemical data showed that ablation of IL-10 in mdx mice reduced the numbers of CD163+ M2c macrophages in 12-week-old mdx muscle by ∼40%, thus decreasing the M2c macrophage numbers to wild-type levels (Fig. 4H). This finding indicates that IL-10-mediated signaling contributes to the M2c macrophage phenotype in regenerative muscle, although IL-10 is not required for activating the M2c phenotype.
IL-10 promotes phagocytosis by mdx muscle macrophages
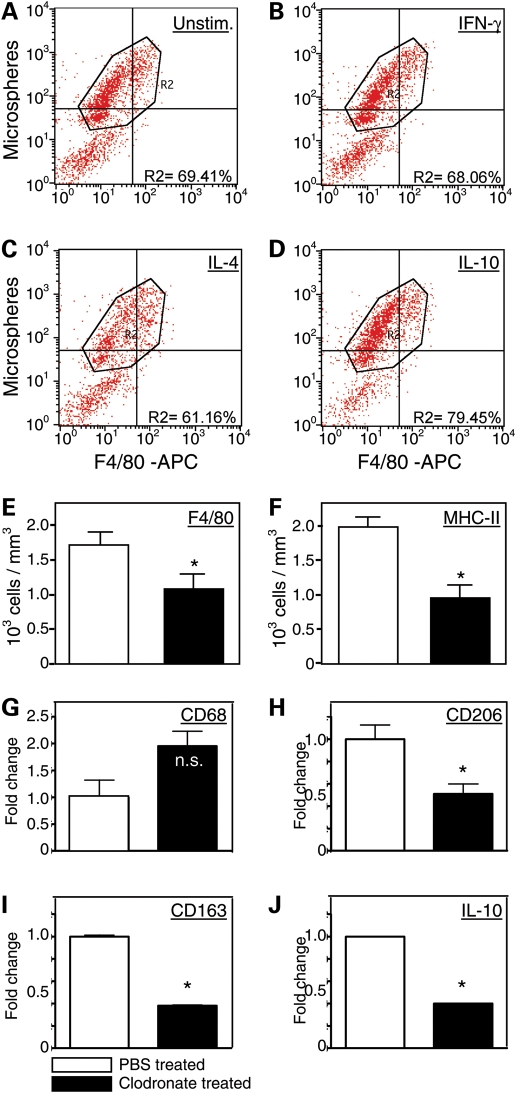
Previous investigators have shown that that shifts in the macrophage phenotype can be affected by phagocytosis (31). We assayed whether cytokines that are present at elevated levels in dystrophic muscle can activate muscle macrophage phagocytosis, which could enable them to influence shifts in the macrophage phenotype. Muscle macrophages were incubated with fluorescent latex particles in the presence or absence of IFN-γ, IL-4 or IL-10. The macrophages were then analyzed by flow cytometry to identify the proportion of the cell population that expressed F4/80 and also contained fluorescent particles. Surprisingly, the Th-1 cytokine IFN-γ did not affect phagocytosis in muscle macrophages (Fig. 5A and B). IL-4, a Th2 cytokine that can reduce phagocytosis by macrophages (32), also reduced phagocytosis by mdx muscle macrophages (Fig. 5C). However, IL-10 caused significant increases in mdx muscle macrophage phagocytosis (Fig. 5D).
Figure 5.
IL-10 induces phagocytosis by macrophages and clodronate depletion of phagocytes from mdx mice selectively reduces M2c macrophages in skeletal muscle. (A–D) Flow cytometric data showing that IL-10 stimulation of macrophages increases macrophage phagocytosis. Macrophages were untreated with exogenous cytokines (A), treated with IFN-γ (B), IL-4 (C) or IL-10 (D) prior to incubation with flourescent microspheres for phagocytosis. Cells were then analyzed by flow cytometry to determine the proportion of cells that contained fluorescent microspheres, as an index of phagocytosis. Neither IFN-γ (68.1%) or IL-4 (61.2%) increased the proportion of cells that were phagocytic, compared with untreated controls (69.4%). However, IL-10 treatments increased the proportion of cells that were phagocytic (79.5%). (E and F) Intraperitoneal injections of clodronate-containing liposomes significantly reduced the numbers of macrophages (E) and MHC-2-presenting cells in the quadriceps muscles of 4-week-old mdx mice. Each experimental group included muscles from six mice. Asterisks indicate a significant difference from PBS-treated controls. Bars represent sem. (G–J) Quantitative, real-time PCR data show that clodronate-mediated depletions of phagocytes did not cause a significant change in the expression of CD68 in the quadriceps muscles of 4-week-old mdx mice (G), but caused large, significant reductions in the expression of CD206 (H), CD163 (I) and IL-10 (J). Each experimental group included muscles from five mice. Asterisks indicate a significant difference from PBS-treated controls. ‘n.s.' indicates no significant effect of the clodronate treatment. Bars represent sem, which is too small to appear for graphs of some data sets.
Depletion of phagocytes from mdx mice diminishes M2c macrophages in muscle
Because phagocytosis can play a role in regulating the shift of macrophages to an M2 phenotype in vitro (31), we assayed whether reductions in phagocytic cells from mdx mice through treatment with clodronate-containing liposomes would affect the macrophage phenotype in mdx muscles in vivo. Clodronate treatments of mdx mice from 2 to 4 weeks of age produced a reduction in F4/80 macrophages in the mdx muscles (Fig. 5E). A nearly identical decrease in cells expressing major histocompatibility complex class 2 (MHC-2) occurred (Fig. 5F), which suggested that the M2 macrophages that were affected by the clodronate treatments were antigen-presenting cells. More specific assays for the macrophage phenotype confirmed that the depletions primarily affected M2 macrophages, especially the M2c phenotype. While there was a strong trend to increase the numbers of CD68+ M1 macrophages in the clodronate-treated mice (Fig. 5G), the treatments caused large reductions of similar magnitudes in the expression of CD206, CD163 and IL-10 (Fig. 5H–J).
IL-10 modulates muscle cell differentiation in vivo and in vitro
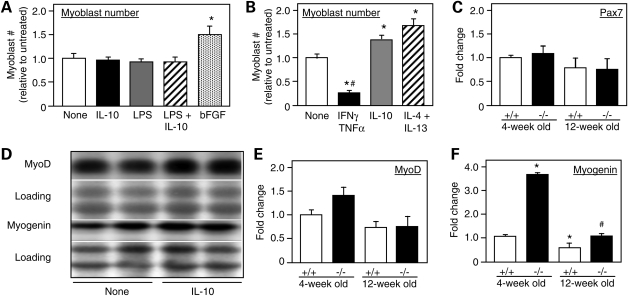
The finding that muscle cells in mdx mice express the IL-10R (Fig. 1D and E) suggested that IL-10 could influence the pathology of muscular dystrophy by direct actions on muscle cells. We first tested whether IL-10 could affect the proliferation of myoblasts in vitro, but found no effect on myoblast cell numbers in cultures treated with IL-10, LPS or IL-10 combined with LPS (Fig. 6A). However, when macrophages were treated with IL-10 or with a combination of IL-4 and IL-13 to drive them to an M2 phenotype, the macrophages were able to stimulate myoblast proliferation when they were subsequently co-cultured with myoblasts (Fig. 6B). In contrast, stimulation of muscle macrophages with IFN-γ to drive them to an M1 phenotype prior to co-culturing with myoblasts reduced numbers of myoblasts in the co-cultures (Fig. 6B). In vivo observations similarly indicate that IL-10 does not play an important role in increasing proliferation of myoblasts because null mutation of IL-10 in mdx mice had no effect on the level of expression of Pax-7 in 4-week-old or 12-week-old mice (Fig. 6C).
