Abstract
Small heat shock proteins (sHSPs) encompass a widespread and diverse class of proteins with molecular chaperone activity. In the present study, two sHSP isoforms (VpsHSP-1 and VpsHSP-2) were cloned from Venerupis philippinarum haemocytes by Rapid Amplification of cDNA Ends (RACE) approaches. The expression profiles of these two genes under Vibrio anguillarum challenge and cadmium exposure were investigated by quantitative real-time reverse transcriptase polymerase chain reaction. The bacterial challenge could significantly up-regulate the mRNA expression of both VpsHSP-1 and VpsHSP-2, with the increase of VpsHSP-2 expression occurred earlier than that of VpsHSP-1. During the cadmium exposure experiment, the expression level of both VpsHSP-1 and VpsHSP-2 decreased significantly with larger amplitude in VpsHSP-2. As time progressed, the expression levels of both genes were up-regulated with more increment in the low-chemical exposure groups. The differences in the response to pathogen stimulation and cadmium exposure indicated that there were functional diversity between the two structurally different molecules, VpsHSP-1 and VpsHSP-2, and they probably played distinct roles in mediating the environmental stress and immune responses in calm.
Keywords: Venerupis philippinarum, Small heat shock protein, Time–course expression, Vibrio anguillarum, Cadmium exposure
Introduction
Exposure of marine bivalves to various stresses elicits a subset of genes expression, in which phylogenetically conserved proteins known as the heat shock proteins (HSPs) are the major group. Heat shock proteins act as molecular chaperones by binding of partially denatured proteins to prevent their degradation, helping to refold these proteins in an ATP-dependent manner after relief of stress and assisting in their elimination if they become irreversibly damaged (Solé et al. 2000; Thomas et al. 2005). In recent years, considerable attention has been paid on the proteins for their immunological roles, because HSPs were potent activators of the innate immune system (Moseley 2000; Wallin et al. 2002; Tsan and Gao 2004a) and have been implicated in the pathogenesis of a number of autoimmune diseases (Pockley 2003; Tsan and Gao 2004b), in antigen presentation, in cross-presentation and in tumor immunity (Li et al. 2002; Srivastava 2002). However, the remote possibility that bacterial contaminants in purified preparations of HSPs may be responsible for the observed effects could not be totally excluded.
Based on their molecular weight, HSPs have been classified into four categories: (I) HSP90 (83–99 kDa), (II) HSP70 (68–80 kDa), (III) HSP60 and (IV) a family of small HSPs (sHSPs, 15–40 kDa) (Lindquist and Craig 1988; Mizrahi et al. 2009). Compared to other types of HSPs, sHSPs are a more diverse and less conserved group of proteins, the number and sequences of which vary between and within species (Morrow et al. 2000). Additionally, the abundance of this kind of protein also varies considerably depending on the cell types and organisms studied (Haslbeck 2002). Functional studies indicate that sHSPs are involved in efficiently binding denatured proteins as molecular chaperones (Haslbeck 2002) and regulation of membrane lipid polymorphism (Tsvetkova et al. 2002). Owing to their response to diverse forms of stress, sHSPs have undergone widespread application in biomonitoring for environmental pollutants (Pomerai 1996).
Up to date, the studies on sHSP mainly focused on mammals and insects. There are rare investigations regarding molecular features and functions of sHSPs in mollusk except scallop (Zhang et al. 2009a, b). To our knowledge, no comparative research has been conducted on different sHSP gene structure and expression profile under heavy metal or pathogen stress in one species of mollusk. In the meantime, taking into account the facts that the marine ecosystem is deteriorating with higher content of heavy metals and frequencies of disease epidemics in aquaculture, here, two full-length cDNAs of clam sHSP were cloned, and their mRNA expression under cadmium and bacteria stress was measured to understand the responses of clams to environmental and pathogen stress.
Materials and methods
Clams, bacterial challenge and cadmium exposure
The clams Venerupis philippinarum (7.5–11 g in weight) were purchased from a local farm and acclimated for a week before commencement of the experiment. The temperature was held at 20 to 22°C throughout the whole experiment. The salinity for the supplied seawater was kept at 30‰.
