Abstract
MicroRNAs (miRNAs) are a class of small RNAs that play a critical role in the coordination of fundamental cellular processes. Recent studies suggest that miRNAs participate in the cellular stress response (CSR), but their specific involvement remains unclear. In this study, we identify a group of thermally regulated miRNAs (TRMs) that are associated with the CSR. Using miRNA microarrays, we show that dermal fibroblasts differentially express 123 miRNAs when exposed to hyperthermia. Interestingly, only 27 of these miRNAs are annotated in the current Sanger registry. We validated the expression of the annotated miRNAs using qPCR techniques, and we found that the qPCR and microarray data was in well agreement. Computational target-prediction studies revealed that putative targets for the TRMs are heat shock proteins and Argonaute-2—the core functional unit of RNA silencing. These results indicate that cells express a specific group of miRNAs when exposed to hyperthermia, and these miRNAs may function in the regulation of the CSR. Future studies will be conducted to determine if other cells lines differentially express these miRNAs when exposed to hyperthermia.
Keywords: MicroRNA, Cellular stress response, Thermal stress, Hyperthermia, Thermally regulated microRNAs (TRMs)
Introduction
Mammalian cells frequently encounter conditions that can lead to stress. A few well-characterized stressors include hyperthermia, hypoxia, ionizing and non-ionizing radiation, ATP depletion, oxidative stress, and pathogenic stimuli (Schreck et al. 1992; Feder and Hofmann 1999; Kabakov et al. 2002; Kultz 2005; Diller 2006; Miller 2006; Millenbaugh et al. 2008). At the cellular level, stressors can inflict strain on intracellular biomolecules (e.g., lipids, proteins, and DNA, and if such strain is appreciable, these biomolecules can undergo structural modifications that can preclude them from functioning properly. In addition, if the degree of strain exceeds the cell’s capacity for repair, such exposures can also lead to cell death.
In order to survive and adapt to stressful conditions, all mammalian cells have evolved a molecular defense reaction called the cellular stress response (CSR). The CSR is rapidly activated in response to stress and primarily involves the following signaling pathways: redox, DNA sensing and repair, molecular chaperones, proteolysis, energy metabolism, and apoptosis (Kultz 2003). Although countless proteins are associated with these pathways, a group of 44 evolutionary conserved proteins have emerged as core mediators (Kultz 2005). These proteins, collectively referred to as minimal stress proteins, are regulated by cells at both the transcriptional and post-transcriptional levels. At the transcription level, the mRNAs for these stress proteins are transcribed for rapid de novo protein synthesis, whereas at the post-transcriptional level, regulation is executed on the pool of existing mRNAs, where some are selected for translation and others are suppressed.
Interestingly, although the pathways and proteins involved in the CSR are well characterized, the role that a recently discovered class of regulatory RNA—microRNAs—play in these mechanisms remains unclear. MicroRNAs (miRNAs) are small (21–23 nt), endogenous, non-coding RNA species that regulate gene expression by targeting mRNAs in a sequence-specific manner (Ambros 2001; Bartel 2004). miRNAs are abundant, and studies suggest that as many as 40,000 molecules may populate a single cell (Lim et al. 2003). The human miRNA gene family consists of 695 genes, and currently, it is regarded as one of the largest gene families (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008).
MicroRNAs suppress protein synthesis at the post-transcriptional level by annealing—through complementary base-pair binding—to the 3′ untranslated region of their target mRNAs. They mediate these repressive effects by associating with a large protein complex called the RNA-induced silencing complex, which includes the Argonaute 2 (Ago2) protein as a core functional component (Pillai et al. 2007). Once associated, miRNAs then guide Ago2 to targeted mRNAs leading to translational repression, either by degrading mRNA (Meister et al. 2004; Bhattacharyya et al. 2006; Valencia-Sanchez et al. 2006; Pillai et al. 2007) or by interfering with translational machinery (Lee et al. 1993; Wightman et al. 1993). Taken together, these mechanisms make miRNAs robust gene suppressors, and estimations predict that they may regulate over 30% of genes in mammalian cells (Lewis et al. 2005; Rajewsky 2006).
Several studies report that miRNAs play critical roles in coordinating many fundamental cellular processes including development, proliferation, differentiation, death, and metabolism (Cimmino et al. 2005; Rougvie 2005; Gu and Iyer 2006; Johnson et al. 2007; Park and Peter 2008). In addition, recent studies suggest that miRNAs participate in the CSR (Cimmino et al. 2005; Leung et al. 2006; Marsit et al. 2006; Kulshreshtha et al. 2007; Leung and Sharp 2007; Babar et al. 2008). However, several fundamental questions remain unanswered concerning the role that miRNAs may play in these mechanisms: First, which specific miRNAs are expressed by cells exposed to stress? Second, what function do these miRNAs play in the context of the CSR? To answer these questions, we identified a group of miRNAs that are expressed by cells exposed to hyperthermia—a prevalent and well-characterized physiologic stressor. In addition, we used computational target-prediction programs to identify putative targets for these specific miRNAs. These answers serve to extend our current understanding of stress pathways and may possibly contribute to the development of clinical techniques, which utilize the inherent healing capacity of the stress response.
Materials and methods
Cell culture conditions and hyperthermia stress protocol Normal adult human dermal fibroblasts (HDF) were cultured as described previously (Wilmink et al. 2006). In brief, HDFs were plated in 60-mm dishes (5,000 cells/cm2) and were incubated overnight. On day 2, the plates were sealed with Para-film® and were heat-shocked in a water bath at 44°C for 40 min (Wilmink et al. 2009). No significant change in pH was observed.