Figure 6.
IL-10 modulates muscle cell differentiation in vitro and in vivo. (A) Myoblasts were cultured without addition of exogenous factors (‘none') or in the presence of IL-10, LPS, LPS and IL-10 together or bFGF, and then assayed for cell number at the end of 3 days of stimulation. Neither IL-10, LPS or IL-10 combined with LPS affected proliferation, although bFGF, which was used as a positive control, increased cell number by nearly 50% under the same culture conditions. (B) Pre-treatment of macrophages with IFN-γ and TNF-α prior to co-culture with myoblasts decreased myoblast numbers at the end of a 3-day period of co-culture, compared with myoblasts cultured alone (‘none'). Pre-treatment of macrophages with IL-10 or IL-4 combined with IL-13 to induce M2c phenotype generated macrophages that promoted myoblast proliferation in co-cultures. n = 6 for each treatment group. Asterisks indicate significant difference from the ‘none' group at P< 0.05. Bars represent sem. (C) Quantitative, real-time PCR results for relative mRNA levels for Pax7 in quadriceps muscles of 4-week-old or 12-week-old mdx mice that expressed IL-10 (+/+) or were IL-10 null mutants (−/−). Expression levels are relative to 4-week-old IL-10+/+/mdx muscles, for which the expression level is set at one unit. Each experimental group included quadriceps from five mice. Bars represent sem. (D) Western blots of extracts of myoblast cultures collected 0–7 days after initiating cultures in the presence of IL-10 or addition of no exogenous cytokine (‘none'). Blots were probed with antibodies to MyoD or myogenin. The membrane used for immunoblotting was stained with Ponceau red (‘Loading') before application of the antibody to verify equal protein loading in each lane. (E and F) Quantitative, real-time PCR results for relative mRNA levels for MyoD (E) or myogenin (F) in quadriceps muscles of 4-week-old or 12-week-old mdx mice that expressed IL-10 (+/+) or were IL-10 null mutants (−/−). Expression levels are relative to 4-week-old IL-10+/+/mdx muscles, for which the expression level is set at one unit. Each experimental group included quadriceps from five mice. Hash symbol indicates significant difference from age-matched IL-10+/+/mdx muscles. Asterisks indicate significant difference from 4-week-old IL-10+/+/mdx muscles. Bars represent sem.
We also assayed whether manipulations of IL-10 could affect differentiation of myoblasts in vitro or in vivo. Although IL-10 had no effect on the level of expression of MyoD or myogenin when myoblasts were treated in vitro (Fig. 6D) and null mutation of IL-10 did not significantly affect MyoD expression in mdx mice at either 4 weeks or 12 weeks of age (Fig. 6E), ablation of IL-10 expression in vivo greatly increased myogenin in mdx muscle (Fig. 6F). However, the reductions in IL-10 expression that resulted from clodronate treatments had no significant effect on the expression of myogenin in muscles of 4-week-old mdx mice (data not shown). This may reflect the failure of the clodronate treatment to eliminate IL-10 expression completely.
DISCUSSION
Membrane damage in dystrophin-deficient muscle fibers can be caused by mechanical stresses placed on the muscle cell membrane (2). However, several investigations have demonstrated that reduction of inflammatory cell involvement in the disease also reduces the number of injured fibers by 50–80%. This was first demonstrated in work that showed an 80% reduction of injured fibers in 4-week-old mdx mice that had been depleted of macrophages starting at 6 days of age, before the early, acute onset of the disease (6). Subsequent findings have shown nearly identical reductions of membrane lesions in mdx mice in which inflammation was reduced either by disrupting NFκB signaling (33), treating with glucocorticoids (34), genetic ablation of metalloproteinases that can activate the inflammatory response (35) or treatment with metalloproteinase inhibitors that reduce the inflammatory response (36). Furthermore, treatment of mdx mice with the free radical scavenger N-acetylcysteine, which reduces NFκB activation and inflammation, causes quantitatively similar reductions in muscle fiber membrane damage (37). Thus, mechanical stress appears to cause directly no more than 20–50% of the muscle fiber damage in mdx muscular dystrophy at the early, acute peak of pathology. The mechanically induced damage is then amplified 2- to 5-fold by inflammatory cells, especially macrophages. This emphasizes the potential value of exploring specific interventions into inflammatory process in dystrophic muscle as a treatment strategy.
The findings of the present investigation contribute to our understanding of processes that regulate macrophage function in mdx muscular dystrophy by demonstrating that expression of IL-10 in dystrophin-deficient mice modulates macrophage activation and lessens the muscle membrane damage that occurs in the disease (Fig. 7). Mdx muscle is enriched in M1 macrophages during the early, acute stage of muscle pathology and these cells are primarily responsible for inflammatory cell-mediated damage to dystrophic muscle fibers (24). Much of the cytolysis of mdx muscle fibers by M1 macrophages is attributable to NO production by iNOS in mdx macrophages, and ablation of iNOS expression in mdx mice significantly reduces damage to the muscle cell membrane (24). Thus, our demonstration that IL-10 deactivates the M1 phenotype in mdx muscle macrophages and specifically inhibits the expression of iNOS by M1 macrophages while reducing muscle damage may indicate a more specific strategy for attenuating the pathology of mdx muscle at the early, acute stage of the disease by targeting the activities of M1 macrophages.
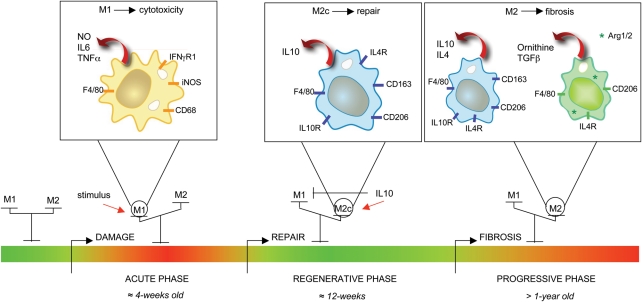
Figure 7.
Model of the influence of IL-10 on macrophage phenotype shift and the progression of muscular dystrophy. The acute onset of pathology in mdx muscle prior to 4 weeks of age involves muscle inflammation, with a bias toward M1 macrophages (6,24). M1 macrophages contribute to oxidative stress and muscle fiber lysis through the production of iNOS-derived NO, and promote inflammation and myoblast proliferation through the production of Th1 cytokines (24). Elevated IL-10 production shifts the macrophage population toward an M2c phenotype and deactivates M1 macrophages, causing reductions in iNOS expression and attenuating muscle damage (present study). IL-10, possibly derived from CD163+ M2c macrophages, can also affect the differentiation of myogenic cells (present study). As mdx muscle reaches the later, progressive stage of the mdx pathology, arginase expression by M2 macrophages is increased (41), which may reflect a shift toward an M2a phenotype. Arginase activity can increase fibrosis in mdx muscle (41) by driving the production of ornithine and thereby increasing collagen deposition (65–67). Green bar, healthy/regenerating muscular tissue; red bar, inflamed tissue; thin red arrow, stimulating cytokines; thick red arrow, macrophage-produced factors.
Our finding that IL-10 stimulation induced phagocytosis by macrophages suggests an unexpected mechanism through which IL-10 may modulate shifts in the macrophage phenotype because previous investigators have shown that phagocytosis by M1 macrophages can induce the M2 phenotype in vitro, and possibly in injured muscle in vivo (31). Thus, the high level of IL-10 production in mdx muscle in 4-week-old mdx mice may promote phagocytosis by macrophages, leading to the phenotype shift. Our data from clodronate-treated mice support this interpretation by showing that depletion of phagocytic cells from mdx mice yields a specific reduction of CD163+ M2 macrophages, which may be attributable to clodronate-mediated induction of apoptosis in macrophages before their shift to an M2 phenotype could occur. Because those phagocytic M1 macrophages would be eliminated from the M1 pool by switching to the M2 phenotype even in the absence of clodronate, no net reduction in the numbers of M1 macrophages would be expected in the muscles of the clodronate-treated mice.