For the bacterial challenge experiment, the clams were randomly divided into six flat-bottomed rectangular tanks with 50-l capacity, each containing 50 clams. One tank served as control, while the other five tanks were immersed with high density of Vibrio anguillarum with final concentration of 107 CFU ml −1. The infected clams were randomly sampled at 6, 12, 24, 48, 72 and 96 h, respectively. For cadmium stress experiment, clams were distributed randomly to two groups, each containing 50 individuals. The clams were exposed with two different final concentrations of CdCl2 (10 and 40 μ g l−1) for 24, 48 and 96 h, respectively. The clams cultured in the normal seawater were used as control group. The haemolymphs from the control and the treated groups were collected using a syringe individually and centrifuged at 2000 × g, 4°C, for 10 min to harvest the haemocytes. During the pathogen stimulation and cadmium exposure experiments, there were five replicates for each treatment and the control group.
cDNA library construction and Expressed Sequence Tag (EST) analysis
One clam was randomly selected for cDNA library construction at 8 h post-V. anguillarum challenge. The cDNA library was constructed using the ZAP-cDNA synthesis kit and ZAP-cDNA GigapackIII Gold cloning kit (Stratagene, Cedar Creek, Texas, USA). Random sequencing of the library using T3 primer yielded 3226 successful sequencing reactions. BLAST analysis of all the 3226 EST sequences revealed that two ESTs of 620 and 548 bp were highly similar to the previously identified sHSPs. Therefore, these EST sequences were selected for further cloning of the full-length cDNA of sHSP from V. philippinarum.
RNA isolation and cDNA synthesis
Total RNA was isolated from the haemocytes of clams using the TRIzol reagent (Invitrogen, Carlsbad, California, USA). First-strand cDNA synthesis was performed according to Promega M-MLV RT Usage information with the RQ1 RNase-Free DNase (Promega, Madison, Wisconsin, USA)-treated total RNA (1 μg) as template and oligo(dT) primer. The reactions were incubated at 42°C for 1 h, terminated by heating at 95°C for 5 min. For rapid amplification of 5′ cDNA ends, terminal deoxynucleotidyl transferase (Takara, Dalian, Liaoning, China) was used to add homopolymer dCTP tails to the 3′ end of the purified first-strand cDNA.
Cloning of the full-length cDNA of VpsHSPs by 5′RACE
Two sets of gene-specific primers, P1 and P2 for VpsHSP-1 and P3 and P4 for VpsHSP-2 (Table 1), were designed based on the two ESTs to clone the full-sequence cDNA of VpsHSP-1 and VpsHSP-2, respectively. Seminested polymerase chain reaction (PCR) approaches were introduced to get the 5′ end of the two genes with 1:50 dilution of the first-round PCR products as templates and oligodG as anchored primer. The PCR programs and PCR product sequencing were performed according to Li et al. (2009). The full-length cDNAs of VpsHSP-1 and VpsHSP-2 were verified of their validity with primer sets of P5, P6 and P6, P7, respectively.
Table 1.