RNA isolation and normalization RNA was harvested from samples 4 h post-exposure, a time point shown to have maximum HSPA1A mRNA levels, using RNeasy Mini-Kit (Qiagen) (Wilmink et al. 2006). The RNA concentration was assessed on a NanoDrop Spectrophotometer (NanoDrop Technologies), and the quality was measured on a 2100 Bioanalyzer™ (Agilent Technologies). We only used samples with an RNA Integrity Number greater than 9.5.
mRNA microarrays and PCR To verify that our hyperthermia stress protocol induced an appreciable CSR, we conducted microarray and real-time comparative Ct RT-PCR studies. Two micrograms of RNA was used for preparation of biotin-labeled targets (cRNA) using MessageAmp™-based protocols (Ambion Inc). Labeled cRNA was fragmented (0.5 µg/µL per reaction) and was used for array hybridization and washing. The cRNA was mixed with a hybridization cocktail, heated to 99°C for 5 min, and then incubated at 45°C for 5 min. Hybridization arrays were conducted for 16 h in an Affymetrix Model 640 hybridization oven (45°C, 60 rpm). Arrays were washed and stained on an FS450 Fluidics station and were scanned on a GeneChip Scanner 3000 7G. Image signal data, detection calls, and annotations were generated for every gene using the Affymetrix Statistical Algorithm MAS 5.0 (GCOS v1.3) algorithm. A log2 transformation was conducted and a Student’s t test was performed for comparison of the two groups. We conducted multiple testing correction—Benjamini and Hochberg—to determine the false discovery rate, and statistical significant genes were identified using Bonferroni correction procedures (−log10pcutoff > 6.04) (Benjamini et al. 2001). The targets identified in the microarray study were validated using qPCR. Runs were performed on a StepOnePlus™ RT PCR system using TaqMan® RNA-to-CT™ 1-Step Kits and TaqMan® Assays for: HSPA1A, HSPA6, HSPA4L, DNAJA4, DNAJB1, HSPH1, and β-actin (Applied Biosystems). Calibrator RNA was used as control (Cell Applications Inc). PCR was conducted using a three-program LightCycler® protocol and analyzed as previously described (Wilmink et al. 2006).
miRNA microarrays and PCR miRNA microarrays were used to identify stress-responsive miRNAs, and qPCR was conducted to verify their expression. MicroRNA was extracted using the miRNeasy mini kit, per manufacturer’s instructions (Qiagen). For the miRNA microarray experiments, we used an Affymetrix GeneChip® (DiscovArray™, Asuragen Services), which includes all miRNAs from the Sanger miRBase, and greater than 12,000 predicted miRNAs (Griffiths-Jones 2004; Bentwich et al. 2005; Berezikov et al. 2005; Xie et al. 2005; Cummins et al. 2006; Griffiths-Jones et al. 2006). From the initial 14,215 probes, a subset of 3,287 probes was selected for statistical analysis. For each probe, we conducted a two-sample t test using empirical Bayes variance estimates. Probes with p values less than 0.05 were determined to be statistically significant. Normalized log2-transformed intensity values were analyzed using JMP Genomics 3.2 (SAS Institute, Cary, NC, USA). To verify and quantify the magnitude of target expression, we conducted a two-step qRT-PCR. PCR was performed on the same pool of miRNA that was used in the microarray study. Reverse transcription was carried out using a TaqMan® miRNA RT Kit, a miRNA Assay RT Probe of interest (Applied Biosystems), and a MasterCycler® Gradient thermal cycler (Eppendorf). The following TaqMan® miRNA Assay probes were used: RNU48 (Control), let-7d, mir-125b, -452, -382, -378, -101, -424, -138, -376a, -196a, and -196b. PCR was performed per manufacturer’s instructions using a StepOnePlus™ Real-Time PCR system, a TaqMan® Universal PCR Master Mix, and No AmpErase® UNG (Applied Biosystems).
The miRBase sequence database The miRBase database is the primary searchable repository for published miRNAs (http://microrna.sanger.ac.uk/). Release 14 of the database contains 10,883 entries for hairpin miRNA precursors and 10,581 mature miRNA products. We used this database to determine which miRNA targets were annotated, to name our unpublished miRNA targets, and to predict putative mRNA targets.
Computational target-prediction algorithms We identified putative miRNA targets using the following computational algorithms: Microcosm’s miRBase target tool, PITA, and Tarbase. Microcosm and PITA were used to quantify the thermodynamic stability for each miRNA–mRNA duplex (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008; Hofacker 2003). The Microcosm target tool is more sophisticated than traditional algorithms, which rely solely on sequence complementarity. miRBase uses a two-step process, where in the first step a miRanda algorithm is used to identify binding sites for miRNAs, and then in the second step, a Vienna RNA folding program is employed to estimate the thermodynamic stability for each predicted duplex. Since this algorithm actually computes the energy gained when a duplex is formed (ΔGduplex), it may provide more accurate target predictions. The second computational tool we used was PITA. PITA is a thermodynamic modeling program that accounts for the fact that in order for miRNAs to bind to their targets, they must first remove their targets 2° structure—a process referred to as the ΔGopen. Thus, in contrast to MiRanda, which only measures half of the binding process (ΔGduplex), PITA actually computes both elements of the process. In addition, PITA can also be used to calculate the ΔΔG, where ΔΔG = ΔGopen − ΔGduplex (Kertesz et al. 2007). Recent studies have demonstrated that ΔΔG values outperform other algorithms and actually correlate quite well with experimentally measured degrees of mRNA suppression. For this study, we used the following PITA settings: minimal seed size, 6; minimum seed conservation, 0; flank settings, no flank. Last, all miRNA targets were examined using the TarBase database. TarBase is a comprehensive database of experimentally supported animal miRNA targets that is available online at http://www.dian.pcbi.upenn.edu/tarbase (Sethupathy et al. 2006).
