Abstract
The Artemis gene encodes a DNA nuclease that plays important roles in non-homologous end-joining (NHEJ), a major double-strand break (DSB) repair pathway in mammalian cells. NHEJ factors repair general DSBs as well as programmed breaks generated during the lymphoid-specific DNA rearrangement, V(D)J recombination, which is required for lymphocyte development. Mutations that inactivate Artemis cause a human severe combined immunodeficiency syndrome associated with cellular radiosensitivity. In contrast, hypomorphic Artemis mutations result in combined immunodeficiency syndromes of varying severity, but, in addition, are hypothesized to predispose to lymphoid malignancy. To elucidate the distinct molecular defects caused by hypomorphic compared with inactivating Artemis mutations, we examined tumor predisposition in a mouse model harboring a targeted partial loss-of-function disease allele. We find that, in contrast to Artemis nullizygosity, the hypomorphic mutation leads to increased aberrant intra- and interchromosomal V(D)J joining events. We also observe that dysfunctional Artemis activity combined with p53 inactivation predominantly predisposes to thymic lymphomas harboring clonal translocations distinct from those observed in Artemis nullizygosity. Thus, the Artemis hypomorphic allele results in unique molecular defects, tumor spectrum and oncogenic chromosomal rearrangements. Our findings have significant implications for disease outcomes and treatment of patients with different Artemis mutations.
INTRODUCTION
Artemis (DCLRE1C, DNA crosslink repair 1C, OMIM# 605988) was initially identified as the gene mutated in a human T−B− severe combined immunodeficiency associated with cellular radiosensitivity (RS-SCID) (1–3). Artemis is a DNA nuclease that plays critical roles in the context of the non-homologous end-joining (NHEJ) pathway of DNA double-strand break (DSB) repair (4,5). The NHEJ factors are required for processing and joining chromosomal ends during general DSB repair as well as V(D)J recombination, a lymphoid-specific DNA rearrangement (6). V(D)J recombination is the process by which the vast array of antigen receptor genes are assembled from component V, D and J coding exons. During early lymphocyte development, the RAG1/RAG2 endonuclease generates DSBs at specific recombination signal (RS) sequences that flank the numerous rearranging segments (7–9). Cleavage by RAG1/RAG2 produces two end structures: covalently closed hairpin coding ends and 5′ phosphorylated, blunt RS ends. Prior to ligation, the hairpin coding ends are nicked open by the Artemis endonuclease which is activated upon interaction with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), a central NHEJ factor (4). In the absence of Artemis function, unopened hairpin coding ends accumulate in developing lymphocytes and remain unjoined (10). Thus, mutations that abrogate or reduce Artemis activity result in defective V(D)J recombination and impaired B and T lymphocyte development.
To date, 48 different mutant Artemis alleles have been identified in association with inherited combined immunodeficiency syndromes, including missense, splice-site and nonsense mutations, gross exonic and smaller deletions and a small insertion (2,11). The majority of mutations are located within a region encoding a highly conserved metallo-β-lactamase/βCASP N-terminal domain (aa 1–385). A smaller subset of Artemis alleles resides within a non-conserved C-terminus (aa 386–692), and these mutations are small nucleotide deletions or insertions resulting in frameshifts followed by premature translation termination. Patients harboring null mutations suffer from an absence of B and T lymphocytes, whereas partial loss-of-function Artemis alleles are associated with immunodeficiency syndromes of varying severity, including B−/lowT−/low SCID, B−/low SCID, chronic inflammatory bowel disease and Omenn syndrome (2,11–24).
The unjoined DNA ends that accumulate in lymphocytes due to defects in the NHEJ pathway can be misrepaired via alternative repair pathways, thereby leading to genome instability and potentially detrimental chromosomal aberrations, including oncogenic translocations (25). In this regard, partial B and T immunodeficiency and aggressive B cell lymphoma was observed in patients harboring a premature translation termination Artemis mutation within exon 14, which encodes the non-conserved C-terminal domain (D451fsX10, referred to as P70, herein) (12). These lymphoid tumors were associated with Epstein-Barr virus (EBV). However, molecular analyses revealed that the lymphomas were of clonal origin, as evidenced by the rearrangement status of the immunoglobulin heavy chain locus, and also harbored chromosomal anomalies and increased genome instability (12). These features suggest that aberrant Artemis activity contributes to oncogenesis; however, it has not yet been established whether hypomorphic Artemis mutations can predispose to tumorigenesis. To date, patients harboring null Artemis alleles have not been reported to exhibit lymphoid malignancies. These findings raise the possibility that partial loss of Artemis alleles that lead to truncation of the non-conserved C-terminus may have greater oncogenic potential compared with complete null alleles.
Artemis forms a complex with DNA-PKcs, a serine–threonine protein kinase, via interactions within the C-terminal domain. The Artemis:DNA-PKcs complex possesses intrinsic endonucleolytic activities that can cleave DNA at single-to-double-strand transitions, including hairpins and 5′ or 3′ overhangs (4,5,26,27), as well as single strands (28). The Artemis C-terminus undergoes extensive phosphorylation by DNA-PKcs (29–31). In vitro biochemical studies with mutant forms of Artemis harboring site-specific mutations revealed that DNA-PKcs-dependent phosphorylation is not required for activation of endonucleolytic activity (29). However, C-terminally truncated forms of Artemis that retain stable interaction with DNA-PKcs but that lack the majority of phosphorylation sites, including the lymphoma-associated Art-P70 protein, exhibit reduced DNA-PKcs-dependent endonucleolytic activity (26,32). In previous studies, we demonstrated that a mouse model harboring the Art-P70 mutation recapitulated the partial B and T immunodeficiency phenotypes observed in patients (32). We determined that lymphocyte development was impaired due to substantially reduced, but not abrogated, hairpin opening activity catalyzed by the Art-P70 mutant protein. Together, these results indicate that the Artemis C-terminal domain plays important roles in modulating biochemical and in vivo Artemis activities, in addition to facilitating DNA-PKcs interaction.
In this study, we examine the impact of the Art-P70 hypomorphic allele on predisposition to tumorigenesis. We observe that loss of a functional region within the non-conserved Artemis C-terminus leads to aberrant intra- and interchromosomal rearrangements within the antigen receptor loci. In addition, we find that the Artemis-P70 allele in the context of p53 inactivation predisposes to a spectrum of B and T lymphoid malignancies that is distinct from that observed in Artemis nullizygosity. The tumors arising in an Art-P70/p53 background are associated with clonal chromosomal translocations involving the rearranging loci due to misrepair of RAG-generated DNA breaks. Together, these findings provide insights into the molecular basis of tumorigenesis associated with defective, but not abrogated, V(D)J recombination activity. In addition, the results uncover potential roles for Artemis function in contributing to DNA end complex stability of RAG1/2-generated chromosomal breaks.
RESULTS
The Artemis-P70 mutation results in elevated interchromosomal V(D)J rearrangements
We previously observed that the Art-P70 homozygous mutation led to an accumulation of hairpin coding ends in developing lymphocytes; however, the levels were notably lower compared with Artemis nullizygosity (32). The presence of coding ends led us to determine whether the unjoined DSB intermediates engage in aberrant chromosomal rearrangements. To this end, we employed a nested polymerase chain reaction (PCR) approach to examine the levels of interchromosomal V(D)J rearrangements between the TCRβ (chr. 6) and TCRγ (chr. 13) loci in ArtP70/P70 and Art−/− thymocytes, followed by Southern blot analysis. Interchromosomal transrearrangement is a global predictor of chromosomal translocation (33,34) and occurs at elevated frequencies in lymphocytes harboring mutations in genes that predispose to lymphoid neoplasia, including ATM (35–39), DNA-PKcs (34), Nbs1 (33,38) and 53BP1 (40).
Initially, we examined levels of normal TCRγ V-to-J and TCRβ D-to-J rearrangements by PCR amplification. As previously reported, we observed that TCRβ D-to-J rearrangements in ArtP70/P70 thymocytes were reduced and were not readily detected in Art−/− thymocytes (10,32) (Fig. 1A). In comparison, products corresponding to TCRγ intrachromosomal V-to-J rearrangements were not significantly decreased in ArtP70/P70 and Art−/− thymocytes compared with controls. These findings indicate that the Artemis P70 and null mutations impair rearrangement at the TCRγ locus to a lesser extent compared with TCRβ rearrangements, as has been observed at the TCRδ locus in NHEJ-deficient backgrounds, including Artemis nullizygosity (41–44).
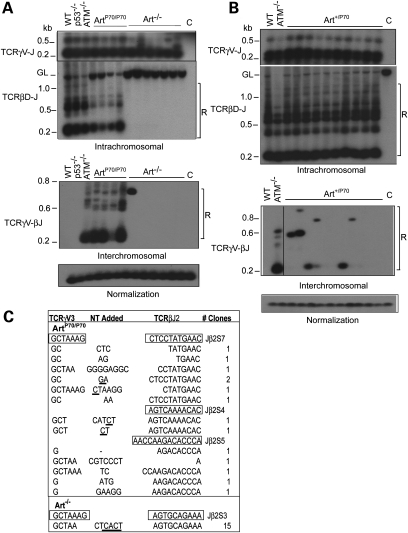
Figure 1.
Aberrant rearrangements in Artemis-P70 lymphocytes. (A) Examination of V(D)J transrearrangements. Intrachromosomal TCRγ V-to-J (chr. 13) and TCRβ Dβ2-to-Jβ2 (chr 6) rearrangements (top) and interchromosomal TCRγV–TCRβJ transrearrangements (bottom) were PCR amplified from genomic thymocyte DNA isolated from wild-type, ArtP70/P70, Art−/−, ATM−/− and p53−/− mice, as indicated. Nested PCR products were detected by Southern blot analysis. Levels of rearrangements were normalized to PCR-amplified non-rearranging locus. GL, germline, unrearranged band; R, bands corresponding to rearrangements; C, PCR amplification of kidney genomic DNA. Representative results are shown. (B) Examination of V(D)J transrearrangements in Artemis-P70 heterozygous mice. Nested PCR analyses of intrachromosomal TCRγ V-to-J, TCRβ Dβ2-to-Jβ2 (top) and interchromosomal TCRγV–TCRβJ transrearrangements (bottom) were performed with genomic DNA isolated from Art+/P70 thymocytes, as described above. Representative results are shown. (C) Sequence analysis of transrearrangement PCR products. Transrearrangement PCR products detected from Southern blot analysis were subcloned and sequenced. Coding sequences are shown in boxes. Nucleotides added include P nucleotides (underlined) as well as N nucleotides.