Oligonucleotide primers used for cloning and expression analysis of VpsHSPs
| Primer | Sequence (5′–3′) | Sequence information |
|---|---|---|
| P1(reverse) | CCATCAGTGAAATGTAAAAGCAA | 5′RACE primer for VpsHSP-1 |
| P2(reverse) | ATTGATGTCAGACGAAGAGGATC | 5′RACE primer for VpsHSP-1 (nested primer) |
| P3(reverse) | TTCTACCTTCCCTTGAGCTATTC | 5′RACE primer for VpsHSP-2 |
| P4(reverse) | TTCTTCTTTACCCTCTTTGGACT | 5′RACE primer for VpsHSP-2 (nested primer) |
| P5(forward) | TTAGAAAGAATCAGAAAGGACAT | Full-length verified primer for VpsHSP-1 |
| P6(reverse) | TTTTTTTTTTTTTTTTTTTTTT | Full-length verified primer for VpsHSP-1 and VpsHSP-2 |
| P7(forward) | TGATCGTCAGCGAGATATGTTCT | Full-length verified primer for VpsHSP-2 |
| P8(forward) | TTTATTTTCGGACTGGGTCAAGG | Real-time primer for VpsHSP-1 |
| P9(reverse) | GGTCTGTCACAAATGGCTTCTC | Real-time primer for VpsHSP-1 |
| P10(forward) | AGGGTACGTGATGAGATGCACAG | Real-time primer for VpsHSP-2 |
| P11(reverse) | GTAACACTCCATCCTCCGACAAG | Real-time primer for VpsHSP-2 |
| P12(forward) | CTCCCTTGAGAAGAGCTACGA | Real-time actin primer |
| P13(reverse) | GATACCAGCAGATTCCATACCC | Real-time actin primer |
Sequence analysis of VpsHSPs
The VpsHSPs gene sequences were analyzed using the BLAST algorithm at NCBI Web site (http://www.ncbi.nlm.nih.gov/blast), and the deduced amino acid sequence was analyzed with the Expert Protein Analysis System (http://www.expasy.org/). Sequence alignment of VpsHSP-1 and VpsHSP-2 was performed with the ClustalW Multiple Alignment program (http://www.ebi.ac.uk/clustalw/) and Multiple Alignment show program (http://www.bio-soft.net/sms/index.html). An unrooted phylogenetic tree was constructed by the neighbor-joining (NJ) algorithm embedded in the Mega3.1 program (Kumar et al. 2004). Bootstrap analysis of 1000 replicates was carried out to determine the confidence of tree branch positions. All the analyzed sequences of sHSPs were retrieved from GenBank and SWISS-PROT database, and their accession numbers were as follows: V. philippinarum (GQ384407, GQ384408), Chlamys farreri (AY362760), Argopecten irradians (AY830445), Angiostrongylus cantonensis-1 (CAR63578), A. cantonensis-2 (CAR63522), Mamestra brassicae (BAF03557), Tribolium castaneum (XP_966780), Glossina morsitans (ADD18867), Ixodes scapularis (XP_002405513), Drosophila ananassae (XP_001963454), Locusta migratoria (ABC84493), Heliconius erato (ABS57447), Bombyx mori (NP_001036985), Homo sapiens (AAC19161), Macaca mulatta (XP_001106498), Mus musculus (NP_034094), Danio rerio (NP_001002670).
Temporal expression profile of VpsHSPs mRNA in haemocytes post-V. anguillarum challenge or cadmium exposure
The expression of VpsHSPs transcript in haemocytes after Vibrio challenge and cadmium exposure was measured by quantitative real-time reverse transcriptase PCR in Applied Biosystem 7500 fast Real-time PCR System. Two sets of gene-specific primers, P8 and P9 for VpsHSP-1 and P10 and P11 for VpsHSP-2 (Table 1), were designed to amplify products of 167 and 298 bp, respectively. These products was purified and sequenced to verify the PCR specificity. Two clam β-actin primers, P12 and P13 (Table 1), were used to amplify a 121-bp fragment as internal control to verify the successful reverse transcription and to calibrate the cDNA template. The reaction component, thermal profile and data analysis were conducted as previously described (Li et al. 2009). All data were given in terms of relative mRNA expression as means ± SE. The results were subjected to analysis of t-test, and the P values less than 0.05 were considered statistically significant.