Results
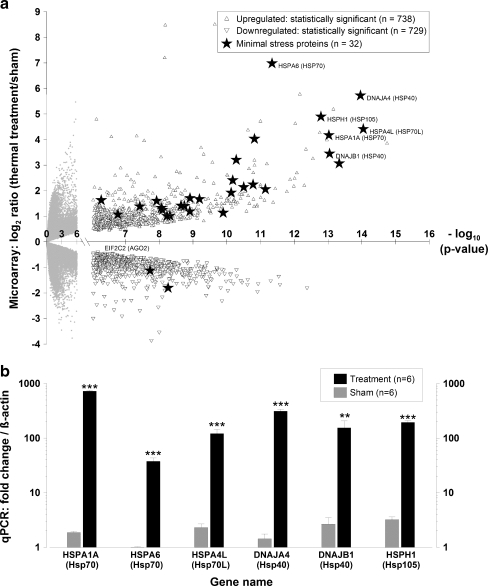
Hyperthermia induces an appreciable cellular stress response We conducted an initial set of experiments to determine whether our hyperthermia stress protocol induced an appreciable transcriptional stress response in dermal fibroblasts. Since a signature feature of an appreciable stress response is the rapid and marked up-regulation of minimal stress proteins (Kultz 2003), we conducted microarray and PCR analyses to examine the genes that dermal fibroblasts express when exposed to hyperthermia (Fig. 1a, b). After applying Bonferroni correction procedures to the microarray data, we found that the treatment group differentially expressed 1,467 genes, and of these, 738 were up-regulated and 729 were down-regulated (Fig. 1a). Additionally, we found that the treatment group expressed transcripts for 32 of the 44 minimal stress proteins, and of these, 23 encoded for molecular chaperone proteins—primarily heat shock proteins (Hsps). We also found that the transcripts encoding for Hsp70 and Hsp40 exhibited the greatest increase in expression and the highest level of statistical significance (p value <10−14) (Fig. 1a). To validate these microarray results, we then conducted qRT-PCR analyses for the gene targets that exhibited statistical significance: HSPA1A (Hsp70), HSPA6 (Hsp70), HSPA4L (Hsp70L), DNAJA4 (Hsp40), DNAJB1 (Hsp40), and HSPH1 (Hsp105) (Fig. 1b). For all of the genes tested, we found that the treatment group exhibited statistically significant increases in expression compared to the sham group. The gene with the greatest increase in expression was HSPA1A, which was induced by ∼728-fold following treatment. Together, the microarray and PCR data both confirm that the hyperthermia protocol induced a considerable stress response.
Fig. 1.
Hyperthermia stress protocol induces an appreciable cellular stress response as evidenced by the marked up-regulation of minimal stress proteins. Dermal fibroblasts were exposed to a hyperthermia protocol (44°C for 40 min), and RNA was extracted 4 h post-exposure. Microarrays and qPCR analyses were conducted for control and treatment groups with n = 6 for each group. a Volcano plot of mRNA microarray data, where the magnitude of the expression is plotted versus the level of statistical significance. Statistical significant genes were identified using Bonferroni correction procedures with a cutoff of −log10p > 6.04. Statistically insignificant targets (empty circles), significant targets (triangles), significant minimal stress proteins (filled stars). Data are expressed as means. b Gene expression for minimal stress proteins (HSPA1A, HSPA6, HSPA4L, DNAJA4, DNAJB1, and HSPH1) using qRT-PCR. The mRNA expression fold values 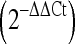 were measured for sham and treatment groups with n = 6 for each group. Values were calculated in relation to β-actin and normalized to a separate RNA calibrator. Sham (gray bar), treatment (black bar). Data are expressed as means ± SD; ***p < 10−5, **p < 10−3; between indicated groups
were measured for sham and treatment groups with n = 6 for each group. Values were calculated in relation to β-actin and normalized to a separate RNA calibrator. Sham (gray bar), treatment (black bar). Data are expressed as means ± SD; ***p < 10−5, **p < 10−3; between indicated groups
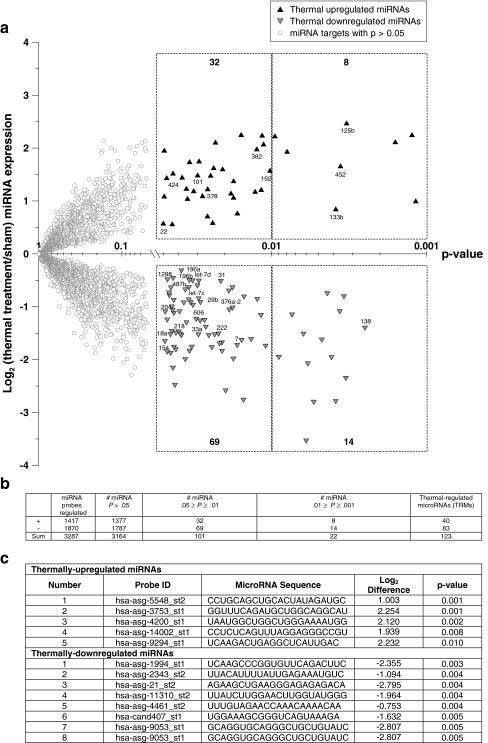
Identification of miRNAs associated with hyperthermia-induced CSR Next, we used miRNA microarrays to identify the miRNAs that dermal fibroblasts express when exposed to our hyperthermia protocol. A summary of the miRNA microarray data is provided as a volcano plot in Fig. 2a. We found that 123 miRNAs—which we refer to as thermally regulated miRNAs (TRMs)—were differentially expressed by the treatment group. Interestingly, of these 123, only 27 are annotated in the current Sanger registry. Further studies are being conducted on the remaining 96 miRNA sequences, and these potential new miRNA genes will be submitted to the registry. A summary of the miRNA microarray data is provided in Fig. 2b. We found that 83 miRNAs were down-regulated and 40 were up-regulated. The predicted miRNA genes exhibiting the highest level of differential expression are provided in Fig. 2c.
Fig. 2.
Identification of miRNAs associated with hyperthermia-induced CSR. Dermal fibroblasts were exposed to a hyperthermia protocol (44°C for 40 min), and RNA was extracted 4 h post-exposure. MicroRNA microarrays were conducted for control and treatment groups with n = 6 for each group. a Volcano plot of miRNA microarray data. The signal difference (log2) is plotted versus the level of statistical significance (p value). Statistically insignificant targets (empty circles), significant up-regulated targets (black triangles), significant down-regulated targets (gray triangles). Data are expressed as means. miRNA targets with published names are provided below each data point. The nonsymmetrical distribution of data points suggests that cells exposed to hyperthermia stress exhibit an overall decrease in miRNA expression. b Summary table for miRNA microarray data. A total of 123 miRNA targets have p values <0.05. These targets are referred to as thermally regulated microRNAs (TRMs). c List of miRNA probes exhibiting the greatest level of differential expression by miRNA microarray analysis. None of these miRNAs are currently annotated in the Sanger Registry
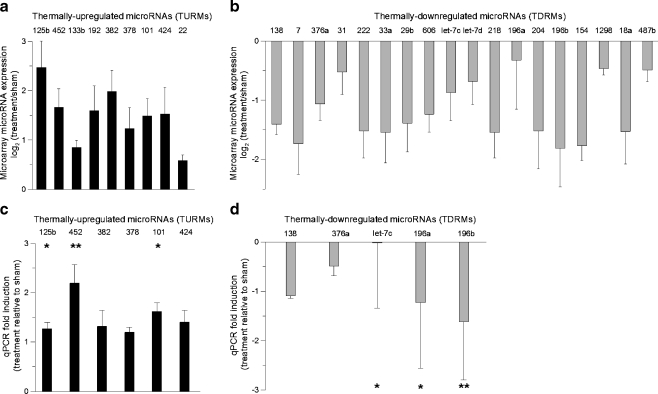
Quantification and validation of annotated thermally regulated miRNAs To examine the consistency of the miRNA microarray data, we quantified the expression of each annotated TRM (Fig. 3a, b). In these plots of the microarray data, the average log2 difference (treatment relative to sham) is provided for each of the 27 annotated TRMs. We found that nine miRNAs were thermally up-regulated (TURMs), and 18 were thermally down-regulated (TDRMs) (Fig. 3a, b). Of the annotated TURMs, miRNA-125b, -382, and -452 showed the greatest increase in expression, and of the annotated TDRMs, miR-138, -7, and -196b showed the greatest decrease in expression. To validate the microarray results, we then applied PCR techniques to quantify the expression for several of the identified miRNAs. Plots of the mean fold-expression changes are provided in Fig. 3c, d. We found that the targets with the highest level of statistical significance were miR-125b, -452, -101, -let-7c, -196a, and -196b. We found that the qPCR and microarray data were consistent for all of the miRNAs tested.