Our results also show that IL-10 induces CD163 expression and modulates activation of the M2c macrophage phenotype in mdx muscles, further influencing the balance between M1 and M2c macrophages within the muscle. Ablation of IL-10 signaling in mdx mice diminishes the shift in macrophage populations from the cytolytic M1 phenotype to the M2c phenotype, which can promote tissue repair. Because M2 macrophages are associated with muscle growth, repair and regeneration following acute damage (38–40), the IL-10-mediated shift in mdx muscle macrophages may contribute to the regenerative process that occurs following the acute peak of mdx pathology at 4 weeks of age. However, M2c macrophages may also contribute to muscle pathology. M2c macrophages express arginase, and arginine metabolism by arginase in mdx muscle promotes fibrosis in skeletal and cardiac muscle (41). Thus, M2c macrophages in mdx muscle may serve beneficial roles in promoting tissue repair, but in the long run, their activities may also worsen pathology by promoting tissue fibrosis.
Although increased expression of CD163 is used primarily in this and other investigations as a marker of the M2c phenotype, upregulation of this antigen is also important in determining the function of macrophages. CD163 is a transmembrane glycoprotein with nine SRCR domains (scavenger receptor cysteine-rich domains) (20) that binds complexed hemoglobin and haptoglobin, leading to internalization and degradation of the complexes by macrophages (20,42). Removal of the hemoglobin–haptoglobin complexes may attenuate muscle pathology because elevated concentrations of hemoglobin are toxic to cells (43), and DMD patients show increased fragility of the red cell membrane (44) that can lead to increased hemolysis and elevated extracellular hemoglobin. In addition, hemoglobin-bound iron can catalyze the synthesis of hydroxyl radicals by the Fenton reaction (45,46), which can promote free-radical-mediated damage to muscle; the importance of free radicals in the pathophysiology of muscular dystrophy has been emphasized in recent demonstrations of the protective effects of free radical scavengers in the pathophysiology of mdx dystrophy (37). CD163 binding and internalization of iron–hemoglobin would decrease the production of free radicals (47) that could promote muscle damage. Indeed, a portion of the protective effect of prednisone in the treatment of DMD may be attributable to its ability to increase CD163 expression by monocytes and macrophages by more than 10-fold over levels of constitutive expression (48).
The presence of IL-10 receptors on the surface of mdx muscle cells in vivo suggests that IL-10 may have direct actions on muscle cell proliferation and differentiation in mdx dystrophy. Rapidly following muscle damage, satellite cells are activated and their population expands as they enter the proliferative phase of regeneration, which is reflected in increased levels of both Pax7 and MyoD (49–51). However, modulating IL-10 levels had no detectible influence on myoblast proliferation or the expression of the transcription factors MyoD or myogenin in muscle cell cultures and neither Pax7 nor MyoD levels were affected by ablation of IL-10 in mdx mice in vivo. Those data indicate that IL-10 does not affect muscle cell proliferation or differentiation at early stages of regeneration. At subsequent stages of muscle regeneration, satellite cells exit the proliferative stage of regeneration and enter the early differentiation phase. During this stage, MyoD expression declines and myogenin expression and activation increase (52). Our finding that there is a large increase in myogenin expression caused by IL-10 mutation in mdx mice without a significant effect on MyoD levels suggests that IL-10 can inhibit exit of muscle cells from the early phase of differentiation to enter the terminal phases of differentiation, when myogenin expression is attenuated.
Although IL-10 mutation appears to slow the transition from the early to terminal phases of differentiation in muscle regeneration and IL-10R is present on the surface of mdx muscle cells in vivo, we are unable to conclude that the effect reflects direct action of IL-10 on muscle cells. An alternative explanation that would be consistent with our data is that IL-10 acts on an intermediate cell type to induce the release of factors that affect muscle differentiation. Macrophages themselves could serve as this intermediary, since IL-10 can affect the production of TNF-α and IFN-γ by myeloid cells (14) and either of those cytokines can affect muscle differentiation. For example, TNF-α can activate p38 (53) and thereby increase the expression myogenin and myosin light chain and promote myocyte fusion (54). An additional alternative is that IL-10 acts directly on muscle cells to affect their differentiation but another factor that is not present in the in vitro assays is required for an IL-10-mediated direct effect on muscle cells. This possibility would be consistent with previous work demonstrating that IL-10 can antagonize signaling pathways that are activated by IFN-γ or TNF-α in vitro (23,24). These possibilities are under current investigation.
Collectively, the findings of the present investigation show that IL-10 reduces the pathology of muscular dystrophy, at least in the short term, and that it has potential value as a therapeutic agent for the treatment of DMD. The data provide strong evidence that the beneficial effects of IL-10 in the treatment of mdx dystrophy are attributable to IL-10's ability to influence the balance between cytolytic M1 macrophages and M2 macrophages that are involved in tissue repair. However, many issues should be carefully explored before IL-10 treatments are provided to DMD patients. Most importantly, whether the shift toward the M2c phenotype has long-term negative or positive effects should be tested. Also, whether the long-term effects of IL-10 treatments on muscle differentiation yield a net, functional benefit remains to be explored.
MATERIALS AND METHODS
Animals
C57BL/6, C57BL/10ScSn-Dmdmdx/J and B10.129P2(B6)-Il10tm1Cgn/J mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and bred in pathogen-free vivaria at the University of California, Los Angeles. Mice carrying null mutation for IL-10 (B10.129P2(B6)-Il10tm1Cgn/J mice) that had been back-crossed onto a C57/BL6 background were crossed with mdx mice to generate mice that lacked both IL-10 and dystrophin using a breeding strategy described previously (55). Null mutation of IL-10 was confirmed by PCR using an upstream primer that was common for both wild-type and mutant DNA (5′-GCC TTC AGT ATA AAA GGG GGA CC-3′), a wild-type downstream primer (5′-GTG GGT GCA GTT ATT GTC TTC CCG-3′) and a downstream primer for the neomycin cassette (5′-AAT CCT GCG TGC AAT CCA TCT TG-3′). Null mutation of the dystrophin gene was confirmed using mdx-amplification-resistant mutation system PCR (56). All animals were handled according to guidelines approved by the Chancellor's Animal Research committee at the University of California, Los Angeles.
Immunohistochemistry
Frozen, serial sections of quadriceps muscles were air-dried for 30 min and fixed in acetone for 10 min at 4oC. Sections were blocked for 1 h with 3% bovine serum albumin (BSA) and 0.05% Tween-20 diluted in 50 mm Tris–HCl (pH 7.6) containing 150 mm sodium chloride (NaCl). Sections were then incubated with anti-IL10R1 (R&D System, 1:50) or anti-MHC-2 (Abcam; 1:100) for 3 h at room temperature. Sections were washed with 50 mm sodium phosphate buffer (pH 7.2) containing 150 mm NaCl (PBS) and then incubated with biotin-conjugated secondary antibodies (Vector Laboratories, 1:200) for 1 h. Sections were subsequently washed with PBS and incubated for 30 min with avidin D-conjugated horseradish peroxidase (Vector Laboratories, 1:1000). Staining was visualized as a red reaction product with peroxidase substrate, 3-amino-9-ethylcarbazole (Vector).