Results
cDNA cloning and analysis of the VpsHSP-1 and VpsHSP-2 genes
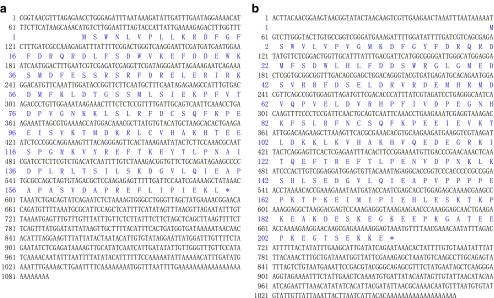
Two nucleotide sequences of 1088 and 1070 bp representing the complete cDNA sequence of VpsHSP-1 (Fig. 1a) and VpsHSP-2 (Fig. 1b) were obtained by overlapping EST and the fragments from 5′RACE, respectively. These sequences were deposited in GenBank under accession no. GQ384407 and GQ384408. The deduced amino acid sequences of VpsHSP-1 and VpsHSP-2 were shown below the corresponding nucleotide acid sequence in Fig. 1.
Fig. 1.
The nucleotide sequence (above) and its deduced amino acid sequence (below) of VpsHSP-1 (a) and VpsHSP-2 (b). Nucleotides were numbered from the first base at the 5′ end. Amino acids were numbered from the initiating methionine. The asterisk indicates the stop codon
The complete sequence of VpsHSP-1 cDNA contained a 5′ untranslated region (UTR) of 76 bp, a 3′ UTR of 490 bp with a polyA tail. Canonical polyadenylation signal sequence AATAAA was substituted by ATTAAA. An open-reading frame (ORF) of 522 bp encoded a polypeptide of 173 amino acids with the predicted molecular weight of 20.29 kDa and the theoretical isoelectric point of 6.19. The typical α-crystalline domain was located from 76aa to 171aa. The full-length cDNA of VpsHSP-2 consisted of a 5′ UTR of 58 bp, a 3′ UTR of 376 bp, and an ORF of 636 bp encoding a polypeptide of 211 amino acids with the predicted molecular weight of 24.66 kDa and the theoretical isoelectric point of 5.25. The typical α-crystalline domain was located from 78aa to 173aa.
Sequences alignment and phylogenetic analysis
Amino acid sequence alignment between VpsHSP-1 and VpsHSP-2 was shown in Fig. 2. It was observed that they were highly conserved, especially in the region of α-crystalline domain. The significant difference was located in the carboxyl ends, where 36 amino acids was absent in VpsHSP-1.
Fig. 2.
Sequences alignment of VpsHSP-1 and VpsHSP-2. The conserved amino acid residues were shaded in dark, and similar amino acids were shaded in grey
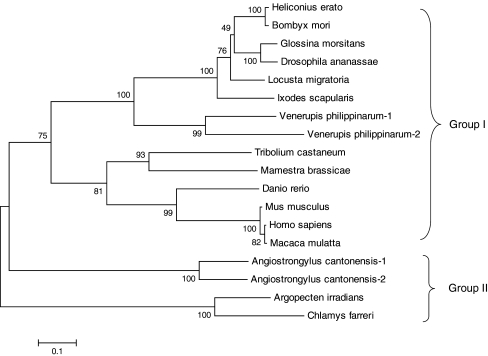
An unrooted phylogenetic tree was constructed based on the protein sequences of 18 sHSPs by the NJ method (Fig. 3). According to the phylogenetic tree, the sHSP members were mainly clustered into two groups. In the branch of group II, scallop sHSPs were clustered with A. cantonensis sHSPs and formed a sister group to the sHSPs of group I. For VpsHSPs, they were first clustered together with other invertebrate sHSP proteins and then grouped with those of vertebrate. Taking together the low similarities between VpsHSPs and scallop counterparts, it was suggested that they belonged to different subtypes of sHSP family.
Fig. 3.
