Abstract
Microcystins (MCs) are cyclic heptapeptides that are the most abundant toxins produced by cyanobacteria in freshwater. The phytoplankton of many freshwater lakes in Eastern Africa is dominated by cyanobacteria. Less is known, however, on the occurrence of MC producers and the production of MCs. Twelve Ugandan freshwater habitats ranging from mesotrophic to hypertrophic conditions were sampled in May and June of 2004 and April of 2008 and were analyzed for their physico-chemical parameters, phytoplankton composition, and MC concentrations. Among the group of the potential MC-producing cyanobacteria, Anabaena (0 - 107 cells ml−1) and Microcystis (103 - 107 cells ml−1) occurred most frequently and dominated in eutrophic systems. A significant linear relationship (n = 31, r2 = 0.38, p < 0.001) between the Microcystis cell numbers and MC concentration (1.3-93 fg of MC cell−1) was observed. Beside [MeAsp3, Mdha7]-MC-RR two new microcystins, [Asp3]-MC-RY and [MeAsp3]-MC-RY were isolated and their constitution assigned by LC-MS2. In order to identify the MC-producing organism in the water samples (i), the conserved aminotransferase domain part of the mcyE gene that is indicative of MC production was amplified by general primers and cloned and sequenced, and (ii), genus-specific primers were used to amplify the mcyE gene of the genera Microcystis, Anabaena, and Planktothrix. Only mcyE genotypes that are indicative of Microcystis sp. were obtained via the environmental cloning approach (337 bp, 96.1%-96.7% similarity to the Microcystis aeruginosa strain PCC7806). Accordingly, only the mcyE primers, which are specific for Microcystis, revealed PCR products. We concluded that Microcystis is the major MC-producer in Ugandan freshwater.
Keywords: eutrophication, Anabaena, Microcystis, toxicity, genetic diversity, mcyE, mcyB gene, microcystin-RY
INTRODUCTION
Uganda, with a total area of 241,000 km2 astride the equator between latitude 1° 30′ S and 4° N and longitude 29° 30′ E and 35° E, has a significant amount of freshwater resources covering approximately 16% of its surface area (UNESCO, 2005). Some of these freshwater bodies are either naturally eutrophic e.g. L. George (Viner and Smith, 1973) or are becoming increasingly eutrophic due to human activities, e.g. L. Victoria (Verschuren et al., 2001). In general, cyanobacteria have often been found to increase under eutrophic conditions.
Blooms formed by cyanobacteria have recently been reported throughout many parts of L. Victoria (Alweny, 2007). These blooms are a cause of concern to local water consumers, policy makers, as well as the National Water and Sewerage Cooperation (NW&SC) that supplies treated water nationally. In their statement to the Ugandan parliament, NW&SC indicated that 11 billion Ugandan shillings (7 million US dollars) would be required in 2008 to treat green water. The fishermen reported that areas covered by cyanobacteria accumulating on the surface could not be used for fishing anymore. Small dead fish were also seen floating close to the shoreline while the larger fish caught within the area appeared weak and stressed. In East Africa, massive fish kills were observed in the Nyanza Gulf of Lake Victoria (Kenya) in 1984, coinciding with the occurrence of cyanobacterial blooms (Ochumba, 1990). The toxins produced by cyanobacteria include neurotoxins, hepatotoxins, and irritant-dermal toxins. All of these cause acute harm to humans, animals, and wildlife after exposure (Chorus et al., 2000; Carmichael, 2001).
The most widely distributed hepatotoxins in freshwater are the microcystins (MCs). They are produced by the planktonic genera Anabaena, Anabaenopsis, Microcystis, Nostoc, and Planktothrix (Sivonen and Jones, 1999). MCs are cyclic heptapeptides and share the common structure cyclo (- D-Ala(1) - X(2) - D-MeAsp(3) - Z(4) - Adda(5) - D-Glu(6) - Mdha(7)), where X and Z are variable L-amino acids (e.g. MC-LR refers to leucine and arginine in the variable positions of this peptide), D-MeAsp is D-erythro-β-iso-methyl-aspartic acid, Adda is (2S, 3S, 8S, 9S)-3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4,6-dienoic acid, and Mdha is N-methyl-dehydroalanine. Structural variation has been reported most frequently in positions 2, 4, and 7 of the molecule resulting in over 60 structural variants that are characterized from field samples or isolated strains (Diehnelt et al., 2006). The elucidation and characterization of the gene cluster that is involved in MC synthesis significantly increased our understanding of the regulation of toxin synthesis in cyanobacteria (Dittmann et al., 1997; Tillett et al., 2000). It was only then that the investigation of the genetic basis of MC production both in the laboratory and in the environment became possible.
Considering the repetitive occurrence of algal blooms reported in Ugandan lakes, understanding the distribution of toxin-producing cyanobacteria in Ugandan freshwaters is essential. This will form the basis of estimating the health risks associated with cyanobacterial occurrence. A previous study reported the isolation of four MC-producing Microcystis sp. strains from Kenya and Uganda (Haande et al., 2007). Since it is generally known that the bias introduced by strain isolation may be substantial (Wilson et al., 2005) we believe that field studies are required to provide the information that is necessary to estimate a potential health risk. The present paper reports the first results on the occurrence of MC-producing cyanobacteria in relation to the environmental conditions and the production of MC in twelve Ugandan freshwaters ranging from mesotrophic to hypertrophic conditions.
MATERIALS AND METHODS
Sampling and nutrient analyses
Depth integrated water samples were taken in May and June 2004 from twelve freshwater bodies (Site no.1-12, Table 1, Fig. 1) using a 2 liter horizontal van Dorn sampler by sampling the water column every meter. To confirm the results obtained during this period the site no. 4, 5, 7, 10, and 11 were re-sampled in April 2008. In parallel, algae were concentrated by taking vertical hauls with a phytoplankton net (30 μm mesh size). Samples were filtered using Whatman GF/C filters. For chlorophyll a analysis, filters were stored frozen (−20°C). For MC and DNA analysis filters were dried overnight (50°C) and stored frozen (−20°C). The filtrates were subsequently filtered through membrane filters (0.45 μm) for the analyses of dissolved nutrients. Soluble reactive phosphorus (SRP), nitrate (NO3-N), and ammonia (NH4-N) were determined by using the ammonium molybdate method (Wetzel and Likens, 2000), sodium-salycilate method (Müller and Wiedemann, 1955), and indophenol blue method (Krom, 1982), respectively. Soluble reactive silica (SRSi) was determined as yellow molybdate-silicic acid (Wetzel and Likens, 2000). The total phosphorus (TP) was determined by persulphate digestion from the aliquots of the samples and analyzed as SRP.
Table 1.
Environmental parameters in Ugandan freshwaters sampled during May and June 2004. SRP, soluble reactive phosphorus, SRSi, soluble reactive silica, NH4-N, ammonia nitrogen, NO3-N, nitrate nitrogen.