In other preparations for double-labeling, frozen muscle sections of 10 µm thickness were fixed in acetone and blocked with 10% normal horse serum and 0.05% Tween-20 diluted in 50 mm Tris–HCl and 150 mm NaCl buffer before probing with anti-F4/80 for 3 h at room temperature. Anti-F4/80 was obtained by ammonium sulfate precipitation from HB-198 hybridoma cultures (ATCC). Sections were incubated with Texas Red-conjugated anti-rat IgG secondary antibody (Vector; 1:200). Sections were then washed, blocked and incubated with rat anti-CD206 (Serotec, 1:50) or rabbit anti-CD163 (Santa Cruz 1:150) for 3 h at room temperature. Sections were subsequently washed and incubated with the species-appropriate, fluorescein-conjugated IgG secondary antibody (Vector, 1:200). Sections were washed with PBS and treated with RNase A (Sigma, 0.2 µg/ml). DAPI diluted 1:100 000 in PBS was applied for 5 min before sections were mounted.
Assays for muscle fiber injury and regeneration
Injuries to muscle fiber membranes were assayed by measuring the relative quantities of the fluorescent extracellular tracer dye procion orange in the cytoplasm of muscle fibers. Muscles that are incubated in procion orange solutions exclude the dye, unless membrane lesions are present. One soleus muscle from each mouse was incubated in 0.5% procion orange dye in Krebs Ringer solution for 1 h followed by washes with Krebs Ringer solution. The muscles were then frozen in isopentane, and mid-belly cross-sections of each muscle were cut. All sections were viewed by epifluorescence. The fluorescence intensity of every individual fiber was measured in a circular area of 8 µm diameter that was sampled at the center of each fiber using a digital imaging system (Bioquant, Nashville, TN, USA). Data were not collected from fibers at the surface of the muscle because some of those fibers may have experienced membrane damage during muscle dissection. Stopping down the field diaphragm prevented photobleaching of fibers outside of the field of signal acquisition. Fluorescence intensity values for each fiber were corrected for background by measuring background signal from an area of the slide that contained no tissue and subtracting that value from the signal recorded within fibers. Data were expressed as a frequency distribution showing the number of fibers at each particular intracellular fluorescence intensity; higher mean values of fluorescence indicated a shift of the population of fibers toward increased membrane injury. In addition, muscle fiber regeneration was assayed by determining the percentage of muscle fibers in cross-sections of entire muscles that showed nuclei located at the center of the fiber. Muscle nuclei translocate from their position at the surface of muscle fibers to the fiber's center during muscle repair and regeneration following injury.
Immunoblotting
The presence of CD163 or iNOS was assayed in whole muscle or macrophage lysates by western blot analysis. Using SDS–PAGE, 30 µg of total, lysate protein were separated and transferred to nitrocellulose membranes. The membranes were blocked with 3% powdered milk and then incubated with antibodies against iNOS (Upstate Biotechnology, 1:300) or CD163 (1:100) for 3 h at room temperature. Equal loading of samples was determined by staining nitrocellulose blots with 0.1% Ponceau S solution (Sigma). Membranes were then washed with PBS containing 0.05% Tween-20 and probed with horseradish peroxidase-conjugated secondary antibodies (Amersham, 1:10 000) for 1 h, followed by PBS washes before signal detection visualized with chemiluminescent substrate and a fluorochem imaging system.
Peritoneal and muscle macrophage isolation
C57BL/6 mice received intraperitoneal injections with 1 ml of 12% sodium caseinate in 0.9% sodium chloride. The mice were euthanized by inhalation of 32% isoflurane at 3 days post-injection and then peritoneal cells were collected sterilely by using peritoneal lavages. Lavage samples were centrifuged and resuspended in 0.85% ammonium chloride to lyze red blood cells. The cells were centrifuged, resuspended with 15 ml of Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin (complete DMEM) and filtered through a 70 µm cell strainer (BD Bioscience). The cell suspensions were overlain on 15 ml of Histopaque 1077 (Sigma-Aldrich) and centrifuged at 1000g for 30 min. Cells at the Histopaque/DMEM interface were collected, centrifuged and resuspended in complete DMEM.
Mdx muscle macrophages were isolated using a modification of a previously described procedure (6). Hindlimb muscles of four to six mice were dissected and cleaned of discernible fat, rinsed with fresh PBS and weighed. The muscles were minced in cold PBS and then transferred to centrifuge tubes containing collagenase Type IV (Sigma) in DMEM (10 mg/ml). Tubes were incubated at 37°C for two 45 min periods, after which the suspension was aspirated, centrifuged at 850g and resuspended in PBS. The cell suspensions were filtered through a 70 µm cell strainer and centrifuged at 850g for 5 min. The filtered cells were then applied to Histopaque 1077, centrifuged and collected as described above for peritoneal macrophages.
The purity of the macrophage preparations was assayed by indirect immunofluorescence of 2×105 peritoneal or muscle-derived cells that were cultured on coverslips and immunolabeled with anti-F4/80 antibody followed by an FITC-conjugated second antibody. Cells were visualized by fluorescence microscopy and macrophage purity was expressed as the percentage of total cells that were F4/80 positive. Data were expressed as the mean and standard error of the mean (sem) of five randomly counted fields in three independent experiments.
RNA isolation
Frozen muscles were homogenized in 1.2 ml of Trizol (Invitrogen) per 100 mg of tissue. The resulting suspension was transferred to 2 ml centrifuge tubes and RNA was extracted according the manufacturer's protocol with minor modification (Invitrogen).
RNA from macrophages was isolated, cleaned and DNase-treated using RNeasy spin columns according the manufacturer's protocol (Qiagen). RNA (500 ng) was loaded on 1.2% agarose gels and RNA quality assessed by determining 28S and 18S ribosomal RNA integrity. Total RNA was reverse transcribed with Super Script Reverse Transcripase II using oligo-dTs to prime extension (Invitrogen).
Real-time PCR
cDNA resulting from reverse-transcription–PCR was used to measure quantitatively the expression of genes associated with M1 and M2 macrophages using SYBR green qPCR master mix according to the manufacturer's protocol (BioRad). Real-time PCR was performed on an iCycler thermocycler system equipped with iQ5 optical system software (BioRad). Primers used for PCR to assay relative levels of expression of muscle-specific and immune-cell-specific genes are shown in Table 1.
Table 1.
Primers used to assay reference genes.
| Gene | Access number | 5′→3′ |
Amplicon size (bp) | Average Ct | |
|---|---|---|---|---|---|
| TPT1 | NM_009429.3 | Fwd | GGAGGGCAAGATGGTCAGTAG | 113 | 16.56 |
| Rev | CGGTGACTACTGTGCTTTCG | ||||
| SRP14 | NM_009273.4 | Fwd | GAGAGCGAGCAGTTCCTGAC | 196 | 19.13 |
| Rev | CGGTGCTGATCTTCCTTTTC | ||||
| RPS4X | NM_009094.1 | Fwd | TGCTGGGTTTATGGATGTCA | 107 | 17.76 |
| Rev | CCTCCTCCGGTGTAATACGA | ||||
| RPL13A | NM_009438.4 | Fwd | CCTGCTGCTCTCAAGGTTGTT | 146 | 17.27 |
| Rev | CGATAGTGCATCTTGGCCTTT | ||||
| RNSP1 | NM_001080127.1 | Fwd | AGGCTCACCAGGAATGTGAC | 196 | 21.72 |
| Rev | CTTGGCCATCAATTTGTCCT | ||||
| EEF1A1 | NM_010106.2 | Fwd | TTGGTTCAAGGGATGGAAAG | 217 | 15.65 |
| Rev | AGCAAAGGTAACCACCATGC | ||||
| GAPDH | NM_008084.2 | Fwd | GCAAATTCAACGGCACAGTCAAG | 248 | 11.98 |
| Rev | GGTACAAACACTACCCACACTTG | ||||
| HPRT1 | NM_013556.2 | Fwd | GCAAACTTTGCTTTCCCTGG | 85 | 22.46 |
| Rev | CCACTTTTCCTGGAGAGCTTCA | ||||
| PPIA | NM_008907 | Fwd | GCAAATGCTGGACCAAACAC | 97 | 17.82 |
| Rev | TACACCAGAAACCCTTCCACT | ||||
Average threshold cycle (Ct) in quadriceps of 4-week-old mdx mice.