Phylogenetic tree constructed by the NJ method in MEGA software based on the sHSP sequences. Bootstrap support values for the NJ tree were shown at the nodes (out of 1000 replicates)
The expression profile of VpsHSP-1 and VpsHSP-2 after Vibrio challenge
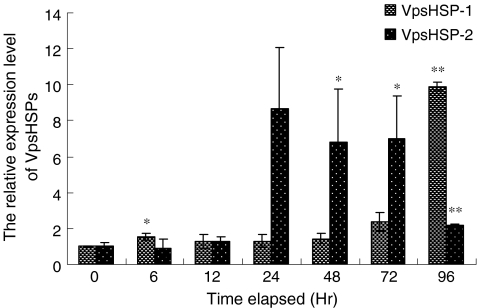
The temporal mRNA expression of the two sHSP genes in the haemocytes post-Vibrio challenge is shown in Fig. 4. During the first 6 h after pathogen challenge, the expression level of VpsHSP-1 mRNA was up-regulated and increased 1.5-fold compared with that in the control group. After that, the expression level kept almost unchanged from 12 to 48 h. A second peak of VpsHSP-1 expression was detected at 96 h postinfection, which was 9.9-fold compared with that in the control group. An unpaired two-tailed t-test with control and challenged groups showed statistically significant difference in VpsHSP-1 gene expression at 6 h (P < 0.05) and 96 h (P < 0.01) postinfection. However, no significant difference was observed in other time points of the challenge group.
Fig. 4.
Time-course expression level of VpsHSP-1 and VpsHSP-2 transcript in haemocytes after V. anguillarum infection measured by quantitative real-time PCR at 0, 6, 12, 24, 48, 72, and 96 h. Each symbol and vertical bar represented the mean±SD (n = 5). Significant differences across control were indicated with an asterisk at P < 0.05 and two asterisks at P < 0.01
The expression profile of VpsHSP-2 was different from that of VpsHSP-1. The expression level of VpsHSP-2 remained at the control level from 6 to 12 h. Then, the expression level was sharply increased and reached 8.7-fold compared with that in the control group at 24 h. After that, VpsHSP-2 mRNA level slightly decreased from 48 to 72 h. At 96 h, VpsHSP-2 mRNA level dropped greatly and only was 2.2-fold compared to the control. t-Test with control and challenged groups indicated statistically significant difference in VpsHSP-2 gene expression at 48 h (P < 0.05), 72 h (P < 0.05) and 96 h (P < 0.01) postinfection.
The expression profile of VpsHSP-1 and VpsHSP-2 after cadmium exposure
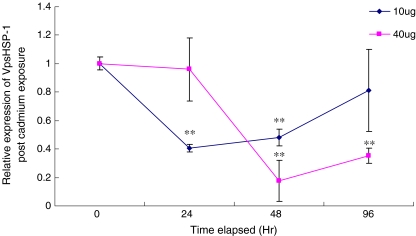
The temporal expression of VpsHSP-1 transcript after cadmium exposure was shown in Fig. 5. At the concentration of 10 μg l−1 Cd2+, the expression level of VpsHSP-1 was obviously down-regulated and reached around 0.4-fold compared with that in the control group from 24 h (P < 0.01) to 48 h (P < 0.01). At 96 h, the expression level almost recovered to the original level. When the concentration increased to 40 μg l−1, no significant change was found during the first 24 h. As time progressed, the expression of VpsHSP-1 mRNA decreased significantly, and the expression level were only 0.18-fold at 48 h (P < 0.01) and 0.35-fold at 96 h (P < 0.01) compared with that in the control group.
Fig. 5.
The VpsHSP-1 mRNA expression levels in clams exposed to different concentrations of Cd2+ after 0, 24, 48 and 96 h. Each symbol and vertical bar represented the mean±SD (n = 5). Significant differences across control were indicated with an asterisk at P < 0.05 and two asterisks at P < 0.01
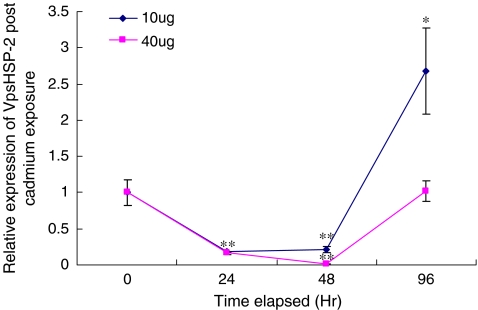
Concerning VpsHSP-2, different expression profile was detected when cadmium concentration came to 40 μg l−1 (Fig. 6). The expression level of VpsHSP-2 was significantly down-regulated from 24 h (P < 0.01) to 48 h (P < 0.01). The minimum expression level was only 1% compared with that in the control group at 48 h. After that, the expression level was sharply recovered and reached the original level at 96 h. When the concentration was 10 μg l−1, similar expression profile was found between VpsHSP-1 and VpsHSP-2. The exclusive difference occurred at 96 h post-cadmium exposure. At this time point, the expression level of VpsHSP-2 sharply up-regulated and arrived to 2.7-fold compared with that in the control group (P < 0.05).