Next, we determined whether transrearrangement occurs between loci on different chromosomes using PCR primers located upstream of TCRγV3S1 and downstream of TCRβJ2. As anticipated, we detected robust levels of transrearrangement between TCRγ and TCRβ in ATM null thymocytes (35–39). PCR products corresponding to interchromosomal V(D)J rearrangements in wild-type or p53−/− thymocytes were not readily observed, as previously reported (33,38,45) (Fig. 1A). Similar to ATM−/− lymphocytes, we detected substantially increased levels of interchromosomal events involving TCRγV3S1 and TCRβJ2 in ArtP70/P70 thymocytes (n= 5) compared with controls, despite harboring lower levels of Dβ2-to-Jβ2 intrachromosomal rearrangements (Fig. 1A, data not shown). On the contrary, we observed a lower frequency of transrearrangement in Art−/− thymocytes. In this regard, two of seven Artemis null mice harbored PCR products corresponding to TCRγ-to-TCRβ interchromosomal rearrangements (Fig. 1A, data not shown), and the events appeared to be clonal as only a single band was observed, compared with multiple bands observed in ArtP70/P70 and ATM−/− thymocytes. Thus, the Art-P70 mutation increases the propensity of unrepaired coding ends to engage in aberrant interchromosomal translocations involving rearranging V(D)J loci. Moreover, this phenotype is distinct from that observed in Artemis null thymocytes which exhibit infrequent transrearrangements despite harboring a higher level of unjoined hairpin coding ends.
We next examined the frequency of interchromosomal rearrangements in Art-P70 heterozygous thymocytes to determine whether the hypomorphic mutation results in a dominant phenotype. TCRγV3S1 to TCRβJ2 transrearrangements were readily detected in Art+/P70 thymocytes in more than half (six of ten) of the mice examined (Fig. 1B). However, fewer PCR products corresponding to distinct rearrangements and lower levels of interchromosomal events were observed in Art+/P70 compared with ArtP70/P70 and ATM−/− thymocytes. The levels of TCRβ D-to-J rearrangements in Art-P70 heterozygous thymocytes were similar to those observed in wild-type controls (Fig. 1B). These findings indicate that the C-terminally truncated Art-P70 protein does not substantially disrupt proper coding end processing and joining, yet increases chromosomal anomalies, even when the wild-type enzyme is present.
The nested PCR products from Art-P70 mutant thymocytes were cloned and sequenced in order to verify that they represent the predicted interchromosomal events. We obtained several unique clones from four ArtP70/P70 mice containing flanking sequence from TCRγV3S1 and TCRβJ2, thereby indicating that the PCR primers indeed amplified V(D)J transrearrangements (Fig. 1C). The junctions contained non-templated (N) and palindromic (P) nucleotides and small deletions, similar to the coding joints analyzed from intrachromosomal V(D)J recombination events in the ArtP70/P70 mice (32). We also cloned and sequenced the PCR products corresponding to the interchromosomal rearrangements obtained from Art−/− thymocytes. Sequencing of multiple clones yielded one unique sequence, thereby indicating that the single PCR product likely represents a clonal event (Fig. 1C). These results support the notion that aberrant end processing due to the hypomorphic Art-P70 mutation generates V(D)J ends that engage in chromosomal translocations at an elevated frequency compared with a complete absence of Artemis.
The Artemis-P70 mutation results in increased deletional chromosomal hybrid joining
Hybrid joint formation occurs between a coding and an RS end and represents an unproductive V(D)J rearrangement. Inversional chromosomal rearrangements require that the two coding and two RS ends generated by the RAG endonuclease are maintained in proximity in order to facilitate coordinated processing and ligation. Increased levels of deletional chromosomal hybrid joining during inversional V(D)J rearrangement are hypothesized to result from inappropriate release of RAG-generated ends from DNA end complexes, thereby leading to the loss of the intervening genomic fragment (41,46,47). Our observations of increased interchromosomal rearrangements in ArtP70/P70, but not Art−/− lymphocytes, led us to examine the frequency of deletional hybrid joining in these mutant backgrounds. To this end, we examined hybrid joint formation during IgL-κ locus rearrangements in splenocytes. Productive rearrangement between Vκ and Jκ segments occurs via inversion; thus, hybrid joining results in deletion of the intervening DNA segment (Fig. 2A).
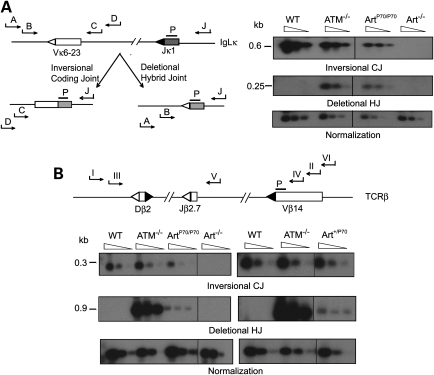
Figure 2.
Deletional hybrid joint formation in Art-P70 mutant lymphocytes. (A) Left panel: Schematic representation of nested PCR strategy for detecting coding and hybrid joints within the IgLκ locus. Relative orientation of Vκ6-23 and Jκ1 coding (rectangles) and RSSs (triangles) within the IgLk locus. Inversional (CJ) and deletional (HJ) products are depicted. Positions of primers (J = pκJa, A = pκ6a, B = pκ6b, C = pκ6c, D = pκ6d) and probe (P) are shown. Right panel: PCR analysis of Vκ6-23 to Jκ1 coding joins (CJs) and hybrid joins (HJs). Genomic splenocyte DNA was isolated from WT, ATM−/−, Art−/− and ArtP70/P70 mice and amplified using primers (pκJa and pκ6d for CJ and pκJa and pκ6a for HJ). Serial 4-fold dilutions of the PCR reaction were amplified using nested primer pairs (pκJa and pκ6c for CJ, and pκJa and pκ6b for HJ). CJ and HJ bands are 0.6 and 0.25 kb, respectively. (B) Deletional hybrid joining and coding joining within the TCRβ locus. Upper panel: Schematic of nested PCR strategy. Relative orientation of Vβ14, Dβ2 and Jβ2.3 coding (rectangles) and RSSs (triangles) within the TCRβ locus. Positions of primers (I = pβa, II = pβb, III = pβc, IV = pβd, V = pβe, VI = pβf) and probe (P = pβg) are shown. Lower panel: PCR analysis of Vβ14 to Jβ2 coding and hybrid joints. Left panel shows CJ and HJ PCR analyses of genomic thymocyte DNA isolated from WT, ATM−/−, Art−/− and ArtP70/P70 mice. Right panel shows CJ and HJ PCR analyses of Art+/P70 and control thymocyte genomic DNA, as indicated. Primers for CJ were pβe and pβf. Primary primers for HJ were pβa and pβb. Serial 4-fold dilutions of this primary HJ reaction were amplified using primers pβc and pβd. CJ and HJ bands are 0.3 and 0.9 kb, respectively. PCR amplification of a non-rearranging locus was performed to normalize the input genomic DNA. Representative results are shown.
Using a nested PCR approach, we readily detected Vκ to Jκ hybrid joints in ArtP70/P70 splenocytes in the majority (three of four) of mutant mice analyzed (Fig. 2A). The levels were lower than those observed in ATM deficiency, but markedly higher compared with wild-type and Art−/− splenocytes. We also observed decreased levels of Vκ to Jκ coding joining in Art-P70 mutant splenocytes compared with controls; therefore, the relative frequency of deletional hybrid joining compared with inversional coding joining within the IgLκ locus is substantially increased by the Art-P70 mutation.
Within the TCRβ locus, Vβ14-to-DJβ2 rearrangements also occur via inversion; thus, we examined deletional hybrid joining between Vβ14 and Dβ2 via nested PCR. We detected increased levels of Vβ14–Dβ2 hybrid joints in ArtP70/P70 thymocytes (two of four), albeit at lower levels compared with ATM deficiency (Fig. 2B). Hybrid joints were not detected in Art−/− thymocytes, as previously reported (47) (Fig. 2B). Given our findings that interchromosomal rearrangements were detected in Art+/P70 thymocytes, we assessed the impact of Art-P70 heterozygosity on Vβ14–Dβ2 hybrid joining (Fig. 2B). We also detected hybrid joints in a subset of Art+/P70 mice examined (two of five), thereby providing additional evidence that the Art-P70 allele may have a dominant effect in promoting aberrant rearrangements.
Examination of ATM- and MRN-dependent DNA damage responses in Artemis-P70 cells
The increased levels of interchromosomal rearrangements and deletional hybrid joining observed in ArtP70/P70 lymphocytes parallel the phenotypes observed in ATM, Mre11 and Nbs1 mutant lymphocytes (41,46,47). These findings raise the possibility that the Art-P70 allele impairs ATM-dependent responses to DNA DSBs and/or disrupts the functional Mre11/Rad50/Nbs1 (MRN) complex. To address these questions, we examined key ATM-dependent cellular responses to ionizing radiation (IR)-induced DSBs in ArtP70/P70 murine embryonic fibroblasts (MEFs). Upon exposure to IR, the ATM protein kinase undergoes autophosphorylation in an MRN-dependent manner (48,49) and subsequently phosphorylates downstream targets, including the histone variant, H2AX (38,50) and the transcriptional corepressor, KAP1 (51). We examined the levels of phospho-ATM (p-ATM), phospho-H2AX (γH2AX) and phospho-KAP-1 (p-KAP1) in ArtP70/P70, Art−/− and wild-type MEFs at 1 h post-irradiation by western blotting. We observed similar levels of IR-induced phosphorylation of ATM, H2AX and KAP1 in wild-type, ArtP70/P70 and Art−/− cells (Fig. 3A). These findings indicate that the Art-P70 and null alleles do not significantly impair ATM-dependent responses to DSBs.