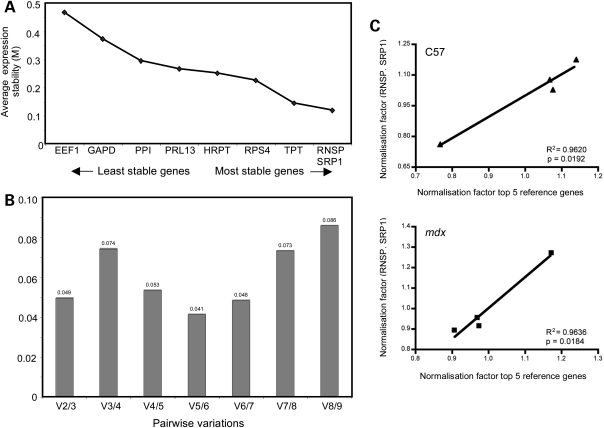
The rigor of quantification of the relative levels of mRNA in samples was maximized by following the guidelines of Bustin et al. (57) and Nolan et al. (58) for sample preparation, experimental design, data normalization and data analysis for quantitative, real-time PCR. Because the expression of reference genes that are used to normalize quantitative, real-time PCR data can vary considerably within and between groups of experimental samples (59), we determined empirically the selection of reference genes that did not vary between the experimental groups in our investigation. Nine reference genes were tested in murine skeletal muscle and analyzed by geometric averaging using geNorm Visual Basic application software version 3.5 (59). Reference genes were chosen based on those previously assayed and the rationale for their selection has been discussed (60). Primers used for PCR to assay expression levels of reference genes are reported in Table 2. The average expression variability of reference genes was evaluated and only the most stable genes between the groups analyzed were used for data normalization. We also assessed the ideal number of reference genes for normalization and determined that as few as two genes were sufficient for reliable normalization. The normalization factors of multiple reference genes were then calculated by geNorm software after transforming Ct values in quantities using the comparative Ct method. For each gene, the highest relative quantity was set to 1 and all the other expression values were scaled relatively to this value. Calculation of normalization factors was based on geometric mean of multiple control genes (Fig. 8).
Table 2.
Primers used to assay expression levels of immune-cell and muscle-cell-specific genes.
| Gene | Access number | 5′→3′ |
Amplicon size (bp) | Average Ct | |
|---|---|---|---|---|---|
| CD206 | NM_008625.1 | Fwd | GGATTGTGGAGCAGATGGAAG | 120 | 22.57 |
| Rev | CTTGAATGGAAATGCACAGAC | ||||
| CD163 | NM_053094 | Fwd | GCAAAAACTGGCAGTGGG | 164 | 24.45 |
| Rev | GTCAAAATCACAGACGGAGC | ||||
| CD68 | NM_009853 | Fwd | CAAAGCTTCTGCTGTGGAAAT | 140 | 21.83 |
| Rev | GACTGGTCACGGTTGCAAG | ||||
| IL10 | NM_010548.1 | Fwd | CAAGGAGCATTTGAATTCCC | 157 | 27.48 |
| Rev | GGCCTTGTAGACACCTTGGTC | ||||
| IL10R1 | NM_008348 | Fwd | CAAGCCCTTCCTATGTGTGG | 166 | 25.23 |
| Rev | AGAAAGGCTCAGGCATTGTC | ||||
| PAX7 | NM_011039 | Fwd | CTCAGTGAGTTCGATTAGCCG | 144 | 26.23 |
| Rev | AGACGGTTCCCTTTGTCGC | ||||
| Myogenin | NM_031189.2 | Fwd | CCAGTACATTGAGCGCCTAC | 163 | 24.34 |
| Rev | ACCGAACTCCAGTGCATTGC | ||||
| MyoD | NM_010866 | Fwd | GAGCGCATCTCCACAGACAG | 178 | 24.55 |
| Rev | AAATCGCATTGGGGTTTGAG | ||||
Average threshold cycle (Ct) in quadriceps of 4-week-old mdx mice.
Figure 8.
Selection of reference genes for real-time, quantitative RT–PCR. (A) The most stable reference genes within and between two different groups of muscle samples were determined using geNorm 3.5 software. The variability in the average expression (M) of each gene was set at 0.5. Between 4-week-old or 12-week-old mdx and C57 quadriceps, RNSP1 and SRP14 had the least variability of the nine genes tested. The expression of EEF1A was recorded as the most unstable relative to the other genes. (B) GeNorm was also used to determine the optimal number of genes required to calculate a reliable normalization factor that was applied to normalized each gene of interest. The chart reports the pairwise variation (V) between two sequential normalization factors (Vn/Vn+1). When the value of variation is below the selected threshold, the addition of additional reference genes is unnecessary for data normalization. The pairwise variation was analyzed starting from the normalization factor generated by the two most stable genes, RNSP1 and SRP14, and adding the following normalization factor generated by adding TPT1, and so on. The cut-off value suggested by Vandesomple et al. (59) was 0.15. Since we reported a very low variation between V2/3, we decided that using RNSP1 and SRP14 was sufficient to generate a reliable normalization factor. (C and D) To confirm the minimum number of genes to use, normalization factors generated by the two most stable genes in C57 (C) and mdx (D) groups were correlated with the normalization factors obtained using the five best genes resulting from picture (A). Correlation between three and five top normalization factors was also tested but not reported. Each point is the mean of five values. Pearson coefficient shows that there is a tight correlation between the two groups of normalization factors and that using two genes for normalization of C57 and mdx 4-week-old and 12-week-old quadriceps data is adequate.
C2C12 myoblast proliferation
C2C12 myoblasts were seeded at 1.0×104 cells/well in six-well plates in complete DMEM and cultured at 37°C and 5% CO2 overnight. The cells were then serum-starved in DMEM with 0.1% FBS for 1 day to arrest the cell cycle and then stimulated with the following cytokines or reagents: IL-10 (10 ng/ml), or IL-10 (10 ng/ml) and LPS (10 ng/ml) or basic fibroblast growth factor (bFGF, 10 ng/ml) in DMEM with 5% FBS for 3 days. Fresh cytokines were replenished to cultures every 24 h. At the end of day 3, C2C12 myoblasts were trypsinized with 0.05% trypsin–EDTA and collected for counting.
The proliferation of C2C12 cells in co-culture with macrophages was determined as reported previously (61). Peritoneal macrophages were seeded at 4.0×105 cells/well in six-well plates and stimulated with mouse recombinant IFN-γ (10 ng/ml, BD Pharmingen) and TNF-α (10 ng/ml, BD Pharmingen) or IL-10 (10 ng/ml, BD Pharmingen) or IL-4 (10 ng/ml, BD Pharmingen) and IL-13 (10 ng/ml, BD Pharmingen) in DMEM containing 0.25% FBS and 1% penicillin–streptomycin (P/S) for 24 h at 37°C and 5% CO2. Following stimulation, macrophages were washed with DMEM twice. C2C12 myoblasts that had been serum-starved in DMEM with 0.1% FBS for 1 day were then plated on top of the stimulated macrophages in DMEM containing 5% FBS and 1% P/S at the density of 1.0×105 cells/well. After 72 h of co-culture, C2C12 cells were dissociated with 0.05% trypsin–EDTA (Invitrogen), collected and counted. Trypsin treatment did not detach the macrophages.