Fig. 3.
Quantification of microarray data and qPCR validation of annotated thermally regulated microRNAs (TRMs). a–b miRNA microarray expression data for thermally up-regulated and thermally down-regulated miRNAs (mean ± SD, n = 6). For both plots, the mean log2 fold change between the treatment and sham groups is plotted for each miRNA. c–d Taqman qRT-PCR validation of thermally regulated miRNA targets. Fold-changes (treatment relative to sham) were calculated using the 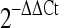 method. Values were calculated against designated calibrators and were normalized to RNU48—endogenous control. TURMs (black bar), TDRMs (gray bar). Data are expressed as means ± SD; *p < 0.05, **p < 0.01; between indicated groups
method. Values were calculated against designated calibrators and were normalized to RNU48—endogenous control. TURMs (black bar), TDRMs (gray bar). Data are expressed as means ± SD; *p < 0.05, **p < 0.01; between indicated groups
Putative target identification To determine putative mRNA targets for the annotated TRMs, we then used several computational target-prediction algorithms. These algorithms were used to generate a list of the five most probable and interesting putative mRNA targets for each miRNA. The first prediction tool we used was the Microcosm miRanda algorithm. This algorithm computes the energy gained when a duplex is formed (ΔGduplex), where more negative ΔGduplex values represent interactions that are more favorable. The second tool we used was the PITA tool (Kertesz et al. 2007). This thermodynamic modeling program accounts for the observation that in order for miRNAs to bind their targets, they must first remove their putative target’s secondary structure. For PITA analyses, negative ΔΔG scores represent more favorable miRNA–mRNA interactions. The last tool we used was the Tarbase database. This database is a collection of empirically validated targets (Papadopoulos et al. 2009).A list of the highest scoring and most interesting mRNA putative targets are provided for each annotated TRM (Tables 1 and 2). Of the 55 putative targets, we found that only eight have been empirically validated: miR-378-SUFU, miR-101-MYCN, miR-138-TERT, miR-196a-HOXB8, miR-196b-HOXB8, miR-196a-HOXC8, miR-196b-HOXC8, and let-7c-TRIM71 (Lewis et al. 2003, 2005; Yekta et al. 2004; Lee et al. 2007; Lin et al. 2007; Mitomo et al. 2008). Interestingly, many of the 47 remaining putative targets have known CSR functions: proteolysis (miR-101-UBE2A); molecular chaperones (miR-125b-DNAJB2, miR-424-HSPA4L, miR-424-DNAJB4, miR-125b-TTC7A, miR-452-TTC7A, miR-378-TTC7A, miR-378-HSP90AB1, miR-138-CCT5, miR-138-HSPA4L, miR-376a-HSPA6, miR-let-7c-HSPB2, and miR-196a-HSPH1) (Kojima et al. 2004; White et al. 2005); protein trafficking (miR-125-ZFYVE1); metabolism (miR-382-KYNU, miR-378-KLK4, miR-376a-MAN1C1, miR-let-7c-GALE, and miR-let-7c-RNF20); cell cycle progression (miR-101-MYCN, miR-196a-HOXC8, and miR-196b-HOXC8) (Deraison et al. 2007; Kamel et al. 2009) (Tables 1 and 2). In addition, several of the annotated TRMs (miR-125b, -138, and -376a) may target AGO2, an integral protein required for miRNA mediated repression (Leung et al. 2006) (Tables 1 and 2).
Table 1.