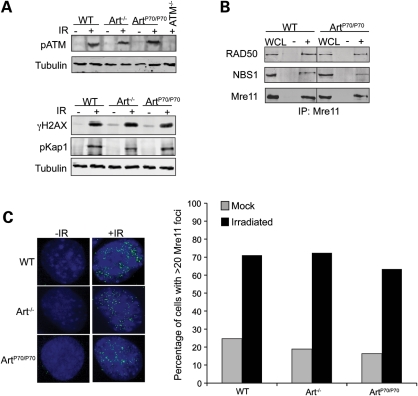
Figure 3.
ATM- and MRN-dependent responses to IR-induced breaks in Art-P70 mutant cells. (A) Art-P70 mutation does not impair ATM-dependent DNA damage responses to IR. Art+/+, Art−/−, ArtP70/P70 and ATM−/− MEFs, as indicated, were unirradiated (−) or exposed to IR (10 Gy, +) and then harvested at 1 h post-irradiation. Whole-cell lysates were analyzed by western blotting using the indicated antibodies. Tubulin was used as a loading control. (B) The MRN complex is intact in Art-P70 mutant cells. The MRN complex was co-immunoprecipitated from whole-cell lysates generated from wild-type or ArtP70/P70 MEFs using α-Mre11 antibodies. The immunoprecipitates were washed with 300 mm KCl, and the proteins were analyzed on a 8% SDS–PAGE followed by western blotting with the indicated antibodies. WCL, whole-cell lysate; −, no antibody; +, α-Mre11 antibody. (C) Mre11 foci formation is not impaired in Art-P70 mutant cells. Art+/+, Art−/− and ArtP70/P70 MEFs were grown on coverslips then irradiated (10 Gy). The irradiated cells and unirradiated controls were fixed in paraformaldehyde and then stained with α-Mre11 antibodies. The number of cells containing >20 Mre11 foci were scored. Left panels, representative images of nuclei. Right panels, quantitative results of Mre11 foci-positive cells. Average of two independent experiments is shown.
We next assessed the impact of the Art-P70 allele on the stability and localization of the MRN complex. We examined the levels of Mre11, Rad50 and Nbs1 in whole-cell lysates and upon immunoprecipitation of the complex using anti-Mre11 antibodies in ArtP70/P70 MEFs and wild-type controls by western blotting. We observed that the overall levels of Mre11, Rad50 and Nbs1 in ArtP70/P70 MEFs were not different from those observed in control cells (Fig. 3B). Furthermore, we observe similar levels of Mre11, Rad50 and Nbs1 upon co-immunoprecipitation of Mre11 from ArtP70/P70 and wild-type cells, thereby indicating that the MRN complex was not disrupted by the Art-P70 allele (Fig. 3B).
During repair of IR-induced DSBs, Mre11 localizes to sites of damage and forms repair foci that can be visualized as punctate staining by immunofluorescence (48,52). DNA damage-induced Mre11 foci formation requires the presence of an intact and functional MRN complex (48). We exposed ArtP70/P70, Art−/− and wild-type MEFs to IR and quantitated the number of untreated and irradiated cells containing Mre11 foci. We observed that the Art-P70 and Art null alleles did not reduce IR-induced Mre11 foci formation (Fig. 3C). These results indicate that the Art-P70 mutation does not impair ATM- and MRN-dependent DNA damage responses. Thus, these findings are consistent with the notion that the aberrant rearrangements observed in ArtP70/P70 lymphocytes are due to defects in Artemis function at RAG-generated DSBs.
Artemis-P70 predisposes to lymphoma in a p53 mutant background
The increased levels of aberrant rearrangements in ArtP70/P70 lymphocytes suggested that the Art-P70 mutation may increase the frequency of RAG-generated DNA ends that engage in oncogenic translocations. To address this question, we examined cohorts of ArtP70/P70, Art−/− and wild-type mice over a period of 12 months to determine whether the Art-P70 mutation predisposes to lymphoid or other tumors. We observed that two of fourteen ArtP70/P70 mice became moribund at 7 and 9 months of age as a result of large thymic masses (Fig. 4A). Flow cytometric analyses revealed that the thymic lymphomas were primarily of a CD4+CD8+TCRβ− origin (Fig. 4B). In comparison, the wild-type and Art−/− control cohorts survived tumor-free within the 12-month period, which is consistent with previous reports (53,54).
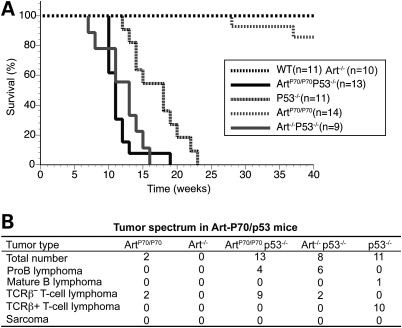
Figure 4.
Art-P70/p53 double-mutant mice are predisposed to thymic lymphomas with chromosomal translocations. (A) Decreased survival of Art-P70/p53 mice. Survival of a cohort of wild-type (n= 11), Art−/− (n= 10), p53−/− (n= 11), ArtP70/P70 (n= 14), ArtP70/P70p53−/− (n= 13) and Art−/−p53−/− (n= 9) mice was observed for a period of 40 weeks. Shown are Kaplan–Meier survival curves representing the percentage of survival of cohort mice versus age in weeks. (B) Distinct tumor spectrum exhibited by Art-P70/p53 mice. Table summarizing the number of the different tumor types observed in mice of the indicated genotypes.
To further assess the oncogenic potential of the Art-P70 hypomorphic allele, we examined the impact of p53 mutation on tumor predisposition through mouse breeding. The experimental cohorts were on a closely matched 129Sv/C57Bl6 background, thereby minimizing potential strain background effects. However, a subtle impact of genetic background cannot be entirely ruled out. Inactivation of the p53-dependent cell-cycle checkpoint in Artemis mutant lymphocytes allows cells harboring unrepaired DSBs or activated oncogenes to survive (55–59). We observed that Art-P70/p53 double mutant mice exhibit significantly decreased survival compared with p53−/− controls (median survival of 11 and 18 weeks, respectively; P = 0.001; Fig. 4A). As previously reported, we found that Art−/−p53−/− mice also exhibited decreased survival compared with p53 null controls, and the median survival (13 weeks) was similar to that observed for Art-P70/p53 double-deficient cohort (Fig. 4A). We found that the Art−/−p53−/− and ArtP70/P70p53−/− mice succumbed to lymphoid tumors. Flow cytometric analysis of the tumors revealed that Art−/−p53−/− mice were predominantly predisposed to disseminated B220+CD43+IgM− pro-B lymphomas, as has been previously reported (54) (Fig. 4B). In contrast, the majority of tumors that arose in the Art-P70/p53 double-mutant background were CD4+CD8+TCRβ− thymic lymphomas (Fig. 4B). B220+CD43+IgM− pro-B lymphomas were also observed in ArtP70/P70p53−/− mice, similar to Art/p53 double null mice (54) (Fig. 4B), albeit at a significantly lower frequency (P = 0.014; two-tailed Fisher's exact test). These findings indicate that the Art-P70 allele predisposes to lymphoid malignancies in a p53-deficient background, and the lymphoma spectrum observed in ArtP70/P70p53−/− mice is distinct from that observed in NHEJ/p53 double null backgrounds, including Art−/−p53−/− mice (54,59).
Distinct chromosomal anomalies associated with Art-P70/p53 lymphoid tumors
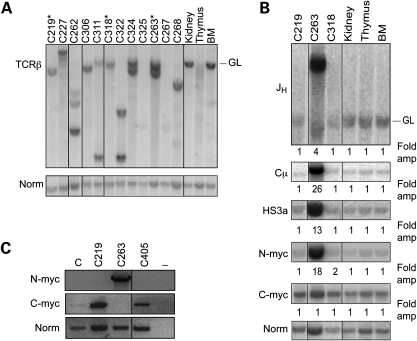
We next examined the status of the rearranging TCRβ and IgH loci in the lymphomas that arose in the ArtP70/P70p53−/− and ArtP70/P70 mice. Genomic DNA isolated from the primary tumors was digested with EcoRI and analyzed by Southern blotting. We used a probe located within the TCR Dβ1 to Jβ1 region to examine rearrangement status of the TCRβ locus. We observed that the ArtP70/P70p53−/− thymic tumors exhibited clonal rearrangements on one or both alleles, as evidenced by hybridization of specific bands that are distinct from the germline, unrearranged band or deletion of the hybridizing region (Fig. 5A). Similarly, the two ArtP70/P70 tumors exhibited clonal DJβ rearrangements (data not shown). These results indicate that the thymic lymphomas emanated from a clonal event within the population of developing Art-P70 mutant thymocytes.
Figure 5.
Clonal rearrangements involving recombining loci in Art-P70/p53 tumors. (A) Analysis of TCR rearrangement status in Art-P70/p53 thymic lymphomas. Genomic DNA isolated from ArtP70/P70p53−/− tumors was digested with EcoRI and then analyzed by Southern blotting. Individual tumors are indicated (top). Thymus, kidney and bone marrow (BM), control tissues; GL, unrearranged, germline band. Amounts of input DNA were normalized to a non-rearranging locus (LR8). *, pro-B lymphomas (C219, C263, C318). (B) Analysis of Art-P70/p53 pro-B lymphomas by Southern blotting. Genomic DNA isolated from Art-P70/p53 double-mutant lymphomas was digested with EcoRI and then analyzed by Southern blotting. Previously characterized probes that hybridized to the JH, Cµ and HS3A regions of the IgH locus, N-myc on chr. 12 and c-myc on chr. 15 were used, as indicated. Individual tumors are indicated, top; Thymus, kidney, and bone marrow (BM), control tissues; GL, unrearranged, germline band. Amounts of input DNA were normalized to a non-rearranging locus (LR8). Fold amplification compared with the non-rearranging locus was calculated as described in Materials and methods. (C) Semi-quantitative RT-PCR of N-myc and c-myc transcript levels. Total RNA was isolated from primary tumor cells, and RT-PCR was performed. cDNAs were PCR amplified using N-myc (exons 2 and 3), c-myc (exons 1–3) and tubulin-specific primers. Tumor numbers, as indicated; C, total RNA from normal, wild-type LN; −, no-RT control. ArtP70/P70p53−/− tumors: thymic lymphomas: C227, C262, C306, C311, C322, C324, C325, C267, C268; pro-B lymphomas: C219, C263, C318. Art−/−p53−/−; pro-B lymphoma: C405.