Cytotoxicity assay
Macrophage-mediated cytotoxicity was assayed as described previously (62). C2C12 myoblasts were cultured in growth medium until they reached confluency, after which they were serum-starved overnight to trigger fusion. Cells were then returned to growth medium in which they differentiated for 3 days. The differentiated myotubes were then incubated with 0.4% 51Cr in Hanks buffered saline solution (HBSS) assay buffer (HBSS containing 0.25% FBS and 400 µm l-arginine) for 2 h. Muscle macrophages were added to the myotube cultures at concentrations of 12 500 macrophages/mm2 in 150 µl of HBSS assay buffer. After 24 h of co-culturing, 75 µl of media were collected from each well and 51Cr was measured using a liquid scintillation counter (Beckman Gamma 5500). Cytotoxicity was expressed as a percentage of total lysis by setting 0% as 51Cr released spontaneously by myotubes cultured without macrophages and 100% cytotoxicity was set as the 51Cr released by myotubes incubated with 0.1% Triton X-100 in HBSS assay buffer.
Phagocytosis assay
Phagocytosis assay was conducted using opsonized 2.0 µm diameter, sulfate-coated, fluorescent latex particles (Molecular Probes; Invitrogen). Latex particles were suspended in sterile PBS to make a 0.5% suspension of particles. Particles were centrifuged at 13 000g and washed twice with 1:1 PBS–sterile water. The pellet was then incubated with 50 mm mouse IgG diluted in 1:1 PBS–water and placed on a tissue rotator for 1 h. The pellet was collected and incubated in 50 µg/ml BSA in 1:1 PBS–water for 1 h. After two more washes, the beads were suspended in PBS or DMEM for use. Muscle macrophages were cultured in low-attachment plates in DMEM conditioned with either IL-10 or IFN-γ and brefeldin A. Opsonized fluorescent particles (150 µl) were added to each plate and incubated for 1 h at 37°C. Cells were then washed and incubated with anti-CD16/32 antibody (eBioscience) to block Fc receptors. Cells were then stained with fluorophore-conjugated antibodies against F4/80 (eBioscience, 1:10). Flow cytometry was performed on an FACSCalibur® analytic flow cytometer (Becton Dickinson) and data analyzed with BD CellQuest Pro (Becton Dickinson).
Clodronate liposomes preparation
Liposomes containing clodronate were prepared following the original protocol described by Van Rooijen and Sanders (63). According to the protocol, phosphatidyl choline and cholesterol were dissolved in a 1:1 solution of methanol and chloroform in a vacuum flask and the organic phase was evaporated. The film that was formed in the flask was dissolved using PBS containing 3.78% clodronate in PBS. Control liposomes preparations were made using PBS that did not contain clodronate. After 90 min at room temperature, both solutions were sonicated for 3 min and then incubated for an additional 90 min. Solutions were then centrifugated at 100 000g for 25 min and each pellet was resuspended in PBS. After three washes with PBS, a drop of each solution was viewed with a microscope to confirm the presence of liposomes. Liposomes were made fresh the day before each injection.
Mice received intraperitoneal injections with 50 µl of clodronate liposomes or PBS control liposomes at 1 week and 2 weeks of age. The mice received 100 µl injections at 3 and 4 weeks of age. One day after the fourth injection, mice were euthanized and muscles collected.
Endurance and strength testing
Running protocol
A treadmill running protocol was used to assess the effect of the IL-10 null mutation on muscle function in mdx mice. Mice were exercised on an Exer 3/6 treadmill (Columbus Instruments) containing a shock grid that stimulated mice to run at a speed of 8 m/min with a 5° incline. The shock stimulus was provided at an intensity of 3.4 mA and a maximum repetition rate of 3 200 ms pulses per second. The end run time was recorded when a mouse rested for a period of 10 continuous seconds on the shock grid. Mice were run for a maximum of 60 min. Data are expressed as the mean maximum running time of 20 mice per group.
Wire-hang test
We used a variation of the wire-hang test described by Gomez et al. (64) to assess the effect of the IL-10 null mutation on muscle function and strength. Mice were allowed to grip a taut, 2 mm diameter wire with their front paws. In pilot experiments, we noted that some mice would also grasp the wire with their hind paws and tails, which contributed to variability in our data. We eliminated this unwanted variable by taping the hind paws individually and preventing mice from wrapping their tails around the wire, which allowed more accurate measurement of forelimb strength. Measurement of hang time was started when a mouse was released and the end times recorded when the mouse's grip failed. Three trials were performed and data expressed as the mean hang time of the averaged trial times of 15–20 mice tested. In order to allow mice to recover and minimize fatigue and distress, mice were allowed to rest for 1min between trials.
Statistical analysis
Multi-factorial comparisons used the Kruskal–Wallis test to determine statistical significance, followed by a post hoc, Student's t-test to determine significance of differences between two groups. One-way ANOVA followed by the Newman–Keuls post hoc test was used for the analyses with more than two sample groups.
Linear regression and Pearson correlation coefficients were used to calculate the correlation between normalization factors generated by two, three and five reference genes. Statistical significance was set at P< 0.05.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by grants from the Muscular Dystrophy Association, USA (157881 to JGT) and the National Institutes of Health (R01 AR40343, R01 AR47721 to JGT and F31 AR054724 to SAV).