Putative mRNA targets for annotated thermally up-regulated microRNAs (TURMs)
| TURM | Putative mRNA targets | Microcosma | PITA: thermodynamic modelb | Tarbasec | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Symbol | Name | Sequence | Function | Score | ΔGduplex | ΔGduplex | ΔGopen | ΔΔG | Validation | |
| miR-125b | OSBPL9 | Oxysterol-binding protein-related protein 9 | NM_024586 | Intracellular lipid receptor | 17.1 | −21.3 | −18.9 | −9.6 | −9.3 | No |
| TTC7A | Tetratricopeptide repeat protein 7A | NM_020458 | Chaperones (White et al.) | 18.9 | −29.1 | −27.4 | −13.6 | −13.8 | No | |
| ZFYVE1 | Zinc finger FYVE domain containing protein | NM_021260 | Recruitment of proteins and membrane trafficking | 16.6 | −28.0 | −25.5 | −9.6 | −15.9 | No | |
| DNAJB2 | DnaJ homolog subfamily B member 2 | NM_006736 | Minimal stress protein (Kultz et al.) | 16.7 | −21.5 | −26.0 | −15.2 | −10.8 | No | |
| AGO2 | Eukaryotic translation initiation factor 2C2 | NM_012154 | RNAinterference, role in CSR (Liu et al.) | 15.9 | −15.7 | −13.7 | −5.7 | −8.0 | No | |
| miR-452 | IGF1R | Insulin-like growth factor 1 receptor | NM_000875 | Resistance to oxidative stress (Holzenberger et al.) | 16.8 | −19.1 | −19.2 | −9.1 | −10.1 | No |
| POLR2B | Polymerase (RNA) II polypeptide B, 140 kDa | NM_000938 | UV sensitive gene (Jung et al.) | 15.8 | −14.8 | NR | NR | NR | No | |
| NOX3 | NADPH oxidase 3 | NM_015718 | Generation reactive oxygen species (Li et al.) | 19.9 | −11.9 | −15.8 | −5.5 | −10.3 | No | |
| MPHOSPH9 | M-phase phosphoprotein 9 | NM_022782 | Cellular physiological process | 16.7 | −13.8 | −13.6 | −6.9 | −6.7 | No | |
| TTC7A | Tetratricopeptide repeat protein 7A | NM_020458 | Chaperones (White et al.) | 18.2 | −15.8 | −15.7 | −10.4 | −5.3 | No | |
| miR-382 | KYNU | Kynureninase (l-kynurenine hydrolase) | NM_001032998 | Biosynthesis of NAD+ | 18.2 | −14.2 | NR | NR | NR | No |
| HMGN3 | High mobility group nucleosomal binding domain 3 | NM_004242 | Ethanol-induced apoptosis (Castenada et al.) | 17.5 | −15.3 | −13.2 | −7.5 | −5.7 | No | |
| DTYMK | Deoxythymidylate kinase | NM_012145 | Metabolism and cytoxicity (Hu et al.) | 18.5 | −21.5 | NR | NR | NR | No | |
| BCL7A | B-cell CLL/lymphoma 7A | NM_001024808 | Hypoxia (van der Meer et al.) | NR | NR | −18.7 | −6.8 | −11.9 | No | |
| PREPL | Prolyl endopeptidase-like | NM_006036 | Metabolism, proteolysis | NR | NR | −16.6 | −6.1 | −10.5 | No | |
| miR-378 | KLK4 | Kallikrein-related peptidase 4 | NM_004917 | Metabolism (Deraison et al.) | 17.6 | −22.5 | −21.0 | −6.2 | −14.8 | No |
| SUFU | Suppressor of fused homolog (Drosophila) | NM_016169 | Enhances cell survival | NR | NR | −14.4 | −8.1 | −6.3 | Lee et al. | |
| AHSA1 | Activator of 90 kDa HSP ATPase | NM_012111 | Response to stress (Amundson et al.) | 17.7 | −24.2 | −21.4 | −14.6 | −6.8 | No | |
| TTC7A | Tetratricopeptide repeat protein 7A | NM_020458 | Chaperones (White et al.) | 17.2 | −25.5 | −21.1 | −16.7 | −4.4 | No | |
| IGF1R | Insulin-like growth factor 1 receptor | NM_000875 | Resistance to oxidative stress (Holzenberger et al.) | 15.8 | −19.4 | −20.1 | −6.0 | −14.2 | No | |
| miR-101 | CDYL | Chromodomain protein, Y-like | NM_004824 | DNA packaging protein | 16.1 | −16.5 | −14.0 | −9.8 | −4.2 | No |
| USP47 | Ubiquitin specific peptidase 47 | NM_017944 | Peptidase activity | 17.6 | −15.1 | −15.9 | −8.4 | −7.5 | No | |
| UBE2A | Ubiquitin-conjugating enzyme E2A | NM_003336 | Proteolysis | 15.4 | −15.4 | −13.3 | −4.3 | −9.0 | No | |
| HSP90AB1 | Heat shock protein 90-beta | NM_007355 | Minimal stress protein | 16.8 | −11.3 | −11.6 | −11.2 | −0.4 | No | |
| MYCN | N-myc proto-oncogene protein | NM_005378 | Positive regulator cellular proliferation | 16.1 | −13.5 | −12.8 | −7.7 | −5.1 | Lewis et al. | |
| miR-424 | STXBP3 | Syntaxin binding protein 3 | NM_007269 | Cellular physiological process | 17.0 | −17.4 | −14.3 | −7.5 | −6.8 | No |
| LRP6 | Lipoprotein receptor-related protein 6 | NM_002336 | Morphogenesis | 18.8 | −18.7 | −20.5 | −11.7 | −8.8 | No | |
| HSPA4L | Heat shock 70 kDa protein 4-like | NM_014278 | Chaperone protein (Kojima et al.) | 16.5 | −12.9 | −13.8 | −9.6 | −4.2 | No | |
| DNAJB4 | DnaJ homolog subfamily B member 4 | NM_007034 | Minimal stress protein | 15.8 | −13.4 | −16.5 | −8.6 | −7.9 | No | |
| VWF | von Willebrand factor precursor | NM_000552 | Stress response | 17.5 | −24.5 | −19.8 | −16.9 | −2.9 | No | |
amiRanda MicroCosm: http://microrna.sanger.ac.uk/index.shtml
bPITA: http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html
cTarBase v.5c: http://diana.cslab.ece.ntua.gr/tarbase/
Table 2.