| Temp (°C) |
Conductivity (μS cm−1) |
pH | Secchi (cm) |
SRP (μgL−1) |
TP (μgL−1) |
NH4-N (μgL−1) |
NO3-N (μgL−1) |
SRSi (mgL−1) |
Chl a (μgL−1) |
Trophic State |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||||||||||||
| Site | Site No | Coordinates | Area (km2) |
depth (m) |
||||||||||||||||||||||
| Latitude | Longitude (E) | May | June | May | June | May | June | May | June | May | June | May | June | May | June | May | June | May | June | May | June | |||||
| Swamp (between Kiranzi and Nabingora) |
1 | 00°29.533′N | 31°09.800′ | 0.001 | 0.5 | 28 | 20 | 66 | 93 | 7 | 10 | 26 | 15 | 167 | 109 | 18 | 17 | 290 | 59 | 27.3 | 7.8 | 0 | 0 | - | ||
| Nyabikere Crater Lake | 2 | 00°29.990′N | 30°19.729′ | 0.04 | 20 | 25 | 24 | 287 | 288 | 7 | 110 | 119 | 70 | 425 | 335 | 1729 | 1983 | 40 | 24 | 28.4 | 17.7 | 8 | 13 | E | ||
| Nkuruba Crater Lake | 3 | 00°31.020′N | 30°18.178′ | 0.03 | 16 | 24 | 24 | 387 | 373 | 7 | 8 | 314 | 41 | 34 | 82 | 71 | 506 | 850 | 20 | 11 | 4.3 | 5.7 | 7 | 9 | E | |
| Lake George (Kahenge) | 4 | 00°03.026′N | 30°03.274′ | 270 | 2.4 | 28 | 25 | 343 | 345 | 10 | 10 | 10 | 20 | 14 | 41 | 138 | 191 | 18 | 52 | 205 | 71 | 34.9 | 27.2 | 128 | 78 | H |
| Lake Edward (Katwe) | 5 | 00°09.096′S | 29°53.073′ | 11.5 | 4 | 28 | 26 | 590 | 592 | 10 | 9 | 30 | 40 | 12 | 8 | 132 | 115 | 26 | 10 | 80 | 64 | 8.6 | 10.5 | 44 | 23 | H |
| Nkugute Crater Lake | 6 | 00°33.086′S | 30°10.362′ | 0.025 | 20 | 25 | 25 | 103 | 120 | 9 | 8 | 16 | 7 | 57 | 23 | 24 | 8 | 20 | 11 | 0.7 | 0.1 | 5 | 5 | M | ||
| Lake Mburo | 7 | 00°39.106′S | 30°56.537′ | 158 | 4 | 26 | 24 | 122 | 136 | 10 | 8 | 30 | 30 | 14 | 12 | 125 | 161 | 19 | 13 | 140 | 46 | 8.3 | 8.0 | 37 | 40 | H |
| Lake Nabugabo | 8 | 00°21.283′S | 31°52.789′ | 214 | 4.5 | 25 | 25 | 20 | 20 | 8 | 8 | 100 | 90 | 2 | 8 | 25 | 29 | 94 | 18 | 80 | 19 | 3.2 | 2.2 | 8 | 11 | M |
| Lake Victoria (Bunjako) | 9 | 00°04.340′S | 32°07.520′ | 60.1 | 5 | 23 | 25 | 82 | 108 | 7 | 9 | 120 | 120 | 3 | 5 | 57 | 51 | 11 | 8 | 25 | 16 | 0.6 | 0.1 | 10 | 8 | E |
| Lake Victoria (Murchison) | 10 | 00°16.911′N | 32°38.398′ | 18 | 4 | 26 | 26 | 100 | 100 | 6 | 80 | 80 | 5 | 8 | 122 | 105 | 28 | 25 | 150 | 199 | 1.0 | 0.4 | 13 | 23 | E | |
| Lake Victoria (Napoleon) | 11 | 00°24.177′N | 33°14.756′ | 66.1 | 18 | 27 | 26 | 94 | 98 | 9 | 100 | 110 | 3 | 7 | 70 | 75 | 19 | 5 | 45 | 14 | 0.3 | 0.1 | 11 | 10 | E | |
| Pond (Jinja) | 12 | 00°29.227′N | 33°12.068′ | 0.001 | 1 | 31 | 26 | 1134 | 1095 | 9 | 11 | 20 | 25 | 12 | 222 | 213 | 24 | 18 | 60 | 46 | 18.2 | 21.8 | 33 | 22 | H | |
M = mesotrophic; E = eutrophic; H = hypertrophic;
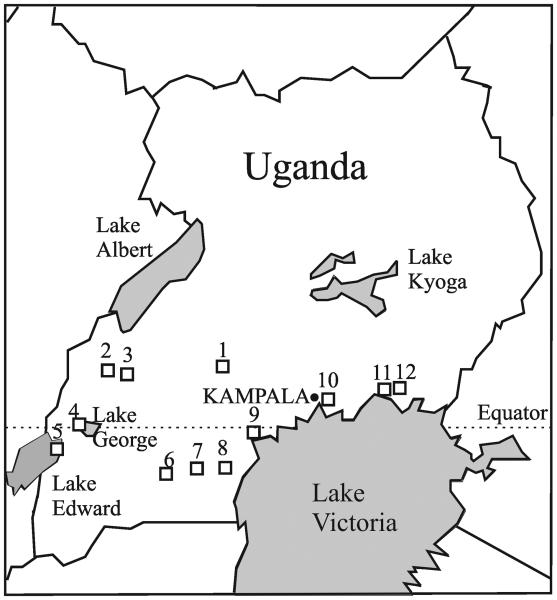
Fig. 1.
Location of the study area and sampling sites in Uganda. Numbering of the sampling sites as in Table 1.
Lake trophic states definition
The trophic state was assigned according to Vollenweider and Kerekes (1982): Mestrophic: TP (10 - 35 μg L−1), chlorophyll a (3 - 8 μg L−1), secchi disc depth (3 - 1.5 m); eutrophic: TP (35 - 100 μg L−1), chlorophyll a (8 - 25 μg L−1), secchi disc depth (1.5 - 0.7 m); hypertrophic: TP (≥ 100 μg L−1), chlorophyll a (≥ 25 μg L−1), secchi disc depth (≤ 0.7 m).
Phytoplankton composition and abundance
Chlorophyll a analysis was based on hot ethanol extraction (ISO, 1992). Algae were counted from Lugol fixed samples by using the inverted microscope, as described (Wetzel and Likens, 2000). Cyanobacteria were identified according to the morphological keys published by Talling (1987), Komárek and Kling (1991), and Komárek and Anagnostidis (1999). At least 400 specimens of the dominant phytoplankton genera were counted at 400× magnification. Most of the genera were counted as single cells (Anabaena, Chroococcus, Merismopedia, Microcystis; the pennate diatom Nitzschia and unidentified centric diatoms, green algae (including desmids) and cryptomonads). Filamentous cyanobacteria were counted as filaments (Planktolyngbia, Pseudanabaena). Aphanocapsa was counted as colonies: cells from 20 of these colonies were counted in order to determine the average number of cells per colony. Microcystis was counted as single cells and could be discriminated from other cyanobacteria due to morphological characters, such as cell size (2.5-3.6±0.2 (SE)-4.9μm) and the formation of small colonies. In order to compare the reproducibility of the Microcystis counting method between two different labs (Kampala vs. Mondsee), a number of depth integrated and plankton net samples (No 4, 5, 7, 11) were repeatedly counted in September 2004 (Kampala) and in November 2006 (Mondsee). On average the Microcystis cell numbers as determined in November 2006 accounted for 76±40% (1 SD, n=13) of the cell numbers as determined in September 2004. This implies that Microcystis cells could be reliably identified among other phytoplankton under an inverted microscope. The biovolume was calculated based on the measured dimensions of cells/filaments (Wetzel and Likens, 2000).
Microcystin analysis
The extraction of MC from filters was performed as described (Kurmayer et al., 2003). One hundred μL of the extracts were injected into high performance liquid chromatography coupled to diode array detection (HPLC-DAD) and MCs were identified by their retention time and characteristic absorption spectra according to Lawton et al., (1994) and Fastner et al., (1999). MCs were quantified at 240 nm and the concentrations of all the MC variants were determined as concentration equivalents of [D-MeAsp, D-Mdha]-MC-LR (Cyanobiotech GmbH, Berlin, Germany). HPLC peaks identified as MC were collected manually and were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) as described in Kurmayer et al., (2004) and identified by the post source decay (PSD) fragment structure analysis as described in Erhard et al., (1997) and Fastner et al., (1999). The constitution of the new MC variants was assigned by ESI-MS and ESI-MS2 experiments that were performed on a Q-TOF Ultima mass spectrometer (Waters, Milford, MA.) equipped with a nanospray source and operated in the positive ionization mode under the control of MassLynx 4.1. The sample was loaded into a PicoTip Emitter (New Objective, Woburn, MA). Single MS measurements were followed by MS2 experiments on the selected precursor ions. Fragmentation was operated by manually increasing the collision energy (4 to 60) to provide optimum fragment ion coverage over the mass range. External calibration was performed with glu-fibinopeptide-B (standard error 50 ppm) and an additional post-calibration was applied when needed. Assignment of fragments by collision-induced dissociation (CID) enhanced PSD was carried out following a published procedure (Diehnelt et al., 2006).
Genetic analysis of the field samples
For DNA extraction, 25 to 200 mL of the depth integrated samples and 25 to 60 mL of the plankton net samples were filtered using Whatman GF/C filters. The filters were dried in the oven at 50°C, overnight. DNA was extracted by the phenol-chloroform method as described (Kurmayer et al., 2003). The DNA was diluted to 2 - 20 ng μL−1 for PCR analyses.
A part of the mcyE gene region was amplified by PCR using the HEPF/HEPR primers (Jungblut and Neilan, 2006). These primers bind to the conserved regions of the aminotransferase domain and, therefore, amplify the mcyE genes of all the MC-producing cyanobacteria (472 bp). Subsequently, the genus-specific primers mcyE-F2/F8 (specific for Microcystis; 370 bp), mcyE-F2/12R (specific for Anabaena; 370 bp) and mcyE-F2/plaR3 (specific for Planktothrix; 370 bp) were used to detect the respective MC-producing species (Rantala et al., 2006). These primers bind to the adenylation domain of the mcyE gene which is responsible for the activation of the strictly conserved glutamic acid residue during MC synthesis (Tillett et al. 2000).