REFERENCES
- 1.Hoffman E.P., Brown R.H., Jr., Kunkel L.M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 2.Petrof B.J., Shrager J.B., Stedman H.H., Kelly A.M., Sweeney H.L. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl Acad. Sci. USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tidball J.G., Wehling-Henricks M. Evolving therapeutic strategies for Duchenne muscular dystrophy: targeting downstream events. Pediatr. Res. 2004;56:831–841. doi: 10.1203/01.PDR.0000145578.01985.D0. [DOI] [PubMed] [Google Scholar]
- 4.Gorospe J.R., Tharp M., Demitsu T., Hoffman E.P. Dystrophin-deficient myofibers are vulnerable to mast cell granule-induced necrosis. Neuromuscul. Disord. 1994;4:325–333. doi: 10.1016/0960-8966(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 5.Hodgetts S., Radley H., Davies M., Grounds M.D. Reduced necrosis of dystrophic muscle by depletion of host neutrophils, or blocking TNFalpha function with Etanercept in mdx mice. Neuromuscul. Disord. 2006;16:591–602. doi: 10.1016/j.nmd.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Wehling M., Spencer M.J., Tidball J.G. A nitric oxide synthase transgene ameliorates muscular dystrophy in mdx mice. J. Cell Biol. 2001;155:123–131. doi: 10.1083/jcb.200105110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wehling-Henricks M., Sokolow S., Lee J.J., Myung K.H., Villalta S.A., Tidball J.G. Major basic protein-1 promotes fibrosis of dystrophic muscle and attenuates the cellular immune response in muscular dystrophy. Hum. Mol. Genet. 2008;17:2280–2292. doi: 10.1093/hmg/ddn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spencer M.J., Montecino-Rodriguez E., Dorshkind K., Tidball J.G. Helper (CD4(+)) and cytotoxic (CD8(+)) T cells promote the pathology of dystrophin-deficient muscle. Clin. Immunol. 2001;98:235–243. doi: 10.1006/clim.2000.4966. [DOI] [PubMed] [Google Scholar]
- 9.Cai B., Spencer M.J., Nakamura G., Tseng-Ong L., Tidball J.G. Eosinophilia of dystrophin-deficient muscle is promoted by perforin-mediated cytotoxicity by T cell effectors. Am. J. Pathol. 2000;156:1789–1796. doi: 10.1016/S0002-9440(10)65050-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manzur A.Y., Kuntzer T., Pike M., Swan A. Glucocorticoid corticosteroids for Duchenne muscular dystrophy. Cochrane DB Syst. Rev. 2008;1 doi: 10.1002/14651858.CD003725.pub3. CD003725. [DOI] [PubMed] [Google Scholar]
- 11.Moxley R.T., III, Ashwal S., Pandya S., Connolly A., Florence J., Mathews K., Baumbach L., McDonald C., Sussman M., Wade C. Practice parameter: corticosteroid treatment of Duchenne dystrophy: report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology. 2005;64:13–20. doi: 10.1212/01.WNL.0000148485.00049.B7. [DOI] [PubMed] [Google Scholar]
- 12.Moore K.W., de Waal Malefyt R., Coffman R.L., O'Garra A. Interleukin-10 and the interleukin-10 receptor. Ann. Rev. Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 13.Fiorentino D.F., Bond M.W., Mosmann T.R. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J. Exp. Med. 1989;170:2081–2095. doi: 10.1084/jem.170.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiorentino D.F., Zlotnik A., Mosmann T.R., Howard M., O'Garra A. IL-10 inhibits cytokine production by activated macrophages. J. Immunol. 1991;147:3815–3822. [PubMed] [Google Scholar]
- 15.Mosser D.M. The many faces of macrophage activation. J. Leukoc. Biol. 2003;73:209–212. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 16.Murray P.J. Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr. Opin. Pharmacol. 2006;6:379–386. doi: 10.1016/j.coph.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Mosser D.M., Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunol. Rev. 2008;226:205–218. doi: 10.1111/j.1600-065X.2008.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 19.Lang R., Rutschman R.L., Greaves D.R., Murray P.J. Autocrine deactivation of macrophages in transgenic mice constitutively overexpressing IL-10 under control of the human CD68 promoter. J. Immunol. 2002;168:3402–3411. doi: 10.4049/jimmunol.168.7.3402. [DOI] [PubMed] [Google Scholar]
- 20.Schaer D.J., Boretti F.S., Hongegger A., Poehler D., Linnscheid P., Staege H., Muller C., Schoedon G., Schaffner A. Molecular cloning and characterization of the mouse CD163 homologue, a highly glucocorticoid-inducible member of the scavenger receptor cysteine-rich family. Immunogenetics. 2001;53:170–177. doi: 10.1007/s002510100304. [DOI] [PubMed] [Google Scholar]
- 21.Sulahian T.H., Hogger P., Wahner A.E., Wardwell K., Goulding N.J., Sorg C., Droste A., Stehling M., Wallace P.K., Morganelli P.M., et al. Human monocytes express CD163, which is upregulated by IL-10 and identical to p155. Cytokine. 2000;12:1312–1321. doi: 10.1006/cyto.2000.0720. [DOI] [PubMed] [Google Scholar]
- 22.Lang R., Patel D., Morris J.J., Rutschman R.L., Murray P.J. Shaping gene expression in activated and resting primary macrophages by IL-10. J. Immunol. 2002;169:2253–2263. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Pomares L., Reid D.M., Brown G.D., Taylor P.R., Stillion R.J., Linehan S.A., Zamze S., Gordon S., Wong S.Y. Analysis of mannose receptor regulation by IL-4, IL-10, and proteolytic processing using novel monoclonal antibodies. J. Leukoc. Biol. 2003;73:604–613. doi: 10.1189/jlb.0902450. [DOI] [PubMed] [Google Scholar]
- 24.Villalta S.A., Nguyen H.X., Deng B., Gotoh T., Tidball J.G. Shifts in macrophage phenotypes and macrophage competition for arginine metabolism affect the severity of muscle pathology in muscular dystrophy. Hum. Mol. Genet. 2009;18:482–496. doi: 10.1093/hmg/ddn376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massimino M.L., Rapizzi E., Cantini M., Libera L.D., Mazzoleni F., Arslan P., Carraro U. ED2+ macrophages increase selectively myoblast proliferation in muscle cultures. Biochem. Biophys. Res. Commun. 1997;235:754–759. doi: 10.1006/bbrc.1997.6823. [DOI] [PubMed] [Google Scholar]
- 26.Strle K., McCusker R.H., Tran L., King A., Johnson R.W., Freund G.G., Dantzer R., Kelley K.W. Novel activity of an anti-inflammatory cytokine: IL-10 prevents TNFalpha-induced resistance to IGF-I in myoblasts. J. Neuroimmunol. 2007;188:48–55. doi: 10.1016/j.jneuroim.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strle K., McCusker R.H., Johnson R.W., Zunich S.M., Dantzer R., Kelley K.W. Prototypical anti-inflammatory cytokine IL-10 prevents loss of IGF-I-induced myogenin protein expression caused by IL-1beta. Am. J. Physiol. Endocrinol. Metab. 2008;294:E709–718. doi: 10.1152/ajpendo.00662.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein M., Keshav S., Harris N., Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J. Exp. Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horsley V., Jansen K.M., Mills S.T., Pavlath G.K. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113:483–494. doi: 10.1016/s0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- 30.Chesrown S.E., Monnier J., Visner G., Nick H.S. Regulation of inducible nitric oxide synthase mRNA levels by LPS, INF-gamma, TGF-beta, and IL-10 in murine macrophage cell lines and rat peritoneal macrophages. Biochem. Biophys. Res. Commun. 1994;200:126–134. doi: 10.1006/bbrc.1994.1424. [DOI] [PubMed] [Google Scholar]