We also analyzed the rearrangement status of the IgH locus in Art-P70/p53 pro-B lymphomas by Southern blotting EcoRI-digested genomic DNA from primary tumors (Fig. 5B). Previous analyses of Art−/−p53−/− pro-B lymphomas by Southern blotting revealed clonal rearrangements and amplification of the JH locus. We observed these similar events in one ArtP70/P70p53−/− pro-B lymphoma (C263). However, pro-B tumors, C219 and C318, did not exhibit clonal IgH rearrangement or amplification as only the germline band was present. It is possible that the region encompassing the probe was deleted from one of the two rearranging IgH alleles. Thus, we further analyzed the molecular events occurring in the Art-P70/p53 double null tumors. One recurrent event within the IgH locus observed in Art−/−p53−/− pro-B lymphomas is amplification of regions downstream of the JH region. We used probes comprised the Cµ constant region and 3′ enhancer regulatory region located approximately 5 and 170 kb, respectively, from the rearranging JH segments. We observed amplification with the more distal probes in tumor C263, but not in the other ArtP70/P70p53−/− pro-B lymphomas analyzed (Fig. 5B).
We next examined the status of the c-myc and N-myc loci, as amplification of either genomic region is associated with Art−/−p53−/− pro-B tumors. Southern blotting revealed genomic amplification of the N-myc locus in tumor C263 (Fig. 5B), and we determined that N-myc expression was elevated by northern blot and semi-quantitative RT-PCR analyses (Fig. 5C, data not shown). Thus, this ArtP70/P70p53−/− pro-B lymphoma harbors the established hallmark events observed in Art/p53 double-mutant tumors, i.e. increased copy number of the IgH and N-myc genomic loci. However, we did not observe genomic amplification of either c-myc or N-myc in the C219 and C318 ArtP70/P70p53−/− pro-B tumors, providing further distinction between lymphomas arising in the Art-P70 versus null backgrounds.
Spectral karyotyping and fluorescence in situ hybridization analyses of Art-P70/p53 double-mutant lymphomas
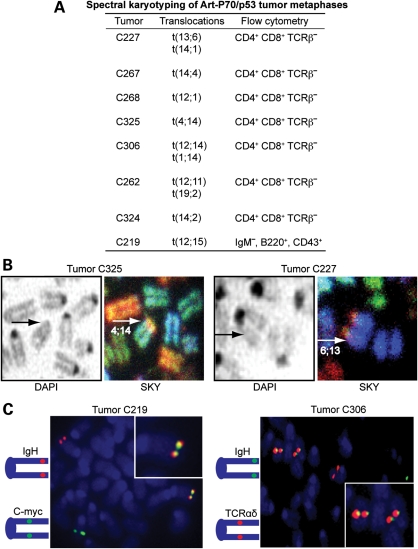
We next examined the cytogenetic events occurring in Art-P70/p53 double-mutant lymphomas using spectral karyotyping (SKY) and fluorescence in situ hybridization (FISH) analyses. We performed SKY on metaphase spreads from seven ArtP70/P70p53−/− thymic lymphomas. We observed that all of the Art-P70/p53 tumors analyzed contained non-reciprocal clonal translocations involving chromosomes harboring rearranging loci (Fig. 6A and B). In this regard, five tumors harbored clonal events involving chr. 14, the location of the TCRαδ locus, translocated to chr. 1, 2, 4 or 12 (Fig. 6A). It is of interest to note that the IgH locus is located on chr. 12, and in addition to the t(12;14) translocation in tumor C306, we also observed t(12;11) and t(12;1) clonal events in tumors C262 and C268, respectively. One tumor, C227, also harbored clonal events involving chr. 13 (TCRγ) translocated to chr. 6 (TCRβ) (Fig. 6A and B). We analyzed one ArtP70/P70p53−/− pro-B lymphoma, C219, by SKY and observed that, similar to other reported NHEJ/p53 double null tumors, it harbored a t(12;15) non-reciprocal translocation (Fig. 6A).
Figure 6.
Art-P70/p53 tumors harbor clonal chromosomal translocations. (A) Table summarizing clonal translocations observed in Art-P70/p53 tumors. Metaphases from primary tumor cells or early passage tumor cell cultures were analyzed by SKY. At least 10 metaphases from each tumor were scored. (B) SKY images of metaphase chromosomes from Art-P70/p53 tumors. DAPI staining (left panels) and SKY analysis (right panels) of tumor C325 and C227 containing clonal t(4;14) and t(6;13) translocations, respectively. Arrows indicate translocated chromosomes. (C) FISH analyses of metaphases from Art-P70/p53 tumors. Representative FISH analyses of metaphases from Art-P70/p53 tumors. Left panel, pro-B lymphoma, C219. Left panel, thymic tumor C306. At least 20 metaphases were scored for each tumor. Diagrams indicate the relative chromosomal positions and fluorescent colors of BAC probes. Inset, enlarged images of co-localized probes.
SKY analyses can effectively identify gross chromosomal anomalies; however, genomic loci that may be amplified or co-localized cannot be accurately detected using this technique. Thus, we further analyzed the metaphases from Art-P70/p53 lymphomas using a two-color FISH approach. We established that thymic lymphomas, C325 and C306, harbored clonal chr. 14 anomalies using bacterial artificial chromosome (BAC) probes comprised of genomic sequences located upstream and downstream of the rearranging TCRαδ locus. In this regard, we observed co-localization of the probes in control metaphases, whereas the two probes were clearly located on different chromosomes in metaphases from the ArtP70/P70p53−/− thymic tumors (Supplementary Material, Fig. S1). In tumor C306, we also observed co-localization of the IgH and TCRαδ BAC probes, as anticipated based on the t(12;14) identified by SKY (Fig. 6C). Likewise, we found co-localization of BAC probes containing TCRβ and TCRγ genomic sequences in tumor C227. Tumor C262 harbored separated single-copy FISH signals using BACs located upstream and downstream of the IgH locus on chr. 12 (Supplementary Material, Fig. S1). Thus, the ArtP70/P70p53−/− thymic lymphomas harbored chromosomal aberrations and translocation events that involved the loci undergoing V(D)J recombination. However, we did not observe amplification of the rearranging loci examined in the thymic lymphomas (Fig. 6C and Supplementary Material, Fig. S1).
FISH analyses of metaphases from Art-P70/p53 pro-B lymphoma, C219, revealed separation of single-copy signals using the upstream and downstream IgH probes, thereby suggesting that RAG-generated DSBs within the rearranging locus initiated the aberrant events (Supplementary Material, Fig. S1). We also observed co-localization of the FISH signals corresponding to c-myc (chr. 15) and IgH loci, as predicted based on the SKY results. However, distinct from the hallmark co-amplification observed in other NHEJ/p53 double null pro-B tumors, no amplification of either signal was found (Fig. 6C). These findings are consistent with the Southern blot analyses that did not detect amplification using probes within the c-myc, JH, Cµ or HS3A loci.
Previous studies of other NHEJ/p53 double null pro-B lymphomas, including Art−/−p53−/− tumors, established that genomic amplification of the c-myc locus was associated with elevated c-myc expression levels in the tumor cells (54,59). As we did not observe amplification of c-myc by Southern or FISH analyses, we sought to determine whether the oncogene may be dysregulated by a distinct mechanism in the ArtP70/P70p53−/− tumor, C219, which harbored the t(12;15) translocation and co-localization of c-myc and IgH FISH probes. Thus, we examined expression levels of c-myc in primary tumor cells compared with control cells by semi-quantitative RT-PCR (Fig. 5C). We observed a substantial increase in c-myc expression in the C219 Art-P70/p53 pro-B lymphoma, comparable to that observed in a control Art−/−p53−/− tumor (C405) which harbored the hallmark c-myc and IgH amplicon (Fig. 5C and Supplementary Material, Fig. S1). These results suggest that elevated c-myc expression observed in the Art-P70/p53 mutant background results from a mechanism independent of genomic amplification and thus distinct from that observed in Art−/−p53−/− tumors.
DISCUSSION
In this study, we demonstrate that a hypomorphic Artemis mutation that results in partial B and T immunodeficiency and EBV-associated lymphoma in patients increases the frequency of aberrant chromosomal rearrangements in primary lymphocytes and predisposes to lymphoid malignancy in a mouse model harboring the human disease allele. The Art-P70 mutation, which truncates the non-conserved C-terminus, results in elevated levels of V(D)J transrearrangements between loci located on different chromosomes and deletional hybrid joining in homozygous and heterozygous mutant lymphocytes. In comparison, undetectable or substantially lower levels of these chromosomal anomalies are present in Artemis null lymphocytes, thereby indicating that loss of functional regions within the C-terminal domain increases the potential for the DSB intermediates to engage in aberrant repair events.
Previously, we demonstrated that a mutant Artemis protein modeled after the P70 allele, Art-D451X, interacted stably with DNA-PKcs and exhibited reduced DNA-PKcs-dependent endonucleolytic activity (32). In addition, we found that loss of the C-terminal 241 amino acids markedly reduced DNA-PKcs-dependent phosphorylation due to deletion of the majority of phosphorylation sites. We hypothesized that these defects may impair the ability of Artemis to associate with and properly act upon DNA ends. Consistent with this notion, hairpin coding ends accumulate in ArtP70/P70 developing lymphocytes, whereas nicked hairpins with blunt or 5′ or 3′ overhanging ends are not detected (32). In the current study, we observed increased levels of interchromosomal V(D)J rearrangements and deletional hybrid joints within the IgLκ and TCRβ loci in ArtP70/P70 primary lymphocytes. The mechanism underlying these events presumably involves inappropriate release of RAG-generated DNA ends from post-cleavage complexes prior to joining (41,46,47). In contrast, these aberrant rearrangements occur infrequently in Artemis null lymphocytes (Figs 1 and 2) (60), despite harboring significantly higher levels of hairpin coding ends compared with Art-P70 mutant lymphocytes (32). Although open hairpins are not detected in ArtP70/P70 lymphocytes, it is possible that a low level of nicked coding ends is present, and these end structures may be more likely to engage in aberrant events. However, interchromosomal rearrangements and deletional hybrid joining occur infrequently in wild-type lymphocytes in which hairpins are efficiently cleaved by Artemis. Likewise, coding ends in normal lymphocytes rarely serve as substrates for oncogenic translocations, even in a p53-deficient background which permits RAG-generated ends to persist throughout the cell cycle (45,54,55,61–66). Thus, the Art-P70 allele likely causes molecular defects in coding end processing and joining beyond impaired hairpin nicking.