Putative mRNA targets for annotated thermally down-regulated microRNAs (TDRMs)
| TDRM | Putative mRNA targets | Microcosma | PITA: thermodynamic modelb | Tarbasec | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Symbol | Gene name | Sequence | Function | Score | ΔGduplex | ΔGduplex | ΔGopen | ΔΔG | Validation | |
| miR-138 | TMTC4 | Transmembrane and tetratricopeptide repeat containing 4 | NM_032813 | Not well characterized | 16.1 | −20.6 | −22.4 | −8.9 | −13.5 | No |
| CCT5 | Chaperonin containing TCP1, subunit 5 (epsilon) | NM_012073 | Chaperonin (TRiC complex)/protein renaturation | 16.4 | −23.8 | −25.4 | −6.0 | −19.4 | No | |
| TERT | Telomerase reverse transcriptase | NM_198253 | Morphogenesis | 16.4 | −26.8 | −28.3 | −9.3 | −19.0 | Mitomo et al. | |
| AGO2 | Eukaryotic translation initiation factor 2C2 | NM_012154 | RNAinterference, role in CSR (Liu et al.) | 15.8 | −29.9 | −28.9 | −10.8 | −18.1 | No | |
| HSPA4L | Heat shock 70 kDa protein 4-like | NM_014278 | Hypertonic and heat stress (Kojima et al.) | 16.9 | −18.4 | NR | NR | NR | No | |
| miR-376a | MAN1C1 | Mannosidase, alpha, class 1C, member 1 | NM_020379 | Metabolic processes | 16.8 | −18.0 | −20.3 | −10.0 | −10.3 | No |
| SLC35F4 | Solute carrier family 35, member F4 | NM_001080455 | Membrane transport | 15.4 | −17.0 | −15.1 | −8.3 | −6.8 | No | |
| ZCCHC7 | Zinc finger, CCHC domain containing 7 | NM_032226 | Ion binding | 17.1 | −13.8 | −11.2 | −8.4 | −2.8 | No | |
| AGO2 | Eukaryotic translation initiation factor 2C2 | NM_012154 | RNAinterference, role in CSR (Liu et al.) | 15.9 | −12.8 | −13.4 | −5.4 | −8.0 | No | |
| HSPA6 | HSP70 protein 6 | NM_002155 | Minimal stress protein (Kultz et al.) | 16.7 | −12.7 | −13.9 | −12.7 | −1.2 | No | |
| miR-let-7c | TRIM71 | Tripartite motif-containing 71 | NM_001039111 | Development | 16.4 | −19.6 | −16.5 | −5.2 | −11.3 | Lin et al. |
| HSPB2 | HSP beta-2 | NM_001541 | Small HSP, minimal stress protein (Kultz et al.) | 17.2 | −15.5 | −18.3 | −11.7 | −6.6 | No | |
| GALE | UDP-galactose-4-epimerase | NM_000403 | Metabolism | 17.6 | −23.2 | −21.4 | −4.3 | −17.1 | No | |
| IGF1R | Insulin-like growth factor 1 receptor | NM_000875 | Resistance to oxidative stress (Holzenberger et al.) | NR | NR | −18.3 | −4.2 | −14.1 | No | |
| RNF20 | Ring finger protein 20 | NM_019592 | Metabolism | 17.0 | −24.8 | −23.5 | −9.0 | −14.5 | No | |
| miR-196a | HOXB8 | Homeobox B8 | NM_024016 | Differentiation | 20.9 | −39.5 | −36.1 | −4.3 | −31.8 | Kawasaki et al. |
| MYC | V-myc myelocytomatosis viral oncogene homolog | NM_002467 | Regulation physiological process | 17.2 | −17.5 | −17.1 | −11.6 | −5.5 | No | |
| HOXC8 | Homeobox C8 | NM_022658 | Cell cycle progression (Kamel et al.) | 18.0 | −34.8 | −31.7 | −9.4 | −22.3 | Yekta et al. | |
| HSPH1 | HSP 105 kDa (HSP 110 kDa) | NM_006644 | Minimal stress protein (Kultz et al.) | 16.3 | −19.3 | NR | NR | NR | No | |
| MAPKAPK5 | MAP kinase-activated protein kinase | NM_139078 | Minimal stress protein | 16.1 | −13.5 | NR | NR | NR | No | |
| miR-196b | HOXB8 | Homeobox B8 | NM_024016 | Differentiation | 20.1 | −34.6 | −31.8 | −4.3 | −27.5 | Yekta et al. |
| MYC | V-myc myelocytomatosis viral oncogene homolog | NM_002467 | Regulation physiological process | 16.8 | −17.2 | −15.0 | −11.6 | −3.4 | No | |
| RNF20 | Ring finger protein 20 | NM_019592 | Metabolism | 15.9 | −16.0 | −17.0 | −9.0 | −8.0 | No | |
| AKR1B1 | Aldo-keto reductase family 1, member B1 | NM_001628 | Response to stress | 15.7 | −15.9 | NR | NR | NR | No | |
| HOXC8 | Homeobox C8 | NM_022658 | Cell cycle progression (Kamel et al.) | 17.2 | −32.0 | −30.0 | −9.4 | −20.6 | Yekta et al. | |
amiRanda MicroCosm: http://microrna.sanger.ac.uk/index.shtml
bPITA: http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html
cTarBase v.5c: http://diana.cslab.ece.ntua.gr/tarbase/
Discussion
MicroRNAs are known to play critical roles in development, cell proliferation, and other fundamental cellular processes (Cimmino et al. 2005; Rougvie 2005; Gu and Iyer 2006; Johnson et al. 2007; Park and Peter 2008). In addition, miRNAs have also been suggested to be involved in the CSR; however, a few fundamental questions remain largely unanswered (Cimmino et al. 2005; Leung et al. 2006; Marsit et al. 2006; Kulshreshtha et al. 2007; Leung and Sharp 2007; Babar et al. 2008). First, which specific miRNAs do cells express when exposed to stress? Second, what function do they play in the context of the stress response? In this study, we show that dermal fibroblasts differentially express a group of 123 miRNAs when exposed to hyperthermia. Interestingly, of these 123, only 27 are annotated in the current Sanger registry. Future validation studies will be conducted on the remaining 96 predictive sequences to determine if they are new miRNA genes. Using computational target-prediction algorithms, we identified a list of putative mRNA targets for the annotated TRMs. Interestingly, the data show that putative targets for the TRMs are the following: DNAJB2, HSPA4L, TTC7A, HSPA6, AGO2, HOXB8, and HOXC8. Thus, the results of this study indicate that dermal fibroblasts do express a specific group of miRNAs when exposed to thermal stress, and these specific miRNAs appear to have putative targets for established components of the stress network.