For three sites (no. 4, 7 and 11) sampled in 2008, the PCR products that were obtained by using the HEPF/HEPR primer pair were cloned using the pDrive Cloning Vector system (Qiagen, VWR, Austria) according to the manufacturer's instructions. For the construction of a clone library, clones were picked for each of the samples and analyzed by using restriction fragment length polymorphism (RFLP) using two restriction enzymes (BsuRI and Tru1I) following standard protocols. Using the NEBcutter program (Vincze et al., 2003), the restriction of mcyE of the genus Microcystis (472 bp, PCC7806, AF183408) resulted in three fragments (restriction type A: 176/160, 92, 18/16/8 bp) that were visualized using ethidium-bromide staining and electrophoresis in 2.0% agarose in 0.5× TBE-buffer. In contrast, the same restriction analysis for mcyE (472 bp) of the genus Anabaena (strain Anabaena 90, AY212249) resulted in two fragments only (352, 110, 8 bp (invisible), denoted as the restriction type B. While the mcyE gene is generally more conserved and useful to detect all MC-producing cyanobacteria, this gene region cannot reveal the genetic diversity within a certain genus on a subpopulation level (Rantala et al., 2004; Jungblut and Neilan, 2006). Consequently the tox4F/tox4R primers (Kurmayer et al., 2002) amplifying the first adenylation domain of the mcyB gene in Microcystis (mcyBA1, 1313 bp) were used to estimate the genetic diversity within the MC-producing Microcystis subpopulation. The tox4F/tox4R PCR products were cloned and the clones were screened by restriction using BsuRI. Following the NEBcutter program, the BsuRI digestion of mcyBA1 (1313 bp) of the Microcystis strain PCC7806 (AF183408, representing mcyB1(B), Mikalsen et al., 2003) resulted in two fragments (restriction type I: 973, 340 bp). In contrast, the recombination type mcyB1(C), (e.g. strain HUB524, Z28338) revealed one much longer and one much shorter fragment (restriction type II: 1236, 77 bp).
From the same sites twelve Microcystis strains were isolated using standard plating procedures as has been described (Okello, 2004). Six strains were found to contain mcyB (strains no: 1B5, 2D6, 18A8, 20A2, 6C5, 20A5, access no. EU014158-EU014163), and this mcyB genotype was not digested by BsuRI (restriction type III). Both mcyE and mcyB environmental sequences that were obtained from various environmental clones identified by RFLP were submitted to the DDBJ/EMBL/GenBank database (access. no. FJ429838-FJ429844).
In order to analyze all of the field samples for the occurrence of potentially MC-producing Microcystis, a PCR test that allows for the detection of lowest Microcystis cell densities was required. In order to check for the quality of the extracted DNA and the presence of potential PCR inhibitors a gene region part of a housekeeping gene was amplified in parallel to the PCR amplifying the mcyB gene. Primers specified to amplify the intergenic spacer region of the phycocyanin operon (PC-IGS) of Microcystis sp. were designed by aligning the sequences of PC-IGS (576 bp) from the twelve freshly isolated Microcystis strains (strains no: R14, 1B5, 2D6, 18A8, R50, 2A3, 20A2, 21A1, 17B7, 6C5, 19G6, 20A5, access no. EU014146-EU014157). In addition, 128 Microcystis sequences of PC-IGS from the DDBJ/EMBL/GenBank (10 June 2007) were included in a multiple sequence alignment (ClustalW2) to design the primer pair: MaPCnewfwd, 5′-GGAGCTTCCGTAGCTGC-3′ (57.6°C), and MaPCnewrev, 5′-TGCAATAAGTTTCCTACGGT-3′ (53.5°C), yielding a total amplification fragment of 192 bp. Pilot experiments showed that these primers were specific to PC-IGS of Microcystis. The six freshly isolated Microcystis strains that were found to contain mcyB (1312 bp) were aligned with eleven sequences from entries in the DDBJ/EMBL/GenBank (10 June 2007) representing the two recombination types that were reported within mcyB of Microcystis sp. (mcyB1 (B), mcyB1(C), Mikalsen et al., 2003). The following primers were designed: MamcyBnewfwd, 5′-GGATATCCTCTCAGATTCGG-3′ (57.3°C), and MamcyBnewrev, 5′-CTGATGTATAAATAACATAGGCTAAA-3′ (55.3°C) yielding a total amplification fragment of 188 bp for both recombination types (Mikalsen et al., 2003). For both PC-IGS and the mcyB primers, the lower limit of detection was equivalent to two cells of Microcystis strain 18A8.
In general, PCR amplifications from field samples were carried out in 20 μL volumes containing 2 μL PCR buffer (Qiagen, Austria), 1.2 μL MgCl2 (25 mM), 0.6 μL deoxynucleotide triphosphates (10 μM each, MBI Fermentas, Germany), 1 μL of each primer (10 pmol), 0.1 μL Taq DNA polymerase (Qiagen), 13.6 μL sterile Millipore water and 0.5 μL of 2 - 20 ng μL−1 DNA. The PCR cycling conditions for the two separate regions of the mcyE gene were performed as described (Jungblut and Neilan, 2006; Rantala et al., 2006). For the PCR amplifying Microcystis specific PC-IGS and mcyB, the thermal cycling protocol included an initial denaturation step at 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min. PCR products (4 μL of the reaction mix) were visualized by ethidium-bromide staining and electrophoresis in 2.0% agarose in 0.5× TBE-buffer.
RESULTS
Trophic characterization of the sampling sites
In the swamp sample (no. 1) the turbidity was high (secchi depth < 10 cm; Table 1) and no chlorophyll a was detected. The three Crater lakes (no. 2, 3, 6) and L. Nabugabo (no. 8) showed low concentrations of chlorophyll a (5-13 μg L−1) resulting in high transparency (> 1 m) suggesting a mesotrophic status. In two of the Crater Lakes (no. 2, 3), however, high TP concentrations were recorded, co-occurring with high SRP and NH4-N concentrations. Therefore, they were classified as eutrophic lakes. All three sites (no. 9, 10, and 11) from L. Victoria were classified as eutrophic. The highest chlorophyll a values were recorded in L. George (no. 4), L. Edward (no. 5), and L. Mburo (no. 7) and in Jinja pond (no. 12), which indicates hypertrophic conditions. Correspondingly, the secchi depths were the lowest (< 0.5 m) and the pH was the highest under hypertrophic conditions.
Phytoplankton species composition and abundance
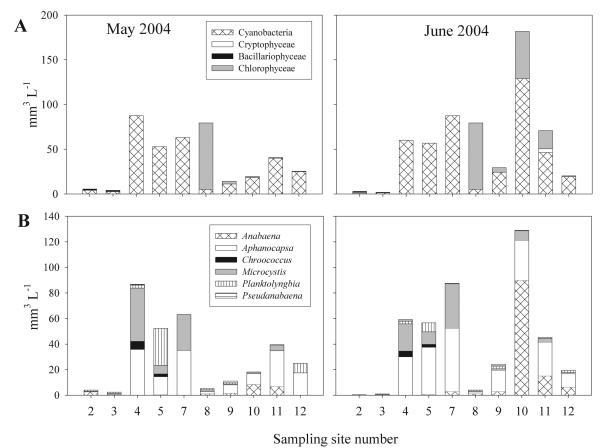
The total biovolume estimated via the microscopical counting correlated significantly with the chlorophyll a concentration (R2 = 0.76, n = 37). Five major phytoplankton classes were observed: Cyanobacteria, green algae and desmids, diatoms, and cryptomonads (Fig 2A). Cyanobacteria occurred in all of the water bodies. The other phytoplankton classes were generally of minor importance. The Crater Lakes (no. 2, 3) and mesotrophic L. Nabugabo (no. 8) had the lowest biovolume of cyanobacteria and mostly contained diatoms, i.e. centric diatoms and Nitzschia (no. 2, 3) or green algae/desmids, i.e. Chlorella, Cosmarium, and Staurastrum (no. 8). Cyanobacterial dominance was only observed under eutrophic and hypertrophic conditions (no. 4, 5, 7, 9, 10, 11, and 12), including the genera Anabaena, Aphanocapsa, Chroococcus, Merismopedia, Microcystis, Planktolyngbia, and Pseudanabaena (Fig. 2B). Anabaena was present in the Crater lakes (no. 2, 3) and in the deep eutrophic L. Victoria (no. 9-11), while Aphanocapsa occurred in both the hypertrophic shallow lakes (no. 4, 5, 7) and the deep eutrophic L. Victoria (no. 9-11). Microcystis was most abundant in the hypertrophic shallow lakes (no. 4, 5, 7). In summary, the dominance of cyanobacteria was positively related with the trophic situation.