Fig. 6.
The VpsHSP-2 mRNA expression levels in clams exposed to different concentrations of Cd2+ after 0, 24, 48 and 96 h. Each symbol and vertical bar represented the mean±SD (n = 5). Significant differences across control were indicated with an asterisk at P < 0.05 and two asterisks at P < 0.01
Discussion
Mollusk is constantly exposed to changeable environmental threats. The HSPs buffer this environmental variation by minimizing the biochemical, physiological and histological alteration of host and are therefore important factors for the maintenance of homeostasis across environmental regimes (Sørensen et al. 2003). On the other hand, mollusks have also been postulated as ideal indicator organisms for assessing levels of environmental pollution because they are ubiquitous, sedentary and filter feeders inhabiting the coastal and estuarine areas (Gao et al. 2007).
In this article, the complete cDNA sequences of two clam sHSP genes VpsHSP-1 and VpsHSP-2 are reported. The two sHSPs had the identical α-crystalline domain not only in size (96 amino acids) but in location. ClustalW analysis revealed that the deduced amino acid sequence of VpsHSP-1 and VpsHSP-2 displayed high identity with other registered sHSPs. For example, VpsHSP-1 shared 56% identities to VpsHSP-2, 48% to Caenorhabditis elegans HSP25 isoform C (AAW57827), 45% to Aedes aegypti sHSP (EDS34135), 43% to I. scapularis HSP20.6 (EEC06453) and 37% to B. mori HSP21.4 (BAD74197). The sequence alignment and structure comparison together favored that these proteins were new members of the sHSP family. However, VPsHSPs shared fewer similarities to scallop counterparts with identities less than 20%, which indicated they belonged to different subtypes of sHSP family.
As stress proteins, HSPs are expressed in response not only to a wide range of abiotic stressors, but also to biological stressors such as infectious pathogens (Song et al. 2006). Usually, HSPs increase dramatically in quantity after viral or bacterial infection (Hickman-Miller and Hildebrand 2004). In scallop, HSP22 transcription was up-regulated and reached a maximal level at 12 h after Vibrio challenge, and then declined progressively to the original level at 48 h (Zhang et al. 2009b). In the present study, the effects of pathogen on VpsHSPs gene expression were investigated and different expression profiles were detected. For VpsHSP-1 expression, only the 6- and 96-h treatments are statistically different from the control with 1.5- and 9.9-fold, respectively. With regard to VpsHSP-2, the expression level increased sharply from 48 to 72 h after “stable phase” at the first 24 h. These different expression trends between the two genes indicated that they were involved in host innate immunity in different manners. To cope with pathogen infection, VpsHSP-1 might be acted as the acute response protein and VpsHSP-2 as the late-stage one. The different expression profile between sHSPs from clam and scallop further elucidated that they belonged to different subtypes of sHSP family.