Thermally regulated miRNAs—identification and implication To our knowledge, this is the first study to identify the miRNAs that are associated with hyperthermia-induced CSR. However, several studies have previously reported that cells do in fact alter their miRNA expression profile when exposed to the following stressors: folate deficiency, arsenic exposure, radiation, hypoxia, and cigarette smoke (Marsit et al. 2006; Kulshreshtha et al. 2007; Weidhaas et al. 2007; Izzotti et al. 2009). Curiously, after comparing our results to those of previous studies, we unveiled three noteworthy findings: (1) several miRNAs (miR-125b, -222, -22, and let-7c) are expressed in response to most stressor types; (2) several TRMs (miR-452, -382, and -378) are only expressed by cells exposed to hyperthermia; and (3) cells primarily down-regulate miRNAs when exposed to stress (Table 3). The above findings have important implications that may serve to extend our current understanding of stress pathways in several ways. First, the observation that a few miRNAs are expressed in response to most types of stress is evidence that these miRNAs are not exclusive to the CSR elicited by hyperthermia. In fact, these particular miRNAs may be part of a larger group of miRNAs that function as integral components of the CSR. Analogous to their protein counterparts, future studies may refer to this group as “minimal stress miRNAs.” Second, the finding that a few of the annotated TRMs (miR-452, -382, and -378) are not expressed by cells exposed to other types of stress suggests that these miRNAs may be stress-type dependent. Since it has been traditionally assumed that cells monitor stress based on general macromolecular damage—without regard for the type of stress imposed—these data may provide evidence that these three miRNAs may in fact be preferentially regulated in response to hyperthermia. In fact, it is possible that these miRNAs are signature thermal biomarkers. Furthermore, since hyperthermia is known to preferentially induce appreciable levels of protein denaturation, it may be possible that these miRNAs are involved in mechanisms responsible for combating this type of perturbation. Additionally, from a more global perspective, this finding may also imply that cells elicit different miRNA mechanisms to sense and respond to different types of stress (Leung and Sharp 2007). Last, we found that a greater number of miRNAs were down-regulated—rather than up-regulated—in cells exposed to stress. Similarly, Izzotti et al. also found that lung cells down-regulate a greater number of miRNAs when exposed to cigarette smoke (Izzotti et al. 2009). Since stressed cells demand rapid protein translation, and miRNAs are known to repress translation, it is possible that cellular mechanisms may have evolved to reduce de novo miRNA synthesis during times of stress (Li et al. 2008). In other words, during times of stress, cells may require miRNA down-regulation to translate minimal stress proteins in a very rapid, robust, and efficient manner. Future studies may discover that miRNA down-regulation may be as critical to the CSR as the up-regulation of minimal stress proteins. Therefore, to provide support for the concept of conservation of hyperthermia-specific TRMs, future studies should be conducted using other cell types.
Table 3.
A summary of microRNAs associated with cellular stressors
| Type of stress | Measurement (treated versus sham) | MicroRNAs | Citation | |||||
|---|---|---|---|---|---|---|---|---|
| 125b | 222 | 22 | 192 | let-7c | let-7d | |||
| Folate deficiency | Fold change | 2.89 | 2.09 | 1.93 | – | – | – | Marsit et al. 2006 |
| Arsenic exposure | Fold change | – | 1.99 | 2.39 | – | – | – | Marsit et al. 2006 |
| Radiation | Log2 ratio | – | – | – | – | −0.8 | −0.5 | Weidhaas et al. 2007 |
| Hypoxia | Fold change | 1.83 | – | – | 1.75 | – | – | Kulshreshtha et al. 2007 |
| Cigarette smoke | Fold change | −5.40 | −3.80 | – | −2.80 | −5.9 | – | Izzotti et al. 2008 |
| Thermal stress | Log2 ratio | 2.47 | −1.52 | 0.58 | 1.59 | −0.87 | – | Figures 2 and 3 |
Delineating the function of miRNAs in the context of the CSR We found that many—32 of the known 44—minimal stress proteins were up-regulated in cells exposed to hyperthermia. Intriguingly, we also found that over 70% of these genes encoded for members of the heat shock protein family. Given that Hsp70 and Hsp40 are the most highly induced and well-characterized targets of heat shock, these findings are not surprising and are consistent with our previous studies (Wilmink et al. 2009). However, one surprising finding is that Hsp70 (HSPA4L and HSPA6) and Hsp40 (DNAJB2) are both putative targets for the TRMs (Tables 1 and 2). Several studies have shown that HSPA4L is a stress-responsive gene that is regulated at a transcriptional level by heat shock factor 1 binding to the 5′ region of the heat shock element (Morimoto 1993; Morimoto et al. 1996; Kojima et al. 2004). However, studies have not shown that HSPA4L may have secondary regulation by miRNAs. Since TRM’s may target HSPA4L, it is possible that cells may have evolved mechanisms to down-regulate the expression of miRNAs that would otherwise bind to and impair the translation of minimal stress proteins, such as HSPA4L.In addition to the HSP targets, we also found Ago2 is a putative target for several TRMs (miR-138, -125b, and -376a). Since miRNAs associate with Ago2 to impair protein translation, it may be possible that cells may require less miRNAs and Ago2 when exposed to stress (Lee et al. 1993; Wightman et al. 1993). In addition, our microarray and qPCR data both confirm that Ago2 mRNA levels are reduced in cells exposed to hyperthermia (Fig. 1a).Although previous studies have suggested a link between Ago2, miRNAs, and cellular stress, the exact mechanisms governing these interactions still remain unclear (Leung et al. 2006; Leung and Sharp 2007). Traditional models contend that Ago2 inhibits protein synthesis in one of two ways: (1) if a miRNA has “perfect” target complementarity then cleavage mechanisms are activated or (2) if a miRNA has “imperfect” target complementarity then impairment mechanisms are activated. In addition to these mechanisms, more recent investigations have shown miRNAs can also directly reduce the concentration of their targets using rapid mRNA deadenylation mechanisms (Lee et al. 1993; Wightman et al. 1993; Moss et al. 1997; Wu et al. 2006). To further complicate matters, it has also been shown that miRNAs can induce gene expression (Place et al. 2008). In addition, miRNAs may actually be able to oscillate between states of protein repression and activation—depending on the state of the cell cycle (Vasudevan et al. 2007). Specifically, such findings showed that during times of proliferation, miRNAs repress translation, but during G1/G0 arrest, they activate translation (Vasudevan et al. 2007). Although this phenomenon was observed using serum-starved conditions, hyperthermic stress may also affect the state of the cell cycle and in turn affect the functional state of the miRNAs. Overall, the role that Ago2 and miRNAs play during cellular stress remains poorly understood. Future investigations need to be conducted to better understand these relationships.In summary, we provide evidence that dermal fibroblasts differentially express a group of 123 miRNAs when exposed to hyperthermia stress. We also provide evidence that minimal stress proteins and AGO-2 are putative mRNA targets for these miRNAs. These results indicate that dermal fibroblasts express a specific group of miRNAs when exposed to hyperthermia, and these miRNAs may function in the CSR. We plan to conduct future studies to determine whether other cell types differentially express these TRMs when exposed to hyperthermic stress.
Acknowledgments
We wish to thank the National Academy of Sciences NRC for providing Dr. Wilmink with a research associateship at the Air Force Research Laboratory. This work was supported by grants provided by HQAF SGRS Clinical Investigation program: “Neurological Impacts of Nanosecond Electric Pulse Exposure” and “Determination of Cellular Bioeffect Thresholds for Terahertz Frequencies.” A special thank-you is also extended to Dr. Susan Opalenik, my eternal lily pad, and Mr. Luisiana X. Cundin.