Fig. 2.
(A) Phytoplankton composition and (B) cyanobacterial composition in various Ugandan freshwater habitats during May and June 2004. Numbering of the sampling sites as in Table 1. No phytoplankton was detected at sites (1) and (6). Only taxa contributing >5% of the total phytoplankton biovolume (upper panel) or cyanobacterial biovolume (lower panel) are shown.
Microcystin analyses of the field samples
MCs were detected in plankton net samples only, i.e. in five plankton net samples in 2004 (no. 4, May; no. 5, May and June; no. 7, May and June) and in all five plankton net samples in 2008. MC 1 [molar weight 1038 (M+H+)] occurred in all samples containing MC and eluted at 15.6 min and was identified by post source decay analysis of the fractionated peak as [D-MeAsp3, Mdha7]-MC-RR. In addition, the MCs extracted from the plankton net samples (L. Edward, L. George, L. Mburo, June 2004, 2008) eluted at 21.7 min [MC 2, molar weight 1031 (M+H+)] and 23.2 min [MC 3, molar weight 1045 (M+H+)]. MC 2 and 3 were also identified in aqueous methanolic extracts of the Microcystis strains 2D6, 6C5, 18A8 isolated from L. Edward and L. Mburo. The original UV spectrum as well as the first order derivative of the peak apex showed a close match with [MeAsp3, Mdha7]-MC-YR, which is available from the spectrum library. However, MC 2 and MC 3 differed in the retention time from [MeAsp3, Mdha7]-MC-YR. The sequence and constitution of MC 2 and MC 3 were investigated by LC-MS2. CID enhanced PSD produced 34 and 33 fragments for MC 2 and MC 3, respectively, detected with a mass accuracy of < 50 ppm (Tables 2 and 3). The amino acid at position 7 was assigned as either Mdha or Dhb. The distinction between these isomers is difficult by MS, as D labelling experiments on MC 2 and MC 3 were inconclusive. The assignment of MC 2 as [Asp3]-MC-RY was based on fragments such as [Ala-Arg-Asp+H]+ (343.1826), [Ala-Arg+H]+ (228.1474), [Mdha or Dhb-Ala-Arg+H]+ (311.1879) and [Tyr-Adda-Glu+H]+] (606.3105), [Tyr-Adda +H]+] (477.2565) and many others (Table 2). The assignment of MC 3 as [MeAsp3]-MC-RY was based on fragments such as Ala-Arg-MeAsp+H]+ (357.2023), [Ala-Arg-NH3+H]+ (211.1247), [Mdha or Dhb-Ala-Arg+H]+ (311.1938) and [MeAsp-Tyr+H]+] (293.1255), [Tyr-Adda +H]+] (477.2628), [Arg-MeAsp-Tyr+H]+ (449.2364) and many others (table 3). These two MC variants have not yet been described in the literature, to the best of our knowledge, and thus constitute new microcystins.
Table 2.
Composition of ions in the MS2 spectrum of the [M+H]+ ion of [Asp3]-MC-RY
| Composition and sequence | Calculated m/z | Measured m/z | Error [ppm] |
|---|---|---|---|
| M (+ H+) | 1031.5197 | 1031.5197 | 0.0 |
| M (− CO) (+ H+) | 1003.5248 | 1003.5244 | −0.4 |
| Glu-(Mdha or Dhb)-Ala-Arg-Asp (+ H+) | 555.2522 | 555.2643 | 21.8 |
| Glu-(Mdha or Dhb)-Ala-Arg (+ H+) | 440.2252 | 440.2386 | 30.4 |
| Glu-(Mdha or Dhb)-Ala-Arg (− COOH) (+ H+) | 395.2276 | 395.2124 | −38.5 |
| Glu-(Mdha or Dhb) (+ H+) | 213.0870 | 213.0879 | 4.2 |
| (Mdha or Dhb)-Ala-Arg-Asp (+ H+) | 426.2096 | 426.2177 | 19.0 |
| (Mdha or Dhb)-Ala-Arg-Asp (− NH3) (+ H+) | 409.1831 | 409.1936 | 25.7 |
| (Mdha or Dhb)-Ala-Arg (+ H+) | 311.1826 | 311.1879 | 17.0 |
| (Mdha or Dhb)-Ala-Arg (− CO) (+ H+) | 283.1877 | 283.1771 | −37.4 |
| (Mdha or Dhb)-Ala-Arg (− NH3) (+ H+) | 294.1561 | 294.1605 | 15.0 |
| (Mdha or Dhb)-Ala (+ H+) | 155.0815 | 155.0824 | 5.8 |
| (Mdha or Dhb)-Ala (− CO) (+ H+) | 127.0866 | 127.0899 | 26.0 |
| Ala-Arg-Asp-Tyr-Adda (+ H+) | 819.4400 | 819.4226 | −21.2 |
| Ala-Arg-Asp (+ H+) | 343.1724 | 343.1826 | 29.7 |
| Ala-Arg (+ H+) | 228.1455 | 228.1474 | 8.3 |
| Arg-Asp-Tyr (+ H+) | 435.1987 | 435.2142 | 35.6 |
| Arg-Asp-Tyr (− COOH) (+ H+) | 390.2010 | 390.1989 | −5.4 |
| Arg-Asp (+ H+) | 272.1353 | 272.1411 | 21.3 |
| Arg (+ H+) | 157.1084 | 157.1098 | 8.9 |
| Asp-Tyr (+ H+) | 279.0975 | 279.1034 | 21.1 |
| Asp-Tyr (− CO) (+ H+) | 251.1026 | 251.1096 | 27.9 |
| Tyr-Adda-Glu-(Mdha or Dhb)-Ala-Arg (+ H+) | 916.4927 | 916.4935 | 0.9 |
| Tyr-Adda-Glu-(Mdha or Dhb)-Ala-Arg (− CO) (+ H+) | 888.4978 | 888.5056 | 8.8 |
| Tyr-Adda-Glu-(Mdha or Dhb) (+ H+) | 689.3545 | 689.3567 | 3.2 |
| Tyr-Adda-Glu (+ H+) | 606.3174 | 606.3105 | −11.4 |
| Tyr-Adda (+ H+) | 477.2748 | 477.2565 | −38.3 |
| Adda-Glu-(Mdha or Dhb)-Ala-Arg-Asp (− NH3) (+ H+) | 851.4298 | 851.4353 | 6.5 |
| Adda-Glu-(Mdha or Dhb)-Ala-Arg-Asp (− NH3) (− 134 Adda) (+ H+) | 717.3565 | 717.3604 | 5.4 |
| Adda-Glu-(Mdha or Dhb) (− NH3) (+ H+) | 509.2647 | 509.2621 | −5.1 |
| Adda-Glu-(Mdha or Dhb) (− NH3) (− 134 Adda) (+ H+) | 375.1915 | 375.1996 | 21.6 |
| Adda-Glu-(Mdha or Dhb) (− CO) (− NH3) (− 134 Adda) (+ H+) | 347.1966 | 347.2035 | 19.9 |
| Adda (− NH3) (− 134 Adda) (+ H+) | 163.1118 | 163.1127 | 5.5 |
| 134Adda (+ H+) | 135.0804 | 135.0824 | 14.8 |
Table 3.