Mutations in ATM, Mre11 or Nbs1 significantly increase the frequency of aberrant chromosomal rearrangements in mutant lymphocytes, including interchromosomal transrearrangements and deletional hybrid joint formation (33,35–39,41,46,47). These observations led to the hypothesis that the ATM kinase and MRN complex function during V(D)J recombination to enhance DNA end complex stability and promote proper joining of RAG-generated breaks (41,46,47). Our findings suggest that truncation of the C-terminus impairs functions of Artemis within post-cleavage DNA end complexes, thereby leading to inappropriate release and altered handling of V(D)J recombination intermediates. It is of interest to note that ATM phosphorylates Artemis at residues S503, S516 and S645 which are located within the C-terminal region that is deleted in the Art-P70 mutant protein (29). ATM does not play a direct role in V(D)J recombination per se as ATM-deficient cells exhibit wild-type levels of V(D)J recombination on extra-chromosomal plasmid substrates (67). However, ATM deficiency in mice leads to accumulation of V(D)J coding end intermediates, impaired lymphocyte development and aberrant, potentially oncogenic, chromosomal rearrangements (39,68–72). Although ATM-dependent phosphorylation of Artemis is not required to activate intrinsic endonucleolytic activity in vitro nor is it required for V(D)J recombination on model plasmid substrates in cells (29), our findings raise the possibility that ATM phosphorylation may modulate Artemis functions during chromosomal V(D)J rearrangements to facilitate the stabilization of DNA end complexes in vivo.
We propose that the Art-P70 mutant protein is recruited to DNA ends via interaction with DNA-PKcs (32), and activation of DNA-PKcs upon autophosphorylation induces large conformational changes to allow the nuclease access to the hairpins (29,73,74). Truncation of the Artemis C-terminal region that contains ATM and DNA-PKcs phosphorylation sites may prevent stable association within conformationally altered DNA end complexes that are poised for further end processing events. Inappropriate release of the RAG-generated ends from aborted post-cleavage complexes would render the ends more susceptible to misrepair, thereby increasing their potential to generate chromosomal aberrations, including oncogenic translocations (41).
We demonstrate that the Art-P70 mutation in a p53 null background accelerates the timing of tumor onset compared with p53 mutation alone. Art-P70/p53 double mutant mice predominantly succumb to CD4+CD8+TCRβ− thymic lymphomas that are associated with clonal chromosomal translocations involving the TCR or IgH loci; however, the majority of tumors do not harbor the hallmark gene amplification events observed in NHEJ/p53 double null lymphomas, including those arising in Art−/−p53−/− mice (54,59,75). We found that the ArtP70/P70p53−/− and Art−/−p53−/− lymphomas arise with a similar latency, despite our observation of substantially higher levels of aberrant interchromosomal rearrangements in Art-P70 mutant lymphocytes. One potential explanation for these observations is that the timing of lymphoma incidence is influenced by the particular oncogenic events associated with tumorigenesis. In this regard, genomic amplification leading to elevated expression of c-myc or N-myc in Art−/−p53−/− lymphomas may lead to a higher proliferative potential compared with oncogenic events in the ArtP70/P70p53−/− background, thereby accelerating tumorigenesis in Art/p53 double null mice.
The frequently arising ArtP70/P70p53−/− thymic lymphomas are associated with clonal translocations involving chr. 6, 12, 13 and 14 which harbor the murine TCRβ, IgH, TCRγ and TCRα/δ loci, respectively, with chr. 14 translocations observed in the majority of the tumors analyzed. Cytogenetic analyses of activated T cells isolated from human lymphoma patients harboring the hypomorphic allele modeled in the Art-P70 mouse and a similar C-terminal truncating mutation (T432SfsX16) revealed a translocation of chr. 7 and 14 and inversion of chr. 7, respectively (12). In humans, the rearranging IgH and TCRα/δ loci reside on chr. 14, whereas TCRγ and TCRβ reside on chr. 7. Thus, Art-P70 hypomorphic allele results in translocations involving chromosomes that undergo V(D)J recombination in both human and murine lymphocytes.
The precise mechanisms underlying the distinct molecular events observed in Art-P70/p53 lymphomas have not yet been elucidated. Inactivation of the p53-dependent cell-cycle checkpoint has been hypothesized to allow unrepaired RAG-induced DNA ends generated during G1 to persist throughout the cell cycle and undergo mis-repair by alternative DSB repair pathways (55,59,75). We speculate that unjoined coding ends in Art-P70 versus Artemis null lymphocytes may be repaired by distinct pathways that function during different cell-cycle phases. Consistent with this notion, the junctional sequences of both intra- and interchromosomal V(D)J rearrangements in ArtP70/P70 lymphocytes are characteristic of joining mediated by the classical NHEJ pathway (32) (Fig. 1). In comparison, an alternative pathway generates aberrant V(D)J junctions containing large deletions and long P nucleotide additions in Art−/− lymphocytes (10) and microhomology mediated translocations in Art−/−p53−/− lymphomas (54). An alternative, though not mutually exclusive, hypothesis is that defects in DNA end complex stability in Art-P70 mutant lymphocytes allow unrepaired breaks to be aberrantly localized in three-dimensional space and engage in translocations that do not require the chromosomal partner to be located in proximity. In this regard, recent studies have provided evidence that loci involved in recurrent oncogenic translocations are located in proximity in lymphocytes (76–79). This hypothesis does not preclude amplification from occurring in the Art-P70 background, and indeed we did observe N-myc amplification in one ArtP70/P70p53−/− pro-B lymphoma (C263). It will be of significant interest to further define the molecular mechanisms underlying the oncogenic translocations in Art-P70/p53 tumors.
Together, these studies provide insight into the consequence of truncation of the Artemis C-terminal domain on the fate of chromosomal DNA ends during endogenous V(D)J rearrangements. Our findings support the notion that loss of the Artemis C-terminus impacts the proper processing and joining of DNA ends via destabilization of end bound complexes that coordinate recombination events. These findings have important clinical implications in the identification and treatment of human immunodeficiency patients harboring similar Artemis mutations that may predispose to aberrant rearrangements and lymphoid malignancy.
MATERIALS AND METHODS
Mice
Gene-targeted ATM null, Artemis null and Art-P70 mice (mixed 129Svev/C57Bl6 genetic background) were previously generated (10,32,80,81). p53 mutant mice (Trp53tm1Tyj) in a 129S2/Sv background were obtained from Jackson Laboratory and bred with Art−/− and ArtP70/P70 animals. Double heterozygous Art+/−p53+/− and Art+/P70p53+/− mice were subsequently interbred to generate progeny of the desired genotypes for the tumorigenesis studies (i.e. Art+/+p53+/+, Art−/−p53+/+, Art−/−p53−/− ArtP70/P70p53+/+, ArtP70/P70p53−/− and Art+/+p53−/−). The single- and double-mutant mice as well as wild-type controls used in this study were approximately 75% 129Sv and 25% C57Bl6; thus, the experimental cohorts are closely strain matched. Mice were housed in a specific pathogen-free facility in a room dedicated to immunocompromised animals.
Interchromosomal V(D)J rearrangements
Nested PCR amplification reactions used to detect TCRγ intrachromosomal and TCRγ-to-TCRβ interchromosomal transrearrangements were modified from methods as described previously (34). Genomic DNA (100 ng) obtained from thymocytes isolated from 4- to 5-week-old mice [Art+/+ (n= 3), Art−/− (n= 7), ArtP70/P70 (n= 5), Art+/P70 (n= 10)] was amplified in 50 µl of reaction mixture containing set ‘a’ primers (10 pmol) (see Supplementary Material). The cycling conditions were: denaturation at 95°C, 30 cycles of amplification at 95°C for 15 s, 55°C for 15 s, 72°C for 30 s with a 6-s increment per cycle followed by 10 min elongation at 72°C. The products from the first reaction (5 µl) were used in a nested PCR reaction with the same conditions using primers set ‘b’. The final PCR products (25 µl) were run on a 1.5% agarose gel followed by Southern blotting using probes (primers set ‘c’) internal to the primers used for PCR amplification (see Supplementary Material). The second-round PCR products were subcloned into pCR 2.1-TOPO (Invitrogen; Carlsbad, CA, USA) and individual clones were sequenced. TCRβ Dβ2-to-Jβ2 intrachromosomal rearrangements were PCR amplified from thymic genomic DNA (100 ng) at an annealing temperature of 62°C (35 cycles). Each experiment was repeated at least thrice independently.
Hybrid join analysis
To analyze the levels of coding and hybrid joints between Vκ6–23 and Jκ1, PCR assays were used as described previously (47). Genomic DNA (0.5 µg) isolated from mouse splenocytes for each genotype (ArtP70/P70; n= 4) was PCR amplified in 50 µl with 15 pmol of each primer. PCR conditions were as follows: 95°C for 5 min followed by 17 cycles of 94°C (30 s), 64°C (30 s), 72°C (30 s). A second PCR reaction was carried out under the same conditions for 25 amplification cycles using 4-fold dilutions of the first PCR reaction and nested primer pairs. The HJ and CJ PCR products were transferred to Zetaprobe membrane and hybridized with pκg oligonucleotide. For normalization, 4-fold dilutions starting with 0.5 µg of genomic DNA were PCR amplified. For Vβ14 coding and hybrid joint analysis, genomic DNA (0.5 µg) was isolated from mouse thymocytes for each genotype [ArtP70/P70 (n= 2), Art+/P70 (n= 5)], and PCR analysis was performed as described (47). The PCR products were analyzed by Southern blotting using the pβg oligonucleotide as a probe (see Supplementary Material).