Footnotes
Contract grant sponsor: DOD- SGRS Clinical Investigation program
References
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–826. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- Babar IA, Slack FJ, et al. miRNA modulation of the cellular stress response. Future Oncol. 2008;4(2):289–298. doi: 10.2217/14796694.4.2.289. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, et al. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1–2):279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37(7):766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, et al. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125(6):1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, He Y, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103(10):3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deraison C, Bonnart C, et al. LEKTI fragments specifically inhibit KLK5, KLK7, and KLK14 and control desquamation through a pH-dependent interaction. Mol Biol Cell. 2007;18(9):3607–3619. doi: 10.1091/mbc.E07-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diller KR. Stress protein expression kinetics. Annu Rev Biomed Eng. 2006;8:403–424. doi: 10.1146/annurev.bioeng.7.060804.100449. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucl Acids Res. 2004;32(suppl_1):D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucl Acids Res. 2006;34(suppl_1):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, et al. miRBase: tools for microRNA genomics. Nucl Acids Res. 2008;36(suppl_1):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Iyer VR. PI3K signaling and miRNA expression during the response of quiescent human fibroblasts to distinct proliferative stimuli. Genome Biol. 2006;7(5):R42. doi: 10.1186/gb-2006-7-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31(13):3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti A, Calin GA, et al. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67(16):7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Kabakov AE, Budagova KR, et al. Stressful preconditioning and HSP70 overexpression attenuate proteotoxicity of cellular ATP depletion. Am J Physiol Cell Physiol. 2002;283(2):C521–C534. doi: 10.1152/ajpcell.00503.2001. [DOI] [PubMed] [Google Scholar]
- Kamel S, Kruger C, et al. Morpholino-mediated knockdown in primary chondrocytes implicates Hoxc8 in regulation of cell cycle progression. Bone. 2009;44(4):708–716. doi: 10.1016/j.bone.2008.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H, Taira K (2009) MicroRNA-196 inhibits HOXB8 expression in myeloid differentiation of HL60 cells. Nucleic Acids Symp Ser 2004 48(1):211–212 [DOI] [PubMed]
- Kertesz M, Iovino N, et al. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kojima R, Randall JD, et al. Regulation of expression of the stress response gene, Osp94: identification of the tonicity response element and intracellular signalling pathways. Biochem J. 2004;380(Pt 3):783–794. doi: 10.1042/BJ20040313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27(5):1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kultz D. Evolution of the cellular stress proteome: from monophyletic origin to ubiquitous function. J Exp Biol. 2003;206(Pt 18):3119–3124. doi: 10.1242/jeb.00549. [DOI] [PubMed] [Google Scholar]
- Kultz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol. 2005;67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, et al. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Lee DY, Deng Z, et al. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. 2007;104(51):20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130(4):581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, et al. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA. 2006;103(48):18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, et al. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, et al. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li L-C, Okino ST, et al. Small dsRNAs induce transcripitional activation in human cells. PNAS. 2008;103(46):17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17(8):991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Hsieh LC, et al. Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Mol Biol Evol. 2007;24(11):2525–2534. doi: 10.1093/molbev/msm195. [DOI] [PubMed] [Google Scholar]
- Marsit CJ, Eddy K, et al. MicroRNA responses to cellular stress. Cancer Res. 2006;66(22):10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Millenbaugh NJ, Roth C, et al. Gene expression changes in the skin of rats induced by prolonged 35 GHz millimeter-wave exposure. Radiat Res. 2008;169(3):288–300. doi: 10.1667/RR1121.1. [DOI] [PubMed] [Google Scholar]
- Miller BA. The role of TRP channels in oxidative stress-induced cell death. J Membr Biol. 2006;209(1):31–41. doi: 10.1007/s00232-005-0839-3. [DOI] [PubMed] [Google Scholar]
- Mitomo S, Maesawa C, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99(2):280–286. doi: 10.1111/j.1349-7006.2007.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Cells in stress: transcriptional activation of heat shock genes. Science. 1993;259(5100):1409–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- Morimoto RI, Kroeger PE, et al. The transcriptional regulation of heat shock genes: a plethora of heat shock factors and regulatory conditions. Exs. 1996;77:139–163. doi: 10.1007/978-3-0348-9088-5_10. [DOI] [PubMed] [Google Scholar]
- Moss EG, Lee RC, et al. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88(5):637–646. doi: 10.1016/S0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Papadopoulos GL, Reczko M, et al. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37(Database issue):D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Peter ME. microRNAs and death receptors. Cytokine Growth Factor Rev. 2008;19(3–4):303–311. doi: 10.1016/j.cytogfr.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, et al. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17(3):118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Place RF, Li LC, et al. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38 Supp:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Rougvie AE. Intrinsic and extrinsic regulators of developmental timing: from miRNAs to nutritional cues. Development. 2005;132(17):3787–3798. doi: 10.1242/dev.01972. [DOI] [PubMed] [Google Scholar]
- Schreck R, Albermann K, et al. Nuclear factor kappa B: an oxidative stress-responsive transcription factor of eukaryotic cells (a review) Free Radic Res Commun. 1992;17(4):221–237. doi: 10.3109/10715769209079515. [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Corda B, et al. TarBase: a comprehensive database of experimentally supported animal microRNA targets. Rna. 2006;12(2):192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, et al. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Weidhaas JB, Babar I, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67(23):11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RA, McNulty SG, et al. Positional cloning of the Ttc7 gene required for normal iron homeostasis and mutated in hea and fsn anemia mice. Genomics. 2005;85(3):330–337. doi: 10.1016/j.ygeno.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, et al. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wilmink GJ, Opalenik SR, et al. Assessing laser-tissue damage with bioluminescent imaging. J Biomed Opt. 2006;11(4):041114. doi: 10.1117/1.2339012. [DOI] [PubMed] [Google Scholar]
- Wilmink GJ, Opalenik SR, et al. Molecular imaging-assisted optimization of hsp70 expression during laser-induced thermal preconditioning for wound repair enhancement. J Invest Dermatol. 2009;129(1):205–216. doi: 10.1038/jid.2008.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, et al. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103(11):4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, et al. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]