Composition of ions in the MS2 spectrum of the [M+H]+ ion of [MeAsp3]-MC-RY
| Composition and sequence | Calculated m/z | Measured m/z | Error [ppm] |
|---|---|---|---|
| M (+ H+) | 1045.5353 | 1045.5353 | 0.0 |
| M (− CO) (+ H+) | 1017.5404 | 1017.5602 | 19.5 |
| Glu-(Mdha or Dhb)-Ala-Arg-MeAsp (+ H+) | 569.2678 | 569.2905 | 39.9 |
| Glu-(Mdha or Dhb)-Ala-Arg (− NH3) (+ H+) or (Mdha or Dhb)-Ala-Arg-MeAsp (− NH3) (+ H+) |
423.1987 | 423.2173 | 44.0 |
| Glu-(Mdha or Dhb)-Ala-Arg (− COOH) (+ H+) | 395.2276 | 395.2204 | −18.2 |
| Glu-Mdha + H | 213.0870 | 213.0916 | 21.6 |
| (Mdha or Dhb)-Ala-Arg-MeAsp-Tyr-Adda (+ H+) or Tyr-Adda-Glu-(Mdha or Dhb)-Ala-Arg (+ H+) |
916.4927 | 916.5134 | 22.6 |
| (Mdha or Dhb)-Ala-Arg-MeAsp-Tyr (− NH3) (+ H+) | 586.2620 | 586.2884 | 45.0 |
| (Mdha or Dhb)-Ala-Arg-MeAsp (+ H+) or Glu-(Mdha or Dhb)-Ala-Arg (+ H+) |
440.2252 | 440.2413 | 36.6 |
| (Mdha or Dhb)-Ala-Arg-MeAsp (− CO) (+ H+) or Glu-(Mdha or Dhb)-Ala-Arg (− CO) (+ H+) |
412.2303 | 412.2466 | 39.5 |
| (Mdha or Dhb)-Ala-Arg (+ H+) | 311.1826 | 311.1938 | 36.0 |
| (Mdha or Dhb)-Ala-Arg (− NH3) (+ H+) | 294.1561 | 294.1656 | 32.3 |
| (Mdha or Dhb)-Ala-Arg (− CO) (+ H+) | 283.1877 | 283.1843 | −12.0 |
| (Mdha or Dhb)-Ala (+ H+) | 155.0815 | 155.0850 | 22.6 |
| (Mdha or Dhb)-Ala (− CO) (+ H+) | 127.0866 | 127.0922 | 44.1 |
| Ala-Arg-MeAsp-Tyr-Adda (+ H+) | 833.4556 | 833.4562 | 0.7 |
| Ala-Arg-MeAsp (+ H+) | 357.1880 | 357.2023 | 40.0 |
| Ala-Arg (− NH3) (+ H+) | 211.1190 | 211.1247 | 27.0 |
| Arg-MeAsp-Tyr (+ H+) | 449.2143 | 449.2364 | 49.2 |
| Arg-MeAsp-Tyr (− COOH) (+ H+) | 404.2167 | 404.2209 | 10.4 |
| Arg-MeAsp (+ H+) | 286.1509 | 286.1632 | 43.0 |
| Arg (+ H+) | 157.1084 | 157.1132 | 30.6 |
| MeAsp-Tyr (+ H+) | 293.1131 | 293.1255 | 42.3 |
| Tyr-Adda-Glu-(Mdha or Dhb)-Ala-Arg (+ H+) | 916.4927 | 916.5134 | 22.6 |
| Tyr-Adda-Glu-(Mdha or Dhb)-Ala-Arg (− CO) (+ H+) | 888.4978 | 888.5134 | 17.6 |
| Tyr-Adda (+ H+) | 477.2748 | 477.2628 | −25.1 |
| Adda-Glu-(Mdha or Dhb)-Ala-Arg-MeAsp (− NH3) (+ H+) | 865.4454 | 865.4628 | 20.1 |
| Adda-Glu-(Mdha or Dhb)-Ala-Arg-MeAsp (− NH3) (− 134 Adda) (+ H+) | 731.3722 | 731.3859 | 18.7 |
| Adda-Glu-(Mdha or Dhb) (− NH3) (+ H+) | 509.2647 | 509.2813 | 32.6 |
| Adda-Glu-(Mdha or Dhb) (− NH3) (− 134 Adda) (+ H+) | 375.1915 | 375.2071 | 41.6 |
| Adda-Glu-(Mdha or Dhb) (− CO) (− NH3) (− 134 Adda) (+ H+) | 347.1966 | 347.2095 | 37.2 |
| Adda (− NH3) (− 134 Adda) (+ H+) | 163.1118 | 163.1154 | 22.1 |
| 134Adda (+ H+) | 135.0804 | 135.0852 | 35.5 |
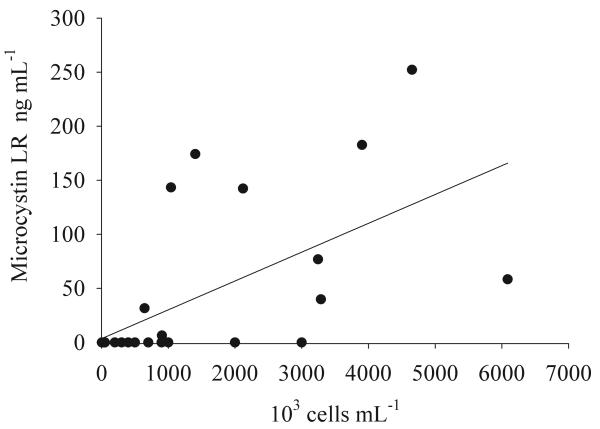
There was a linear relationship between the Microcystis cell concentrations and the total MC concentrations: y = 0.03x + 3.54 (R2 = 0.38, n = 31, p < 0.001, Fig. 3), where y is the MC concentration in ng ml−1 and x is the cell concentration ml−1. In contrast, the MC concentrations were found to vary independently from the Anabaena cell concentrations.
Fig. 3.
Relationship between the MC concentrations (in ng ml−1 of MC-LR equivalents) and the concentration of cells ml−1 of Microcystis during May and June 2004 and April 2008. For details on the regression curve, see the text.
Identification of microcystin producers and genetic diversity
With the exception of a plankton net sample from L. Mburo (June 2004), all of the samples that contained detectable MCs revealed PCR products for mcyE by using HEPF/HEPR primers. In order to identify the MC-producing genus, further PCR analyses were performed using the Microcystis (mcyE-F2/F8) and Anabaena (mcyE-F2/12R) and Planktothrix (mcyE-F2/plaR3) specific primers. In all of the cases, the mcyE products that are indicative of Microcystis were obtained from the plankton net samples (Table 4), while no PCR products that are indicative of mcyE of Anabaena were obtained.
Table 4.
Field samples of twelve Ugandan freshwater sites analyzed for cyanobacterial biovolume, Microcystis cell numbers, by PCR for the presence (+) or absence (−) of microcystin (MC) producers (HEP), mcyE, PC-IGS, mcyB of Microcystis and for MCs by HPLC. Only the plankton net samples from the site nos. 4, 5, 7, 10, and 11 were analyzed in April 2008.
| Sample ID | Site no. |
Filtered volume (mL) |
Cyanobacterial Biovolume (mm3 L−1) |
Microcystis (cells mL−1) |
HEP-PCR product |
Microcystis mcyE-F2/F8* PCR product |
PC-IGS PCR product |
McyB PCR product |
fg MC-LR equivalent cell−1 |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M4 | J4 | A8 | M4 | J4 | A8 | M4 | J4 | A8 | M4 | J4 | A8 | M4 | J4 | A8 | M4 | J4 | A8 | M4 | J4 | A8 | M4 | J4 | A8 | ||
|
| |||||||||||||||||||||||||
| Depth Integrated samples | |||||||||||||||||||||||||
| Kiranzi swamp | 1 | 250 | 250 | − | − | − | − | − | − | − | − | − | − | − | − | ||||||||||
| Nyabikere Crater Lake | 2 | 400 | 700 | 4 | 0.4 | − | − | − | − | − | − | − | − | − | − | − | − | ||||||||
| Nkuruba Crater Lake | 3 | 500 | 500 | 3 | 1 | 5×103 | 5×103 | − | − | − | − | + | + | − | − | − | − | ||||||||
| Lake George | 4 | 25 | 50 | 88 | 60 | 4×105 | 7×105 | + | + | + | + | + | + | + | + | − | − | ||||||||
| Lake Edward (Katwe) | 5 | 150 | 150 | 53 | 57 | 2×105 | 2×105 | + | + | + | + | + | + | + | + | − | − | ||||||||
| Nkugute Crater Lake | 6 | 700 | 500 | ||||||||||||||||||||||
| Lake Mburo | 7 | 100 | 200 | 63 | 88 | 9×105 | 9×105 | + | + | + | + | + | + | + | + | − | − | ||||||||
| Lake Nabugabo | 8 | 250 | 250 | 5 | 5 | 5×104 | 2×104 | − | − | − | − | + | + | + | + | − | − | ||||||||
| Lake Victoria (Bunjako Bay) | 9 | 800 | 500 | 11 | 24 | 2×104 | 4×104 | − | − | − | − | + | + | + | + | − | − | ||||||||
| Lake Victoria (Murchison Bay) | 10 | 500 | 500 | 18 | 129 | 4×104 | 2×105 | + | + | + | + | + | + | + | + | − | − | ||||||||
| Lake Victoria (Napoleon Gulf) | 11 | 500 | 500 | 40 | 46 | 3×104 | 3×104 | + | + | + | + | + | + | + | + | − | − | ||||||||
| Jinja Pond | 12 | 200 | 19 | 3×104 | − | − | − | − | − | ||||||||||||||||
| Plankton Net samples | |||||||||||||||||||||||||
| Kiranzi swamp | 1 | 40 | 30 | Not applicable | − | − | − | − | − | − | − | − | − | − | − | − | |||||||||
| Nyabikere Crater Lake | 2 | 50 | 50 | ″ | − | − | − | − | − | − | − | − | − | − | − | − | |||||||||
| Nkuruba Crater Lake | 3 | 50 | 50 | ″ | − | − | − | − | − | − | − | − | − | − | |||||||||||
| Lake George | 4 | 25 | 50 | 60 | ″ | 3×106 | 3×106 | 6×106 | + | − | + | + | − | + | + | + | + | + | + | + | 8.1 | − | 9.6 | ||
| Lake Edward (Katwe) | 5 | 35 | 40 | 60 | ″ | 1×106 | 4×106 | 2×106 | + | + | + | + | + | + | + | + | + | + | + | + | 92.9 | 31.2 | 67.1 | ||
| Lake Mburo | 7 | 35 | 50 | 60 | ″ | 3×106 | 3×106 | 4×106 | + | − | + | + | − | + | + | + | + | + | + | + | 5.2 | 15.8 | 52.4 | ||
| Lake Nabugabo | 8 | 40 | 40 | ″ | 3×105 | 5×105 | − | − | − | − | + | + | + | + | − | − | |||||||||
| Lake Victoria (Bunjako Bay) | 9 | 50 | 50 | ″ | 2×105 | 2×105 | − | − | − | − | + | + | + | + | − | − | |||||||||