Western blot analysis of ATM-dependent responses to IR
Art+/+, Art−/−, ArtP70/P70 and ATM−/− MEFs (SV40 large T-antigen immortalized) were plated at a density of 3.5 × 106 cells per 10-cm dish then exposed to 10 Gy of γ-rays from a 137Cs source. The cells were allowed to recover for 1 h and then harvested in Laemmli buffer (4% sodium dodecyl sulfate, 20% glycerol, 120 mm Tris–HCl, pH 6.8). Equivalent amounts of whole-cell lysates were resolved on either a 12 or 6% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel and transferred to polyvinylidene fluoride membrane. Primary antibodies used were: γH2AX S139 (1:1000, Millipore; Billerica, MA, USA); pKap1 S824 (1:500, Bethyl Laboratories; Montgomery, TX, USA); pATM S1981 (1:500, Rockland; Gilbertsville, PA, USA). This experiment was repeated thrice independently.
Co-immunoprecipitation of the MRN complex
Art+/+ and ArtP70/P70 MEFs were grown to confluency, harvested and then lysed in a buffer containing 25 mm HEPES, pH 7.4, 150 mm KCl, 10 mm MgCl2, 10% glycerol, 2 mm DTT and protease inhibitors (Roche; Basel, Switzerland). Protein concentrations were determined using the Bradford assay. Lysates (6 mg) were pre-cleared for 1 h with protein G beads (GE Healthcare) at 4°C, then incubated with α-Mre11 antibody (4.5 µg; Cell Signaling) and protein G beads overnight at 4°C with constant rotation. The beads were washed twice with lysis buffer followed by two washes in lysis buffer containing 300 mm KCl. The immunoprecipitates were analyzed by 8% SDS–PAGE followed by western blotting with α-Mre11 (Cell Signaling; Danvers, MA, USA), α-Nbs1 (Novus Biologicals; Littleton, CO, USA) and α-Rad50 (Bethyl Laboratories; Montgomery, TX, USA) antibodies. The protein bands were visualized using IRDye800CW-conjugated goat anti-rabbit secondary antibody (LiCor Biosciences; Lincoln, NE, USA). The co-IPs were repeated thrice independently.
Immunofluorescence analysis of Mre11 foci
Art+/+, Art−/− and ArtP70/P70 MEFs (SV40 large T-antigen immortalized) were plated at a density of 2 × 105 cells per well of a 12-well dish and then exposed to 10 Gy of γ-rays from a 137Cs source. Cells were allowed to recover for 8 h and then fixed in 4% paraformaldehyde solution (4% paraformaldehyde, 2% sucrose, pH 7.5) followed by treatment with a permeabilization solution (50 mm NaCl, 3 mm MgCl2, 200 mm sucrose, 10 mm HEPES, pH 7.9, 0.5% Triton X-100), as previously described (82). Fixed cells were incubated for 1 h in phosphate buffered saline (PBST), 0.1% Tween-20 incubated with primary antibody Mre11 (1:500, Cell Signaling; Danvers, MA, USA) for 1 h and then incubated with secondary antibody for 1 h (AlexaFlour 488, donkey anti-rabbit IgG; Invitrogen). Images were visualized using an Olympus BX-61 microscope. Cells containing greater than 20 Mre11 foci were considered foci-positive, and approximately 40–50 cells were scored for each genotype. The slides were scored blinded, and the experiment was repeated twice independently.
Characterization of tumors
All mice were regularly monitored for tumors and analyzed when moribund. Lymphoid tumors were analyzed by flow cytometry with antibodies against surface B-cell (CD43, B220, IgM) and T-cell (CD4, CD8, CD3, TCRβ, CD44, CD25) markers. Thymic and pro-B lymphomas were cultured in RPMI medium 1640 supplemented with 15% fetal calf serum, 25 U/ml IL-2 (BD Biosciences) and 25 ng/ml of IL-7 (PeproTech, Rocky Hill, NJ, USA). The proportion of pro-B and thymic lymphomas in the ArtP70/P70p53−/− cohort was calculated to be statistically significantly different from that observed for Art−/−p53−/− lymphomas (P = 0.014, two-tailed Fisher's exact test). Data for Art−/−p53−/− tumors also in a mixed 129Sv/C57Bl6 genetic background from a previous publication (54) were included in the calculation (total of 13 lymphomas: 10 pro-B and 3 thymic.)
Chromosomal analyses of tumor metaphases
SKY was performed on metaphases from cells derived from the primary tumor or early passage cultured tumor cells using an interferometer (Applied Spectral Imaging; Vista, CA, USA) and SkyView software. For FISH analyses, early passage tumor cultures were exposed to 100 ng/ml Colcemid for 5.5 h. BAC probes for FISH analysis were obtained from the RPCI-23 library (Children's Hospital Oakland Research Institute; Oakland, CA, USA) and nick-translated using biotin-11-dUTP or digoxigenin-16-dUTP by standard procedures (Roche; Basel, Switzerland). BAC probes hybridizing to TCRα/δ are as follows: RPCI-23 204N18 (centromeric to TCRα/δ region) and RPCI-23 269E2 (telomeric to TCRα/δ region). BAC probe hybridizing to TCRβ are as follows: RPCI-23 216J19 (spans TRBD1–TRBV31). BAC probe hybridizing to TCRγ are as follows: RPCI-23 212N5 (within TCRγ). BAC probes hybridizing to IgH are as follows: N-myc BAC A-10-1 (54), Bac199 (hybridizes to Cα), Bac 207 (hybridizes to V region). c-myc BAC probe was previously described (83). At least 10 metaphases for each tumor were analyzed by SKY and at least 20 metaphases by FISH.
Southern blot and RT-PCR analyses
Genomic DNA (20 µg) isolated from control tissues (tail or kidney) or ArtP70/P70 p53−/− tumor masses was digested with EcoRI. Southern blotting was performed with previously characterized probes hybridizing within the TCRβ locus (Drd1), JH region, HS3a, Cµ, N-myc and c-myc loci. Southern blots were visualized using a Phosphorimager. Band intensities were quantitated using Image Quant TL v2005 software, and relative levels were normalized to a non-lymphoid locus (LR8). Fold amplification was calculated compared with the intensities of bands in the kidney controls on the same membrane.
Reverse transcription of total RNA (1 µg) isolated from primary ArtP70/P70p53−/− (C219, C263) and Art−/−p53−/− (C405) pro-B lymphomas and wild-type LN was performed using a poly-dT (20) primer and MLV-reverse transcriptase (Invitrogen). PCR amplification of cDNAs was performed using gene-specific primers to c-myc (exons 1 and 3) and N-myc (exons 2 and 3). cDNA levels were normalized to tubulin. Bands were quantitated using AlphaImager 2200 (AlphaInnotech; Santa Clara, CA, USA). RT-PCR reactions were repeated at least four times.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by NIH grant AI063058 (NIAID), Pew Scholar's Award (Pew Charitable Trusts), Munn IDEA award (UM Cancer Center) (to J.M.S.) and by the NIH UM Cancer Center Support Grant (5 P30 CA46592). C.J. received support from the NIH Genetics Training grant T32-GM07544 (NIGMS), T.M. is a Fulbright Scholar and W.L. received support from the Cellular and Molecular Biology Training Grant (T32-GM007315).
ACKNOWLEDGEMENTS
We thank Drs David Ferguson and John Moran for helpful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Moshous D., Callebaut I., de Chasseval R., Corneo B., Cavazzana-Calvo M., Le Deist F., Tezcan I., Sanal O., Bertrand Y., Philippe N., et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. doi:10.1016/S0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 2.Le Deist F., Poinsignon C., Moshous D., Fischer A., de Villartay J.P. Artemis sheds new light on V(D)J recombination. Immunol. Rev. 2004;200:142–155. doi: 10.1111/j.0105-2896.2004.00169.x. doi:10.1111/j.0105-2896.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- 3.de Villartay J.P. V(D)J recombination deficiencies. Adv. Exp. Med. Biol. 2009;650:46–58. doi: 10.1007/978-1-4419-0296-2_4. doi:10.1007/978-1-4419-0296-2_4. [DOI] [PubMed] [Google Scholar]
- 4.Ma Y., Pannicke U., Schwarz K., Lieber M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. doi:10.1016/S0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 5.Ma Y., Schwarz K., Lieber M.R. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. doi:10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;23:23–39. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fugmann S.D. RAG1 and RAG2 in V(D)J recombination and transposition. Immunol. Res. 2001;23:23–39. doi: 10.1385/IR:23:1:23. doi:10.1385/IR:23:1:23. [DOI] [PubMed] [Google Scholar]
- 8.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. doi:10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 9.Sekiguchi J., Alt F.W., Oettinger M. Honjo T. In: Molecular Biology of B Cells. Alt F.W., editor; USA: Elsevier Science; 2004. pp. 57–78. [Google Scholar]
- 10.Rooney S., Sekiguchi J., Zhu C., Cheng H.L., Manis J., Whitlow S., DeVido J., Foy D., Chaudhuri J., Lombard D., et al. Leaky SCID phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol. Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. doi:10.1016/S1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 11.Pannicke U., Honig M., Schulze I., Rohr J., Heinz G.A., Braun S., Janz I., Rump E.M., Seidel M.G., Matthes-Martin S., et al. The most frequent DCLRE1C (ARTEMIS) mutations are based on homologous recombination events. Hum. Mutat. 2010;31:197–207. doi: 10.1002/humu.21168. doi:10.1002/humu.21168. [DOI] [PubMed] [Google Scholar]
- 12.Moshous D., Pannetier C., Chasseval Rd R., Deist Fl F., Cavazzana-Calvo M., Romana S., Macintyre E., Canioni D., Brousse N., Fischer A., et al. Partial T and B lymphocyte immunodeficiency and predisposition to lymphoma in patients with hypomorphic mutations in Artemis. J. Clin. Invest. 2003;111:381–387. doi: 10.1172/JCI16774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ege M., Ma Y., Manfras B., Kalwak K., Lu H., Lieber M.R., Schwarz K., Pannicke U. Omenn syndrome due to ARTEMIS mutations. Blood. 2005;105:4179–4186. doi: 10.1182/blood-2004-12-4861. doi:10.1182/blood-2004-12-4861. [DOI] [PubMed] [Google Scholar]
- 14.van der Burg M., Verkaik N.S., den Dekker A.T., Barendregt B.H., Pico-Knijnenburg I., Tezcan I., vanDongen J.J., van Gent D.C. Defective Artemis nuclease is characterized by coding joints with microhomology in long palindromic-nucleotide stretches. Eur. J. Immunol. 2007;37:3522–3528. doi: 10.1002/eji.200737624. doi:10.1002/eji.200737624. [DOI] [PubMed] [Google Scholar]