There is growing evidence that environmental contaminants may be partially responsible for the disease outbreak of marine organisms by adversely affecting their immune system. As a major toxic metal in sea water, Cd2+ is responsible for many toxic effects such as generating abnormal or denatured proteins and causing oxidative stress in various living organisms (Gao et al. 2007). Heat shock proteins were a promising candidate marker for monitoring the marine heavy mental pollution. In scallop, HSP90 responded to various heavy metal stresses (Cd2+, Cu2+, Pb2+) with a dose-dependent expression pattern as well as exposure time (Gao et al. 2007). A clearly time-dependent expression pattern of HSP70 in bay scallop was also observed by Song et al. (2006). In the present study, the expression of VpsHSPs did not change linearly along with the rise of Cd2+ concentration. However, many previous studies have showed that the expression of sHSPs at transcriptional level was increased responding to Cd2+ stress. In scallops, the mRNA level of AiHSP22 was up-regulated after a 10-day exposure to Cd2+ (Zhang et al. 2009a). In Xenopus laevis A6 kidney epithelial cells, the HSP30 mRNA was elevated when exposed to cadmium (Woolfson and Heikkila 2009). The down-regulation of VpsHSP-1 and VpsHSP-2 expression level after Cd2+ exposure was similar to the previous study by Huang et al. (2008), who reported that HSP21 gene expression in shrimp Penaeus monodon was inactivated after white spot syndrome virus (WSSV) infection. Recently, sHSPs have been discovered to have the function in blocking apoptosis (Mehlen et al. 1996; Oshita et al. 2010). The decrease of VpsHSPs mRNA was perhaps consistent with their roles as negative regulators in apoptosis. Further investigation should be conducted to reveal the explicit roles of VpsHSPs involved in the responses to pathogen and cadmium stress.
Acknowledgements
The project was supported by the Key Laboratory of Experimental Marine Biology (Institute of Oceanology, CAS) and Chinese Academy of Sciences Innovation Program (kzcx2-yw-225, KZCX2-YW-Q07-04), and grants from NSFC (30901115) and SDSFC (ZR2009CZ008).
References
- Gao Q, Song L, Ni D, Wu L, Zhang H, Chang Y. cDNA cloning and mRNA expression of heat shock protein 90 gene in the haemocytes of Zhikong scallop Chlamys farreri. Comp Biochem Physiol. 2007;147B:704–715. doi: 10.1016/j.cbpb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Haslbeck M. sHsps and their role in the chaperone network. Cell Mol Life Sci. 2002;59:1649–1657. doi: 10.1007/PL00012492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman-Miller HD, Hildebrand WH. The immune response under stress: the role of HSP-derived peptides. Trends Immunol. 2004;25:427–433. doi: 10.1016/j.it.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Huang P, Kang S, Chen W, Hsu T, Lo C, Liu K, Chen L. Identification of the small heat shock protein, HSP21, of shrimp Penaeus monodon and the gene expression of HSP21 is inactivated after white spot syndrome virus (WSSV) infection. Fish Shellfish Immunol. 2008;25:250–257. doi: 10.1016/j.fsi.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Li Z, Menoret A, Srivastava P. Roles of heat-shock proteins in antigen presentation and cross-presentation. Curr Opin Immunol. 2002;14:45–51. doi: 10.1016/S0952-7915(01)00297-7. [DOI] [PubMed] [Google Scholar]
- Li C, Sun H, Chen A, Ning X, Wu H, Qin S, Xue Q, Zhao J. Identification and characterization of an intracellular Cu, Zn-superoxide dismutase (icCu/Zn-SOD) gene from clam Venerupis philippinarum. Fish Shellfish Immunol. 2009 doi: 10.1016/j.fsi.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Mehlen P, Schulze-Osthoff K, Arrigo AP. Small stress proteins as novel regulators of apoptosis. J Biol Chem. 1996;271:16510–16514. doi: 10.1074/jbc.271.28.16510. [DOI] [PubMed] [Google Scholar]