| Lake Victoria (Murchison Bay) | 10 | 50 | 25 | 60 | ″ | 4×105 | 4×106 | 1×107 | + | + | + | + | + | + | + | + | + | + | + | + | − | − | 1.3 | ||
| Lake Victoria (Napoleon Gulf) | 11 | 50 | 50 | 60 | ″ | 1×106 | 7×105 | 6×105 | + | + | + | + | + | + | + | + | + | + | + | + | − | − | 49.1 | ||
| Jinja Pond | 12 | 50 | ″ | ||||||||||||||||||||||
M4 = May 2004; J4 = June 2004; A8 = April 2008;
using mcyE-F2/12R for Anabaena and plaR3 for Planktothrix no PCR products were obtained
Three samples (lakes George, Mburo and Victoria-Napoleon, no. 4, 7, 11) revealed abundant Microcystis but also showed a frequent occurrence of Anabaena (Fig. 2B). Using the HEPF/HEPR primers, the mcyE gene was amplified and the clones were analyzed by RFLP. In the vast majority of the clones (n=165), only restriction type A, which is indicative of Microcystis, was found (Table 5). No restriction type B, which is indicative of mcyE of Anabaena, was obtained. Only in L. Mburo three clones were not digested (restriction type C). Those sequences could not be assigned and consequently restriction type C was considered an unspecific PCR product. Eleven clones of restriction type A were sequenced (337 bp) and revealed a 96.1-96.7% similarity to the M. aeruginosa strain PCC7806, AF183408.
Table 5.
Number of restriction types as observed in clone libraries obtained from HEP-PCR products (mcyE, restriction types A-C) and tox4F/4R PCR products (mcyB, restriction types I-III). For each restriction type the respective genus (A, B) or Microcystis strain (I, II, III) is indicated in parentheses.
| HEPF/HEPR-PCR (mcyE, 470 bp) | Tox4f/4r-PCR (mcyB, 1330 bp) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Sites/Restriction type |
Site No |
Date | No of clones |
A (Microcystis) |
B (Anabaena) |
C undigested |
Not assigned |
No of clones |
I (PCC7806) | II (HUB524) | III (Strain 18A8)2 |
Not assigned |
| L. George | 4 | 26 April 2008 |
20 | 20 | 0 | 0 | 0 | 20 | 0 | 0 | 20 | 0 |
| L. Mburo | 7 | 27 April 2008 |
75 | 71 | 0 | 3 | 11 | 24 | 8 | 0 | 15 | 1 |
| L. Victoria (Napoleon) |
11 | 26 April 2008 |
70 | 70 | 0 | 0 | 0 | 35 | 11 | 0 | 20 | 4 |
Sequencing revealed a 382 bp (FJ429839) with a 96% similarity to Microcystis strain PCC7806
In addition, twelve Microcystis strains were isolated from the same sites (Okello, 2004). Six strains were found to contain mcyB (strain no.: 1B5, 2D6, 18A8, 20A2, 6C5, 20A5, access no. EU014158-EU014163).
In order to characterize the genetic diversity within MC-producing Microcystis, the first adenylation domain of the mcyB gene was amplified by PCR, cloned, and then the clones were screened by RFLP. Restriction type I, which is indicative of mcyB1(B) and restriction type III (undigested), occurred, while restriction type II, which is indicative of mcyB1(C), was not detected. Three environmental clones that were assigned to restriction type I had the highest sequence similarity to the strain PCC7806 (1190 bp, 97.2-98.1% similarity). In contrast, clones that were assigned to restriction type III had the highest sequence similarity to the strains 18A8, 20A2, 20A5, and 2D6 (1190 bp, 97.4-98.4%). Restriction type I differed by 24.7-25.7% of the nucleotides (1190 bp) from restriction type III.
Distribution of microcystin-producing Microcystis
Using the PC-IGS primers that were designed in the present study, the distribution of Microcystis was studied in all of the field samples that have been analyzed for phytoplankton composition under the microscope (Table 4). PCR products were obtained from all of the samples containing Microcystis as inferred from the microscope. Vice versa in the swamp (no. 1) and in Nyabikere Crater Lake (no. 2), no PC-IGS of Microcystis was detected, which was in accordance with the microscopical analysis. Except for the depth integrated sample from the Nkuruba Crater Lake, all of the samples containing PC-IGS, indicative of Microcystis, were found to contain mcyB.
DISCUSSION
Environmental parameters and phytoplankton composition
We recorded high concentrations of nutrients, but low concentrations of chlorophyll a and phytoplankton in the two Crater lakes (no. 2 and 3). Correspondingly, Kizito et al., 1993 reported low chlorophyll a, but high nutrient concentrations for Lake Nkuruba (no. 3). The highest NH4-N concentration that was observed in the Crater lakes may indicate significant vertical stratification, since the nitrification of ammonia to nitrate requires oxygen. During a two-year study, Chapman et al., 1998 reported that the average anoxia (0 mg L−1 O2) in Lake Nkuruba (no. 3) was down to nine meters. In this study, it is possible that a part of the anoxygenic water column with the highest ammonium concentrations was sampled resulting in a higher average ammonium concentration due to the vertical integration down to 15 m. Despite its shallowness, L. Nabugabo (no. 8) also had the lowest phytoplankton density, which was dominated by green algae and desmids. The soils of the catchment area of Lake Nabugabo have a very low salt content, possibly resulting in low calcium carbonate levels and the lowest conductivity (Beadle, 1981). A belt of mosses (Sphagnum, Miscanthidium violaceum) that is surrounding the lake has been suggested to indicate more acidic conditions (Kateyo, 2006). The general low ionic content in combination with a low nutrient concentration might be responsible for the dominance of green algae and desmids.
The cyanobacterial dominance in L. Victoria (no. 9-11) as well as in Jinja Pond was linked to a high concentration of nutrients. Kling et al., 2001 reported that the eutrophic condition in L. Victoria supported a high algal biomass that has risen by a factor of 4 to 5 since the 1960s. Hecky (1993), Lipiatou et al., (1996) and Verschuren et al., (2001) reported a shift in dominance from diatoms and green algae to cyanobacteria. Mugidde et al., (2003) suggested that because of their ability to fix nitrogen cyanobacteria may particularly increase in L. Victoria in response to phosphorus loading and increasing nitrogen limitation.
In all the hypertrophic shallow lakes (no. 4, 5, and 7) Microcystis was abundant. These three lakes situated within the national park had a high pH ranging from 8 to 10. Cyanobacteria are better competitors due to their efficient carbon-concentrating mechanism at a higher pH (Shapiro 1984). In addition these shallow waters are polymictic while at calm conditions a high insolation will penetrate the whole water column. In general these physical conditions favour the genus Microcystis sp. at the expense of other genera and very often Microcystis sp. is able to dominate phytoplankton for long times (Reynolds et al., 2002). The dominance of Microcystis sp. probably remained unaltered in Lake George for decades (Ganf, 1974).