- 15.Musio A., Marrella V., Sobacchi C., Rucci F., Fariselli L., Giliani S., Lanzi G., Notarangelo L.D., Delia D., Colombo R., et al. Damaging-agent sensitivity of Artemis-deficient cell lines. Eur. J. Immunol. 2005;35:1250–1256. doi: 10.1002/eji.200425555. doi:10.1002/eji.200425555. [DOI] [PubMed] [Google Scholar]
- 16.de Villartay J.P., Shimazaki N., Charbonnier J.B., Fischer A., Mornon J.P., Lieber M.R., Callebaut I. A histidine in the beta-CASP domain of Artemis is critical for its full in vitro and in vivo functions. DNA Repair (Amst) 2009;8:202–208. doi: 10.1016/j.dnarep.2008.10.010. doi:10.1016/j.dnarep.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Darroudi F., Wiegant W., Meijers M., Friedl A.A., van der Burg M., Fomina J., van Dongen J.J., van Gent D.C., Zdzienicka M.Z. Role of Artemis in DSB repair and guarding chromosomal stability following exposure to ionizing radiation at different stages of cell cycle. Mutat. Res. 2007;615:111–124. doi: 10.1016/j.mrfmmm.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 18.Lagresle-Peyrou C., Benjelloun F., Hue C., Andre-Schmutz I., Bonhomme D., Forveille M., Beldjord K., Hacein-Bey-Abina S., De Villartay J.P., Charneau P., et al. Restoration of human B-cell differentiation into NOD-SCID mice engrafted with gene-corrected CD34+ cells isolated from Artemis or RAG1-deficient patients. Mol. Ther. 2008;16:396–403. doi: 10.1038/sj.mt.6300353. doi:10.1038/sj.mt.6300353. [DOI] [PubMed] [Google Scholar]
- 19.Evans P.M., Woodbine L., Riballo E., Gennery A.R., Hubank M., Jeggo P.A. Radiation-induced delayed cell death in a hypomorphic Artemis cell line. Hum. Mol. Genet. 2006;15:1303–1311. doi: 10.1093/hmg/ddl050. doi:10.1093/hmg/ddl050. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi N., Agematsu K., Sugita K., Sako M., Nonoyama S., Yachie A., Kumaki S., Tsuchiya S., Ochs H.D., Fukushima Y., et al. Novel Artemis gene mutations of radiosensitive severe combined immunodeficiency in Japanese families. Hum. Genet. 2003;112:348–352. doi: 10.1007/s00439-002-0897-x. [DOI] [PubMed] [Google Scholar]
- 21.Li L., Moshous D., Zhou Y., Wang J., Xie G., Salido E., Hu D., de Villartay J.P., Cowan M.J. A founder mutation in Artemis, an SNM1-like protein, causes SCID in Athabascan-speaking Native Americans. J. Immunol. 2002;168:6323–6329. doi: 10.4049/jimmunol.168.12.6323. [DOI] [PubMed] [Google Scholar]
- 22.Noordzij J.G., Verkaik N.S., van der Burg M., van Veelen L.R., de Bruin-Versteeg S., Wiegant W., Vossen J.M., Weemaes C.M., de Groot R., Zdzienicka M.Z., et al. Radiosensitive SCID patients with Artemis gene mutations show a complete B-cell differentiation arrest at the pre-B-cell receptor checkpoint in bone marrow. Blood. 2003;101:1446–1452. doi: 10.1182/blood-2002-01-0187. doi:10.1182/blood-2002-01-0187. [DOI] [PubMed] [Google Scholar]
- 23.van Zelm M.C., Geertsema C., Nieuwenhuis N., de Ridder D., Conley M.E., Schiff C., Tezcan I., Bernatowska E., Hartwig N.G., Sanders E.A., et al. Gross deletions involving IGHM, BTK, or Artemis: a model for genomic lesions mediated by transposable elements. Am. J. Hum. Genet. 2008;82:320–332. doi: 10.1016/j.ajhg.2007.10.011. doi:10.1016/j.ajhg.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohr J., Pannicke U., Doring M., Schmitt-Graeff A., Wiech E., Busch A., Speckmann C., Muller I., Lang P., Handgretinger R., et al. Chronic inflammatory bowel disease as key manifestation of atypical ARTEMIS deficiency. J. Clin. Immunol. 2010;30:314–320. doi: 10.1007/s10875-009-9349-x. [DOI] [PubMed] [Google Scholar]
- 25.Brandt V.L., Roth D.B. Recent insights into the formation of RAG-induced chromosomal translocations. Adv. Exp. Med. Biol. 2009;650:32–45. doi: 10.1007/978-1-4419-0296-2_3. doi:10.1007/978-1-4419-0296-2_3. [DOI] [PubMed] [Google Scholar]
- 26.Niewolik D., Pannicke U., Lu H., Ma Y., Wang L.C., Kulesza P., Zandi E., Lieber M.R., Schwarz K. DNA-PKcs dependence of Artemis endonucleolytic activity, differences between hairpins and 5′ or 3′ overhangs. J. Biol. Chem. 2006;281:33900–33909. doi: 10.1074/jbc.M606023200. doi:10.1074/jbc.M606023200. [DOI] [PubMed] [Google Scholar]
- 27.Yannone S.M., Khan I.S., Zhou R.Z., Zhou T., Valerie K., Povirk L.F. Coordinate 5′ and 3′ endonucleolytic trimming of terminally blocked blunt DNA double-strand break ends by Artemis nuclease and DNA-dependent protein kinase. Nucleic Acids Res. 2008;36:3354–3365. doi: 10.1093/nar/gkn205. doi:10.1093/nar/gkn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu J., Li S., Zhang X., Wang L.C., Niewolik D., Schwarz K., Legerski R.J., Zandi E., Lieber M.R. DNA-PKcs regulates a single-stranded DNA endonuclease activity of Artemis. DNA Repair (Amst) 2010;9:429–437. doi: 10.1016/j.dnarep.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodarzi A.A., Yu Y., Riballo E., Douglas P., Walker S.A., Ye R., Harer C., Marchetti C., Morrice N., Jeggo P.A., et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. Embo J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. doi:10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soubeyrand S., Pope L., de Chasseval R., Gosselin D., Dong F., de Villartay J.P., Hache R.J. Artemis phosphorylated by DNA-dependent protein kinase associates preferentially with discrete regions of chromatin. J. Mol. Biol. 2006;358:1200–1211. doi: 10.1016/j.jmb.2006.02.061. doi:10.1016/j.jmb.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 31.Ma Y., Pannicke U., Lu H., Niewolik D., Schwarz K., Lieber M.R. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J. Biol. Chem. 2005;280:33839–33846. doi: 10.1074/jbc.M507113200. doi:10.1074/jbc.M507113200. [DOI] [PubMed] [Google Scholar]
- 32.Huang Y., Giblin W., Kubec M., Westfield G., St Charles J., Chadde L., Kraftson S., Sekiguchi J. Impact of a hypomorphic Artemis disease allele on lymphocyte development, DNA end processing, and genome stability. J. Exp. Med. 2009;206:893–908. doi: 10.1084/jem.20082396. doi:10.1084/jem.20082396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang J., Bronson R.T., Xu Y. Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair. Embo J. 2002;21:1447–1455. doi: 10.1093/emboj/21.6.1447. doi:10.1093/emboj/21.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lista F., Bertness V., Guidos C.J., Danska J.S., Kirsch I.R. The absolute number of transrearrangements between the TCRγ and TCRβ loci is predictive of lymphoma risk: a severe combined immune deficiency (SCID) murine model. Cancer Res. 1997;57:4408–4413. [PubMed] [Google Scholar]
- 35.Lipkowitz S., Stern M.H., Kirsch I.R. Hybrid T cell receptor genes formed by interlocus recombination in normal and ataxia-telangiectasis lymphocytes. J. Exp. Med. 1990;172:409–418. doi: 10.1084/jem.172.2.409. doi:10.1084/jem.172.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stern M.H., Lipkowitz S., Aurias A., Griscelli C., Thomas G., Kirsch I.R. Inversion of chromosome 7 in ataxia telangiectasia is generated by a rearrangement between T-cell receptor beta and T-cell receptor gamma genes. Blood. 1989;74:2076–2080. [PubMed] [Google Scholar]
- 37.Kobayashi Y., Tycko B., Soreng A.L., Sklar J. Transrearrangements between antigen receptor genes in normal human lymphoid tissues and in ataxia telangiectasia. J. Immunol. 1991;147:3201–3209. [PubMed] [Google Scholar]
- 38.Kang J., Ferguson D., Song H., Bassing C., Eckersdorff M., Alt F.W., Xu Y. Functional interaction of H2AX, NBS1, and p53 in ATM-dependent DNA damage responses and tumor suppression. Mol. Cell. Biol. 2005;25:661–670. doi: 10.1128/MCB.25.2.661-670.2005. doi:10.1128/MCB.25.2.661-670.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liyanage M., Weaver Z., Barlow C., Coleman A., Pankratz D.G., Anderson S., Wynshaw-Boris A., Ried T. Abnormal rearrangement within the alpha/delta T-cell receptor locus in lymphomas from Atm-deficient mice. Blood. 2000;96:1940–1946. [PubMed] [Google Scholar]
- 40.Ward I.M., Difilippantonio S., Minn K., Mueller M.D., Molina J.R., Yu X., Frisk C.S., Ried T., Nussenzweig A., Chen J. 53BP1 cooperates with p53 and functions as a haploinsufficient tumor suppressor in mice. Mol. Cell. Biol. 2005;25:10079–10086. doi: 10.1128/MCB.25.22.10079-10086.2005. doi:10.1128/MCB.25.22.10079-10086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deriano L., Stracker T.H., Baker A., Petrini J.H., Roth D.B. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol. Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. doi:10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogue M.A., Jhappan C., Roth D.B. Analysis of variable (diversity) joining recombination in DNA-dependent protein kinase (DNA-PK)-deficient mice reveals DNA-PK-independent pathways for both signal and coding joint formation. Proc. Natl Acad. Sci. USA. 1998;95:15559–15564. doi: 10.1073/pnas.95.26.15559. doi:10.1073/pnas.95.26.15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bogue M.A., Wang C., Zhu C., Roth D.B. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7:37–47. doi: 10.1016/s1074-7613(00)80508-7. doi:10.1016/S1074-7613(00)80508-7. [DOI] [PubMed] [Google Scholar]
- 44.Carroll A.M., Bosma M.J. T-lymphocyte development in SCID mice is arrested shortly after the initiation of T-cell receptor delta gene recombination. Genes Dev. 1991;5:1357–1366. doi: 10.1101/gad.5.8.1357. doi:10.1101/gad.5.8.1357. [DOI] [PubMed] [Google Scholar]