- 31.Arnold L., Henry A., Poron F., Baba-Amer Y., van Rooijen N., Plonquet A., Gherardi R.K., Chazaud B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varin A., Mukhopadhyay S., Herbein G., Gordon S. Alternative activation of macrophages by IL-4 impairs phagocytosis of pathogens but potentiates microbial-induced signalling and cytokine secretion. Blood. 2010;115:353–362. doi: 10.1182/blood-2009-08-236711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acharyya S., Villalta S.A., Bakkar N., Bupha-Intr T., Janssen P.M., Carathers M., Li Z.W., Beg A.A., Ghosh S., Sahenk Z., et al. Interplay of IKK/NF-kappaB signaling in macrophages and myofibers promotes muscle degeneration in Duchenne muscular dystrophy. J. Clin. Invest. 2007;117:889–901. doi: 10.1172/JCI30556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wehling-Henricks M., Lee J.J., Tidball J.G. Prednisolone decreases cellular adhesion molecules required for inflammatory cell infiltration in dystrophin-deficient skeletal muscle. Neuromuscul. Disord. 2004;14:483–490. doi: 10.1016/j.nmd.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Li H., Mittal A., Makonchuk D.Y., Bhatnagar S., Kumar A. Matrix metalloproteinase-9 inhibition ameliorates pathogenesis and improves skeletal muscle regeneration in muscular dystrophy. Hum. Mol. Genet. 2009;18:2584–2598. doi: 10.1093/hmg/ddp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar A., Bhatnagar S., Kumar A. Matrix metalloproteinase inhibitor batimastat alleviates pathology and improves skeletal muscle function in dystrophin-deficient mdx mice. Am. J. Pathol. 2010;177:248–260. doi: 10.2353/ajpath.2010.091176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitehead N.P., Pham C., Gervasio O.L., Allen D.G. N-acetylcysteine ameliorates skeletal muscle pathophysiology in mdx mice. J. Physiol. 2008;586:2003–2014. doi: 10.1113/jphysiol.2007.148338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krippendorf B.B., Riley D.A. Distinguishing unloading- versus reloading-induced changes in rat soleus muscle. Muscle Nerve. 1993;16:99–108. doi: 10.1002/mus.880160116. [DOI] [PubMed] [Google Scholar]
- 39.St Pierre B.A., Tidball J.G. Differential response of macrophage subpopulations to soleus muscle reloading after rat hindlimb suspension. J. Appl. Physiol. 1994;77:290–297. doi: 10.1152/jappl.1994.77.1.290. [DOI] [PubMed] [Google Scholar]
- 40.Tidball J.G., Wehling-Henricks M. Macrophages promote muscle membrane repair and muscle fibre growth and regeneration during modified muscle loading in mice in vivo. J. Physiol. 2007;578:327–336. doi: 10.1113/jphysiol.2006.118265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wehling-Henricks M., Jordan M.C., Gotoh T., Grody W.W., Roos K.P., Tidball J.G. Arginine metabolism by macrophages promotes cardiac and muscle fibrosis in mdx muscular dystrophy. PLoS ONE. 2010;5:e10763. doi: 10.1371/journal.pone.0010763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kristiansen M., Graversen J.H., Jacobsen C., Sonne O., Hoffman H.J., Law S.K., Moestrup S.K. Identification of the haemoglobin scavenger receptor. Nature. 2001;409:198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 43.Moestrup S.K., Moller H.J. CD163: a regulated hemoglobin scavenger receptor with a role in the anti-inflammatory response. Ann. Med. 2004;36:347–354. doi: 10.1080/07853890410033171. [DOI] [PubMed] [Google Scholar]
- 44.Kim H.D., Luthra M.G., Watts R.P., Stern L.Z. Factors influencing osmotic fragility of red blood cells in Duchenne muscular dystrophy. Neurology. 1980;30:726–731. doi: 10.1212/wnl.30.7.726. [DOI] [PubMed] [Google Scholar]
- 45.Lim S.K., Kim H., Lim S.K., bin Ali A., Lim Y.K., Wang Y., Chong S.M., Costantini F., Baumman H. Increased susceptibility in Hp knockout mice during acute hemolysis. Blood. 1998;92:1870–1877. [PubMed] [Google Scholar]
- 46.Sadrzadeh S.M., Graf E., Panter S.S., Hallaway P.E., Eaton J.W. Hemoglobin. A biologic Fenton reagent. J. Biol. Chem. 1984;259:14354–14356. [PubMed] [Google Scholar]
- 47.Philippidis P., Mason J.C., Evans B.J., Nadra I., Taylor K.M., Haskard D.O., Landis R.C. Hemoglobin scavenger receptor CD163 mediates interleukin-10 release and heme oxygenase-1 synthesis: antiinflammatory monocyte-macrophage responses in vitro, in resolving skin blisters in vivo, and after cardiopulmonary bypass surgery. Circ. Res. 2004;94:119–126. doi: 10.1161/01.RES.0000109414.78907.F9. [DOI] [PubMed] [Google Scholar]
- 48.Schaer D.J., Boretti F.S., Schoedon G., Schaffner A. Induction of the CD163-dependent haemoglobin uptake by macrophages as a novel anti-inflammatory action of glucocorticoids. Br. J. Haematol. 2002;119:239–243. doi: 10.1046/j.1365-2141.2002.03790.x. [DOI] [PubMed] [Google Scholar]
- 49.Cornelison D.D., Wold B.J. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev. Biol. 1997;191:270–283. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- 50.Fuchtbauer E.M., Westphal H. MyoD and myogenin are coexpressed in regenerating skeletal muscle of the mouse. Dev. Dynam. 1992;193:34–39. doi: 10.1002/aja.1001930106. [DOI] [PubMed] [Google Scholar]
- 51.Yablonka-Reuveni Z., Rivera A.J. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Dev. Biol. 1994;164:588–603. doi: 10.1006/dbio.1994.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lluis F., Perdiguero E., Nebreda A.R., Munoz-Canoves P. Regulation of skeletal muscle gene expression by p38 MAP kinases. Trends Cell Biol. 2006;16:36–44. doi: 10.1016/j.tcb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Raingeaud J., Gupta S., Rogers J.S., Dickens M., Han J., Ulevitch R.J., Davis R.J. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J. Biol. Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 54.Zerria K., Jerbi E., Hammami S., Maaroufi A., Boubaker S., Xiong J.P., Arnaout M.A., Fathallah D.M. Recombinant integrin CD11b A-domain blocks polymorphonuclear cells recruitment and protects against skeletal muscle inflammatory injury in the rat. Immunology. 2006;119:431–440. doi: 10.1111/j.1365-2567.2006.02454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spencer M.J., Walsh C.M., Dorshkind K.A., Rodriguez E.M., Tidball J.G. Myonuclear apoptosis in dystrophic mdx muscle occurs by perforin-mediated cytotoxicity. J. Clin. Invest. 1997;99:2745–2751. doi: 10.1172/JCI119464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amalfitano A., Chamberlain J.S. The mdx-amplification-resistant mutation system assay, a simple and rapid polymerase chain reaction-based detection of the mdx allele. Muscle Nerve. 1996;19:1549–1553. doi: 10.1002/(SICI)1097-4598(199612)19:12<1549::AID-MUS4>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 57.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 58.Nolan T., Hands R.E., Bustin S.A. Quantification of mRNA using real-time RT–PCR. Nat. Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 59.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. research/0034.1-0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pilbrow A.P., Ellmers L.J., Black M.A., Moravec C.S., Sweet W.E., Troughton R.W., Richards A.M., Frampton C.M., Cameron V.A. Genomic selection of reference genes for real-time PCR in human myocardium. BMC Med. Genomics. 2008;1:64. doi: 10.1186/1755-8794-1-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chazaud B., Sonnet C., Lafuste P., Bassez G., Rimaniol A.C., Poron F., Authier F.J., Dreyfus P.A., Gherardi R.K. Satellite cells attract monocytes and use macrophages as a support to escape apoptosis and enhance muscle growth. J. Cell Biol. 2003;163:1133–1143. doi: 10.1083/jcb.200212046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen H.X., Tidball J.G. Null mutation of gp91phox reduces muscle membrane lysis during muscle inflammation in mice. J. Physiol. 2003;553:833–841. doi: 10.1113/jphysiol.2003.051912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Rooijen N., Sanders A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. J. Immunol. Methods. 1994;174:83–93. doi: 10.1016/0022-1759(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 64.Gomez C.M., Maselli R., Gundeck J.E., Chao M., Day J.W., Tamamizu S., Lasalde J.A., McNamee M., Wollmann R.L. Slow-channel transgenic mice: a model of postsynaptic organellar degeneration at the neuromuscular junction. J. Neurosci. 1997;17:4170–4179. doi: 10.1523/JNEUROSCI.17-11-04170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lumeng C.N., DelProposto J.B., Westcott D.J., Saltiel A.R. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes. 2008;57:3239–3246. doi: 10.2337/db08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martinez F.O., Sica A., Mantovani A., Locati M. Macrophage activation and polarization. Front. Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 67.Odegaard J.I., Ricardo-Gonzalez R.R., Red Eagle A., Vats D., Morel C.R., Goforth M.H., Subramanian V., Mukundan L., Ferrante A.W., Chawla A. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008;7:496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]