Microcystin analyses
In the present study, MCs were found in plankton net samples that were obtained from five sites (no. 4, 5, 7, 10, and 11). In contrast no MCs were found in aqueous-methanolic extracts obtained from depth-integrated samples. In general, the high silt content that was clogging the filters during the filtration process resulted in low filtration volumes (50-200 ml) increasing the lower limit of detection for MC by HPLC significantly above 1 μg L−1. It is expected that microcystin concentrations would have been in the measurable range, if more phytoplankton biomass would have been extracted. Indeed in the phytoplankton net samples with higher Microcystis cell numbers ranging from 2×105 - 1×107 cells mL−1, MCs were recorded more frequently. Nevertheless, in plankton net samples, the cellular MC content was found to be within the range that was also observed in other field studies previously (1.3 - 93 fg MC-LR equivalent cell−1, Kurmayer et al. 2003). It is, therefore, concluded that the net samples were useful for analyzing the mean cellular MC content. These results are in agreement with other studies that document, by means of plankton net samples, that whole water conditions can be adequately represented (Rogalus and Watzin, 2008).
Genetic diversity and microcystin net production
In most of the samples, the genera Anabaena, Aphanocapsa, Chroococcus, Merismopedia, Microcystis, Planktolyngbia, and Pseudanabaena were identified under the microscope. Among those, Anabaena, Chroococcus, Microcystis, and Pseudanabaena have been reported to produce MCs (Jungblut and Neilan, 2006; Sivonen and Börner, 2008). We used the conserved HEPF/HEPR primer pair to amplify all the potential MC-producing cyanobacteria. Surprisingly, only PCR products that are indicative of mcyE of Microcystis were obtained. Correspondingly, the independent application of the genus specific mcyE primers revealed the occurrence of MC-producing Microcystis only. Consequently, Anabaena and other taxa, although rather abundant, did not have the potential to produce MC. In other studies, the populations of Microcystis from all five continents typically have been found to contain mcy genotypes as well as MCs (Sivonen and Börner, 2008). In contrast, other taxa show a more irregular pattern of MC production, for example MC-producing Anabaena have been reported from Europe, Canada, and North Africa, but not from Australia (Sivonen and Jones, 1999; Sivonen and Börner, 2008).
Using the more sensitive primers, all of the Microcystis populations were found to be able to produce MCs. This result is in agreement with other studies showing that Microcystis populations always contain the mcy genotype. For example, in a European survey including nine water bodies in seven countries, mcyA and mcyB genes as a part of the Microcystis population were always detected (Via-Ordorika et al., 2004). Within mcyBA1, the highest genetic diversity was found (24.7-25.7%), which has also been reported for populations sampled in Europe, e.g. Lake Wannsee (Kurmayer et al., 2002) and in Scandinavia (Mikalsen et al., 2003). It is generally accepted that this highest genetic diversity resulted from frequent recombination processes involving the exchange of shorter fragments (< 1000 bp) of DNA. Besides for the mcyBA1 restriction type I showing the highest similarity to the Microcystis strain PCC7806, a new mcyBA1 restriction type III that is indicative of a mcyBA1 genotype, which was so far unknown from the strains of the northern temperate hemisphere (Mikalsen et al., 2003), was found. In contrast, restriction II mcyB (C) was not detected. Notably, from the same mcyBA1 restriction type two new microcystins, [Asp3]-MC-RY and [MeAsp3]-MC-RY, were isolated. The genetic variation that is causing the biosynthesis of MC-RY remains to be elucidated.
Although the linear relationship between the Microcystis cell numbers and the total MC concentration was highly significant, as much as 62% of the variation remained unexplained (Fig. 3). Other studies documenting a significant linear relationship between the Microcystis cell concentration and the total MC concentration, also report a relatively high proportion of unexplained variation (Kotak et al., 2000; Ozawa et al. 2005; Hotto et al., 2008). It is emphasized that so far no effort has been made to identify the contribution of systematic errors in cell counting and/or analysis of MC concentrations to the unexplained part of variation that is observed. However, during a consecutive field study sampling the same water bodies during one year consistent lake specific differences in MC cell quotas were observed, e.g. MC cell quotas obtained from Lake George were on average 20-fold lower when compared with MC cell quotas obtained from Lake Mburo. We are currently investigating this phenomenon, however, lake specific differences in MC cell quotas could explain the relatively large part of unexplained variation observed.
CONCLUSION
The trophic conditions and water depth were of profound importance for the occurrence of MC producing cyanobacteria. Seven of the sampling sites were nutrient rich. However, only the shallow sites had favorable environmental conditions that enabled MC-producing cyanobacteria to flourish. From the extensive genetic analysis, it is concluded that Microcystis sp. constitute the major MC producing taxon in Ugandan freshwaters. Beside MC-RR the new MC-RY variants constitute abundant structural variants in the phytoplankton dominated by Microcystis sp. in Uganda. However, the annual MC production rate in tropical freshwater systems will exceed that of comparable systems in the temperate region due to the generally higher primary production rate.
ACKNOWLEDGEMENTS
We are most grateful to Gerold Winkler and Sabine Wanzenböck for their administrative work throughout the International Postgraduate Training Course in Limnology (IPGL). Johanna Schmidt provided assistance in field sampling and strain isolation. Guntram Christiansen (FWF grant P18185) assisted in cloning and sequencing. This study was funded by the North-South-Dialogue program through a Ph.D. fellowship awarded to William Okello. The study (EC 650) was approved by the Uganda National Council for Science and Technology in conjunction with the Uganda Wildlife Authority. The sampling during 2004 was financed by the IPGL, while sampling during 2008 was partly supported by the British Ecological Society and International Foundation for Sciences. K.G. is a European Young Investigator (EURYI) and thanks the Swiss National Science Foundation for support (PE002—117136/1). We thank Dr. Laure Menin (MS Service ISIC EPFL) for skilful technical assistance.
REFERENCES
- Alweny S. Lake Victoria is critically sick. The Daily Monitor; Uganda: 2007. (Online) [cited 2008 Sept 1] Available from: http://www.monitor.co.ug/artman/publish/insights/Lake_Victoria_is_critically_sick_52775.shtml. [Google Scholar]
- Beadle LC. The Inland Waters of Tropical Africa. An introduction to tropical limnology. 2nd Edition Longman; London: 1981. p. 475. [Google Scholar]
- Carmichael WW. Health effects of toxin-producing cyanobacteria: “The cyanoHABs”. Hum Ecol Risk Assess. 2001;7:1393–1407. [Google Scholar]
- Chapman LJ, Chapman CA, Crisman TL, Nordlie FG. Dissolved oxygen and thermal regimes of a Ugandan crater lake. Hydrobiologia. 1998;385:201–211. [Google Scholar]
- Chorus I, Falconer IR, Salas HJ, Bartram J. Health risks caused by freshwater cyanobacteria in recreational waters. J Toxicol Environ Health, Part B. 2000;3:323–347. doi: 10.1080/109374000436364. [DOI] [PubMed] [Google Scholar]
- Diehnelt CW, Dugan NR, Peterman SM, Budde WL. Identification of microcystin toxins from a strain of Microcystis aeruginosa by liquid chromatography introduction into a hybrid linear ion trap-fourier transform ion cyclotron resonance Mass Spectrometer. Anal Chem. 2006;78:501–512. doi: 10.1021/ac051556d. [DOI] [PubMed] [Google Scholar]
- Dittmann E, Neilan BA, Erhard M, von Döhren H, Börner T. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol Microbiol. 1997;26:779–787. doi: 10.1046/j.1365-2958.1997.6131982.x. [DOI] [PubMed] [Google Scholar]
- Erhard M, von Döhren H, Jungblut P. Rapid typing and elucidation of new secondary metabolites of intact cyanobacteria using MALDI-TOF mass spectrometry. Nature Biotechnol. 1997;15:906–909. doi: 10.1038/nbt0997-906. [DOI] [PubMed] [Google Scholar]
- Fastner J, Erhard M, Carmichael WW, Sun F, Rinehart KL, Rönicke H, Chorus I. Characterization and diversity of microcystins in natural blooms and strains of the genera Microcystis and Planktothrix from German freshwaters. Arch Hydrobiol. 1999;145:147–163. [Google Scholar]