- 45.Giblin W., Chatterji M., Westfield G., Masud T., Theisen B., Cheng H.L., DeVido J., Alt F.W., Ferguson D.O., Schatz D.G., et al. Leaky severe combined immunodeficiency and aberrant DNA rearrangements due to a hypomorphic RAG1 mutation. Blood. 2009;113:2965–2975. doi: 10.1182/blood-2008-07-165167. doi:10.1182/blood-2008-07-165167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helmink B.A., Bredemeyer A.L., Lee B.S., Huang C.Y., Sharma G.G., Walker L.M., Bednarski J.J., Lee W.L., Pandita T.K., Bassing C.H., et al. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J. Exp. Med. 2009;206:669–679. doi: 10.1084/jem.20081326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bredemeyer A.L., Sharma G.G., Huang C.Y., Helmink B.A., Walker L.M., Khor K.C., Nuskey B., Sullivan K.E., Pandita T.K., Bassing C.H., et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. doi:10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 48.Buis J., Wu Y., Deng Y., Leddon J., Westfield G., Eckersdorff M., Sekiguchi J.M., Chang S., Ferguson D.O. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. doi:10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J.H., Paull T.T. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. doi:10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 50.Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. doi:10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 51.Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D.C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. doi:10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 52.Maser R.S., Monsen K.J., Nelms B.E., Petrini J.H. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woo Y., Wright S.M., Maas S.A., Alley T.L., Caddle L.B., Kamdar S., Affourtit J., Foreman O., Akeson E.C., Shaffer D., et al. The nonhomologous end joining factor Artemis suppresses multi-tissue tumor formation and prevents loss of heterozygosity. Oncogene. 2007;26:6010–6020. doi: 10.1038/sj.onc.1210430. doi:10.1038/sj.onc.1210430. [DOI] [PubMed] [Google Scholar]
- 54.Rooney S., Sekiguchi J., Whitlow S., Eckersdorff M., Manis J.P., Lee C., Ferguson D.O., Alt F.W. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc. Natl Acad. Sci. USA. 2004;101:2410–2415. doi: 10.1073/pnas.0308757101. doi:10.1073/pnas.0308757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dujka M.E., Puebla-Osorio N., Tavana O., Sang M., Zhu C. ATM and p53 are essential in the cell-cycle containment of DNA breaks during V(D)J recombination in vivo. Oncogene. 2010;29:957–965. doi: 10.1038/onc.2009.394. doi:10.1038/onc.2009.394. [DOI] [PubMed] [Google Scholar]
- 56.Guidos C.J., Williams C.J., Grandal I., Knowles G., Huang M.T., Danska J.S. V(D)J recombination activates a p53-dependent DNA damage checkpoint in SCID lymphocyte precursors. Genes Dev. 1996;10:2038–2054. doi: 10.1101/gad.10.16.2038. doi:10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- 57.Zhu C., Mills K.D., Ferguson D.O., Lee C., Manis J., Fleming J., Gao Y., Morton C.C., Alt F.W. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. doi:10.1016/S0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 58.Negrini S., Gorgoulis V.G., Halazonetis T.D. Genomic instability—an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. doi:10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 59.Mills K.D., Ferguson D.O., Alt F.W. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. doi:10.1034/j.1600-065X.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 60.Bredemeyer A.L., Huang C.Y., Walker L.M., Bassing C.H., Sleckman B.P. Aberrant V(D)J recombination in ataxia telangiectasia mutated-deficient lymphocytes is dependent on nonhomologous DNA end joining. J. Immunol. 2008;181:2620–2625. doi: 10.4049/jimmunol.181.4.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Difilippantonio M.J., Petersen S., Chen H.T., Johnson R., Jasin M., Kanaar R., Ried T., Nussenzweig A. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J. Exp. Med. 2002;196:469–480. doi: 10.1084/jem.20020851. doi:10.1084/jem.20020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao M.J., Zhang X.X., Hill R., Gao J., Qumsiyeh M.B., Nichols W., Van Dyke T. No requirement for V(D)J recombination in p53-deficient thymic lymphoma. Mol. Cell. Biol. 1998;18:3495–3501. doi: 10.1128/mcb.18.6.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haines B.B., Ryu C.J., Chang S., Protopopov A., Luch A., Kang Y.H., Draganov D.D., Fragoso M.F., Paik S.G., Hong H.J., et al. Block of T cell development in P53-deficient mice accelerates development of lymphomas with characteristic RAG-dependent cytogenetic alterations. Cancer Cell. 2006;9:109–120. doi: 10.1016/j.ccr.2006.01.004. doi:10.1016/j.ccr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 64.Bassing C.H., Suh H., Ferguson D.O., Chua K.F., Manis J., Eckersdorff M., Gleason M., Bronson R., Lee C., Alt F.W. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. doi:10.1016/S0092-8674(03)00566-X. [DOI] [PubMed] [Google Scholar]
- 65.Celeste A., Difilippantonio S., Difilippantonio M.J., Fernandez-Capetillo O., Pilch D.R., Sedelnikova O.A., Eckhaus M., Ried T., Bonner W.M., Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. doi:10.1016/S0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vanasse G.J., Halbrook J., Thomas S., Burgess A., Hoekstra M.F., Disteche C.M., Willerford D.M. Genetic pathway to recurrent chromosome translocations in murine lymphoma involves V(D)J recombinase. J. Clin. Invest. 1999;103:1669–1675. doi: 10.1172/JCI6658. doi:10.1172/JCI6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsieh C.L., Arlett C.F., Lieber M.R. V(D)J recombination in ataxia telangiectasia, Bloom's syndrome, and a DNA ligase I-associated immunodeficiency disorder. J. Biol. Chem. 1993;268:20105–20109. [PubMed] [Google Scholar]
- 68.Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D., et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. doi:10.1016/S0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 69.Elson A., Wang Y., Daugherty C.J., Morton C.C., Zhou F., Campos-Torres J., Leder P. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl Acad. Sci. USA. 1996;93:13084–13089. doi: 10.1073/pnas.93.23.13084. doi:10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu Y., Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. doi:10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- 71.Borghesani P.R., Alt F.W., Bottaro A., Davidson L., Aksoy S., Rathbun G.A., Roberts T.M., Swat W., Segal R.A., Gu Y. Abnormal development of Purkinje cells and lymphocytes in Atm mutant mice. Proc. Natl Acad. Sci. USA. 2000;97:3336–3341. doi: 10.1073/pnas.050584897. doi:10.1073/pnas.050584897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Callen E., Jankovic M., Difilippantonio S., Daniel J.A., Chen H.T., Celeste A., Pellegrini M., McBride K., Wangsa D., Bredemeyer A.L., et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. doi:10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 73.Mahaney B.L., Meek K., Lees-Miller S.P. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 2009;417:639–650. doi: 10.1042/BJ20080413. doi:10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hammel M., Yu Y., Mahaney B.L., Cai B., Ye R., Phipps B.M., Rambo R.P., Hura G.L., Pelikan M., So S., et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. doi:10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rooney S., Chaudhuri J., Alt F.W. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. doi:10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 76.Zha S., Bassing C.H., Sanda T., Brush J.W., Patel H., Goff P.H., Murphy M.M., Tepsuporn S., Gatti R.A., Look A.T., et al. ATM-deficient thymic lymphoma is associated with aberrant tcrd rearrangement and gene amplification. J. Exp. Med. 2010;207:1369–1380. doi: 10.1084/jem.20100285. doi:10.1084/jem.20100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Osborne C.S., Chakalova L., Mitchell J.A., Horton A., Wood A.L., Bolland D.J., Corcoran A.E., Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. doi:10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang J.H., Gostissa M., Yan C.T., Goff P., Hickernell T., Hansen E., Difilippantonio S., Wesemann D.R., Zarrin A.A., Rajewsky K., et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. doi:10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nussenzweig A., Nussenzweig M.C. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. doi:10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zha S., Sekiguchi J., Brush J.W., Bassing C.H., Alt F.W. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proc. Natl Acad. Sci. USA. 2008;105:9302–9306. doi: 10.1073/pnas.0803520105. doi:10.1073/pnas.0803520105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rooney S., Alt F.W., Sekiguchi J., Manis J.P. Artemis-independent functions of DNA-dependent protein kinase in Ig heavy chain class switch recombination and development. Proc. Natl Acad. Sci. USA. 2005;102:2471–2475. doi: 10.1073/pnas.0409857102. doi:10.1073/pnas.0409857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Theunissen J.W., Petrini J.H. Methods for studying the cellular response to DNA damage: influence of the Mre11 complex on chromosome metabolism. Methods Enzymol. 2006;409:251–284. doi: 10.1016/S0076-6879(05)09015-4. doi:10.1016/S0076-6879(05)09015-4. [DOI] [PubMed] [Google Scholar]
- 83.Gao Y., Ferguson D.O., Xie W., Manis J., Sekiguchi J., Frank K.M., Chaudhuri J.J.H., DePinho R.A., Alt F.W. Interplay of p53 and DNA-repair protein XRCC4 in tumourigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. doi:10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 84.Wright S.M., Woo Y.H., Alley T.L., Shirley B.J., Akeson E.C., Snow K.J., Maas S.A., Elwell R.L., Foreman O., Mills K.D. Complex oncogenic translocations with gene amplification are initiated by specific DNA breaks in lymphocytes. Cancer Res. 2009;69:4454–4460. doi: 10.1158/0008-5472.CAN-08-4515. doi:10.1158/0008-5472.CAN-08-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.