- Mizrahi T, Heller J, Goldenberg S, Arad Z. Heat shock proteins and resistance to desiccation in congeneric land snails. Cell Stress Chaperon. 2009 doi: 10.1007/s12192-009-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G, Inaguma Y, Kato K, Anguay RM T. The small heat shock protein hsp22 of Drosophila melanogaster is a mitochondrial protein displaying oligomeric organization. J Biol Chem. 2000;275:31204–31210. doi: 10.1074/jbc.M002960200. [DOI] [PubMed] [Google Scholar]
- Moseley P. Stress proteins and the immune response. Immunopharmacology. 2000;48:299–302. doi: 10.1016/S0162-3109(00)00227-7. [DOI] [PubMed] [Google Scholar]
- Oshita SE, Chen F, Kwan T, Yehiely F, Cryns VL. The small heat shock protein HspB2 is a novel anti-apoptotic protein that inhibits apical caspase activation in the extrinsic apoptotic pathway. Breast Cancer Res Treat. 2010 doi: 10.1007/s10549-010-0735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pockley AG. Heat shock proteins as regulators of the immune response. Lancet. 2003;362:469–476. doi: 10.1016/S0140-6736(03)14075-5. [DOI] [PubMed] [Google Scholar]
- Pomerai D. Heat-shock proteins as biomarkers of pollution. Hum Exp Toxicol. 1996;15:279–285. doi: 10.1177/096032719601500401. [DOI] [PubMed] [Google Scholar]
- Solé M, Morcillo Y, Porte C. Stress-protein response in tributyltin-exposed clams. Bull Environ Contam Toxicol. 2000;64:852–858. doi: 10.1007/s001280000081. [DOI] [PubMed] [Google Scholar]
- Sørensen JG, Kristensen TN, Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecol Lett. 2003;6:1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x. [DOI] [Google Scholar]
- Song L, Wu L, Ni D, Chang Y, Xu W, Xing K. The cDNA cloning and mRNA expression of heat shock protein 70 gene in the haemocytes of bay scallop (Argopecten irradians, Lamarck 1819) responding to bacteria challenge and naphthalin stress. Fish Shellfish Immunol. 2006;21:335–345. doi: 10.1016/j.fsi.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Srivastava PK. Role of heat-shock proteins in innate and adaptive immunity. Nat Rev Immunol. 2002;2:185–194. doi: 10.1038/nri749. [DOI] [PubMed] [Google Scholar]
- Thomas X, Campos L, Le Q, Guyotat D. Heat shock proteins and acute leukemias. Hematology. 2005;10:225–235. doi: 10.1080/10245330500093120. [DOI] [PubMed] [Google Scholar]
- Tsan MF, Gao B. Heat shock protein and innate immunity. Cell Mol Immunol. 2004;1:274–279. [PubMed] [Google Scholar]
- Tsan MF, Gao B. Cytokine function of heat shock proteins. Am J Physiol. 2004;286:C739–C744. doi: 10.1152/ajpcell.00364.2003. [DOI] [PubMed] [Google Scholar]
- Tsvetkova NM, Horváth I, Zs T, Wolkers WF, Balogi Z, Shigapova N, Crowe LM, Tablin F, Vierling E, Crowe JH, Vígh L. Small heat-shock proteins regulate membrane lipid polymorphism. Proc Natl Acad Sci USA. 2002;99:13504–13509. doi: 10.1073/pnas.192468399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin RP, Lundqvist A, More SH, Bonin A, Kiessling R, Ljunggren HG. Heat shock proteins as activators of the innate immune system. Trends Immunol. 2002;23:l30–135. doi: 10.1016/S1471-4906(01)02168-8. [DOI] [PubMed] [Google Scholar]
- Woolfson JP, Heikkila JJ. Examination of cadmium-induced expression of the small heat shock protein gene, hsp30, in Xenopus laevis A6 kidney epithelial cells. Comp Biochem Physiol. 2009;152A:91–99. doi: 10.1016/j.cbpa.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang L, Song L, Zhao J, Qiu L, Dong C, Li F, Zhang H, Yang G. The involvement of HSP22 from bay scallop Argopecten irradians in response to heavy metal stress. Mol Biol Rep. 2009 doi: 10.1007/s11033-009-9603-6. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang L, Zhao J, Qiu L, Song L, Dong C, Li F. The responsive expression of heat shock protein 22 gene in Zhikong scallop Chlamys farreri against a bacterial challenge. Aquac Res. 2009;41:257–266. doi: 10.1111/j.1365-2109.2009.02328.x. [DOI] [Google Scholar]