- Ganf GG. Diurnal mixing and the vertical distribution of phytoplankton in a shallow equatorial lake (Lake George, Uganda) J Ecol. 1974;62:611–629. [Google Scholar]
- Haande S, Ballot A, Rohrlack T, Fastner J, Wiedner C, Edvardsen B. Diversity of Microcystis aeruginosa isolates (Chroococcales, Cyanobacteria) from East-African water bodies. Arch Microbiol. 2007;188:15–25. doi: 10.1007/s00203-007-0219-8. [DOI] [PubMed] [Google Scholar]
- Hecky RE. The eutrophication of Lake Victoria. Verh Internat Verein Limnol. 1993;25:39–48. [Google Scholar]
- International Organisation for Standardisation . Water quality. Measurement of biochemical parameters. International Organisation for Standardisation: ISO 10260; Geneva (Switzerland): 1992. [Google Scholar]
- Hotto AM, Satchwell MF, Berry DL, Gobler CJ, Boyer GL. Spatial and temporal diversity of microcystins and microcystin-producing genotypes in Oneida Lake, NY. Harmful Algae. 2008;7:671–681. [Google Scholar]
- Jungblut AD, Neilan BA. Molecular identification and evolution of the cyclic peptide hepatotoxins, microcystin and nodularin, synthetase genes in three orders of cyanobacteria. Arch Microbiol. 2006;185:107–114. doi: 10.1007/s00203-005-0073-5. [DOI] [PubMed] [Google Scholar]
- Kateyo E. Biodiversity of an interface zone of a nutrient-deficient lake (Nabugabo) in Uganda: macrophytes. Afr J Ecol. 2006;45:130–134. [Google Scholar]
- Kizito YS, Nauwerck A, Chapman LJ, Koste W. A limnological survey of some western Uganda crater lakes. Limnologica. 1993;23:335–347. [Google Scholar]
- Kling HJ, Mugidde R, Hecky RE. Recent changes in phytoplankton community of Lake Victoria in response to eutrophication. In: Munawar M, Hecky RE, editors. The Great Lakes of the World (GLOW): Food-web, health and Integrity. Backhuys Publisher; Leiden: 2001. pp. 47–65. [Google Scholar]
- Komárek J, Anagnostidis K. Cyanoprokaryota, 1. Teil Chroococcales. Gustav Fischer Verlag; Jena: 1999. pp. 225–236. [Google Scholar]
- Komárek J, Kling H. Variation in six planktonic cyanophyte genera in Lake Victoria (East Africa) Algol Stud. 1991;61:21–45. [Google Scholar]
- Kotak BG, Lam AKY, Prepas EE, Hrudey SE. Role of chemical and physical variables in regulating microcystin-LR concentration in phytoplankton of eutrophic lakes. Can J Fish Aquat Sci. 2000;57:1584–1593. [Google Scholar]
- Krom MD. Spectrophotometric determination of ammonia: a study of a modified Bertholet reaction using salicylate and dichlorisocyanurate. Analyst. 1982;105:305–316. [Google Scholar]
- Kurmayer R, Dittmann E, Fastner J, Chorus I. Diversity of microcystin genes within a population of the toxic cyanobacterium Microcystis spp. in Lake Wannsee (Berlin, Germany). Microb Ecol. 2002;43:107–118. doi: 10.1007/s00248-001-0039-3. [DOI] [PubMed] [Google Scholar]
- Kurmayer R, Christiansen G, Chorus I. The abundance of microcystin-producing genotypes correlates positively with colony size in Microcystis and determines its microcystin net production in Lake Wannsee. Appl Environ Microbiol. 2003;69:787–795. doi: 10.1128/AEM.69.2.787-795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmayer R, Christiansen G, Fastner J, Börner T. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ Microbiol. 2004;6:831–841. doi: 10.1111/j.1462-2920.2004.00626.x. [DOI] [PubMed] [Google Scholar]
- Lawton LA, Edwards C, Codd GA. Extraction and high-performance liquid chromatographic method for the determination of microcystins in raw and treated waters. Analyst. 1994;119:1525–1530. doi: 10.1039/an9941901525. [DOI] [PubMed] [Google Scholar]
- Lipiatou E, Hecky RE, Eisenreich, Lockhart L, Wilknson P. Recent Ecosystem changes in Lake Victoria reflected in sedimentary natural rocks and anthropogenic organic compounds. In: Johnson TC, Odada E, editors. The limnology, climatology and paleoclimatology of the East African lakes. Gordon and Breach; Toronto: 1996. pp. 523–541. [Google Scholar]
- Mikalsen B, Boison G, Skulberg OM, Fastner J, Davies W, Gabrielsen TM, Rudi K, Jakobsen KS. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J Bacteriol. 2003;185:2774–2785. doi: 10.1128/JB.185.9.2774-2785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugidde R, Hecky RE, Hendzel L, Taylor WD. Pelagic nitrogen fixation in Lake Victoria, Uganda. J Great Lakes Res. 2003;29:76–88. [Google Scholar]
- Müller R, Wiedemann O. Die Bestimmung des Nitrations im Wasser. Vom Wasser. 1955;22:247–271. [Google Scholar]
- Ochumba PBO. Massive fish kills within the Nyanza Gulf of lake Victoria, Kenya. Hydrobiologia. 1990;208:93–99. [Google Scholar]
- Okello W. Toxic cyanobacteria in Ugandan freshwater habitats. UNESCO-IHE; 2004. p. 60. MSc Thesis ES 04.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa K, Fujioka H, Muranaka M, Yokoyama A, Karagami Y, Homma T, Ishikawa K, Tsujimura S, Kumagai M, Watanabe MF, Park H-D. Spatial distribution and temporal variation of Microcystis species composition and microcystin concentration in Lake Biwa. Environ Toxicol. 2005;20:270–276. doi: 10.1002/tox.20117. [DOI] [PubMed] [Google Scholar]
- Rantala A, Rajaniemi-Wacklin P, Lyra C, Lepistö L, Rintala J, Mankiewicz-Boczek J, Sivonen K. Detection of microcystin-producing cyanobacteria in Finnish lakes with genus-specific microcystin synthetase Gene E (mcyE) PCR and associations with environmental factors. Appl Environ Microbiol. 2006;72:6101–6110. doi: 10.1128/AEM.01058-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CS, Huszar V, Kruk C, Naselli-Flores L, Melo S. Towards a functional classification of the freshwater phytoplankton. J Plankton Res. 2002;24:417–428. [Google Scholar]
- Rogalus MK, Watzin MC. Evaluation of sampling and screening techniques for tiered monitoring of toxic cyanobacteria in lakes. Harmful Algae. 2008;7:504–514. [Google Scholar]
- Shapiro J. Blue-green dominance in lakes: The role and management significance of pH and CO2. Internat Rev Ges Hydrobiol. 1984;64:766–779. [Google Scholar]
- Sivonen K, Jones G. Cyanobacterial toxins. In: Chorus I, Bartram J, editors. Toxic cyanobacteria in water. A guide to their public health consequences, monitoring and management. E & FN Spon; London: 1999. pp. 41–112. [Google Scholar]
- Sivonen K, Börner T. Bioactive compounds produced by cyanobacteria. In: Herrero A, Flores E, editors. The Cyanobacteria: Molecular Biology, Genomics and Evolution. Caister Academic Press; Norfolk, UK: 2008. pp. 159–197. [Google Scholar]
- Talling JF. The phytoplankton of Lake Victoria (East Africa) Arch Hydrobiol Beih Ergebn Limnol. 1987;25:229–256. [Google Scholar]
- Tillett D, Dittmann E, Erhard M, von Döhren H, Börner T, Neilan BA. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem Biol. 2000;7:753–764. doi: 10.1016/s1074-5521(00)00021-1. [DOI] [PubMed] [Google Scholar]
- UNESCO, World Water Assessment Programme Uganda National Development Report. 2nd United Nations World Water Development Report: Water, a shared responsibility. 2006:220. [Google Scholar]
- Verschuren D, Johnson TC, Kling HJ, Edgington DN, Leavitt PR, Brown ET, Talbot MR, Hecky RE. History and timing of human impact on Lake Victoria, East Africa. Proc Roy Soc Lond B. 2001;269:289–294. doi: 10.1098/rspb.2001.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via-Ordorika L, Fastner J, Kurmayer R, Hisbergues M, Dittmann E, Komarek J, Erhard M, Chorus I. Distribution of microcystin-producing and non-microcystin-producing Microcystis sp. in European freshwater bodies: detection of microcystins and microcystin genes in individual colonies. System Appl Microbiol. 2004;27:592–603. doi: 10.1078/0723202041748163. [DOI] [PubMed] [Google Scholar]
- Vincze T, Posfai J, Roberts RJ. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003;31:3688–3691. doi: 10.1093/nar/gkg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viner A, Smith I. Geographical, historical and physical aspects of Lake George. Proc R Soc Lond B. 1973;184:235–270. [Google Scholar]
- Vollenweider RA, Kerekes J. Eutrophication of waters. Monitoring, assessment and control. OECD; Paris: 1982. pp. 75–85. [Google Scholar]
- Wetzel RG, Likens GE. Limnological analyses. 3rd edition Springer-Verlag; New York: 2000. p. 429. [Google Scholar]
- Wilson AE, Sarnelle O, Neilan BA, Salmon TP, Gehringer MM, Hay ME. Genetic variation of the bloom-forming cyanobacterium Microcystis aeruginosa within and among lakes: Implications for harmful algal blooms. Appl Environ Microbiol. 2005;71:6126–6133. doi: 10.1128/AEM.71.10.6126-6133.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]