Abstract
The mitochondrial Hsp60 chaperonin plays an important role in sustaining cellular viability. Its dysfunction is related to inherited forms of the human diseases spastic paraplegia and hypomyelinating leukodystrophy. However, it is unknown whether the requirement for Hsp60 is neuron specific or whether a complete loss of the protein will impair mammalian development and postnatal survival. In this study, we describe the generation and characterization of a mutant mouse line bearing an inactivating gene-trap insertion in the Hspd1 gene encoding Hsp60. We found that heterozygous mice were born at the expected ratio compared to wild-type mice and displayed no obvious phenotype deficits. Using quantitative reverse transcription PCR, we found significantly decreased levels of the Hspd1 transcript in all of the tissues examined, demonstrating that the inactivation of the Hspd1 gene is efficient. By Western blot analysis, we found that the amount of Hsp60 protein, compared to either cytosolic tubulin or mitochondrial voltage-dependent anion-selective channel protein 1/porin, was decreased as well. The expression of the nearby Hspe1 gene, which encodes the Hsp10 co-chaperonin, was concomitantly down regulated in the liver, and the protein levels in all tissues except the brain were reduced. Homozygous Hspd1 mutant embryos, however, died shortly after implantation (day 6.5 to 7.5 of gestation, Theiler stages 9–10). Our results demonstrate that Hspd1 is an essential gene for early embryonic development in mice, while reducing the amount of Hsp60 by inactivation of one allele of the gene is compatible with survival to term as well as postnatal life.
Keywords: Chaperonin 60; Chaperonin 10; Embryonic development; Gene knockout techniques; Insertional mutagenesis, OmniBank®
Introduction
The mitochondrial chaperonin Hsp60 is a highly conserved protein during evolution and plays a crucial role in cell maintenance and survival (Horwich et al. 2007). It belongs to the chaperonin subgroup of the molecular chaperones, which are characterized by their ability to form high-molecular weight oligomers that mediate adenosine triphosphate (ATP)-dependent folding of proteins that are resistant to the actions of more simple chaperones like Hsp70 (Bukau and Horwich 1998). Together with its co-chaperonin Hsp10, Hsp60 forms a large multisubunit complex that is one of the most important factors in the facilitation of the proper folding and assembly of proteins in the mitochondrial matrix. In addition to its chaperone activity, Hsp60 appears to have anti- or proapoptotic roles (Arya et al. 2007; Chandra et al. 2007) and immunoregulatory properties (Habich and Burkart 2007; Pockley et al. 2008). The homologous complex in Escherichia coli, GroEL/GroES has been extensively studied (Hartl and Hayer-Hartl 2009).
Inactivation and mutations have demonstrated that the orthologous genes encoding Hsp60 in E. coli, Saccharomyces cerevisiae, and Drosophila melanogaster (Cheng et al. 1989; Hemmingsen et al. 1988; Perezgasga et al. 1999) are essential genes, indicating that Hsp60 plays an important role both in prokaryotic organisms and as a component of the mitochondria in eukaryotes.
Mutations in the nuclear gene that encodes Hsp60 in humans (HSPD1) have been associated with autosomal dominantly inherited hereditary spastic paraplegia (Hansen et al. 2002, 2007) and with an autosomal recessively inherited hypomyelinating leukodystrophy termed MitCHAP-60 disease (Magen et al. 2008). In its pure form, hereditary spastic paraplegia is characterized by a progressive development of weakness and spasticity of the lower limbs as well as a subtle impairment in vibratory sense (i.e., impaired perception of a vibratory stimulus). More than 30 hereditary spastic paraplegia gene loci have been mapped, and specific mutations that result in disease have been identified in 17 different genes. Mutations in the HSPD1 gene are among the more rare causes of the disease (Svenstrup et al. 2009).
On the basis of studies of several genes that cause hereditary spastic paraplegia, it has been hypothesized that a defect in the genes leads to impairment of the axonal transport of macromolecules, organelles, and other cargoes primarily affecting the longest neurons in the spinal cord (Salinas et al. 2008). That the development of spastic paraplegia symptoms is caused by degeneration of primarily the spinal cord motorneurons is supported by post-mortem findings in patients (Deluca et al. 2004) and the evidence of dying-back axonopathy in paraplegin-deficient (Ferreirinha et al. 2004) and spastin-deficient mouse models (Kasher et al. 2009; Tarrade et al. 2006).
On the basis of these observations, it is clear that mitochondrial Hsp60 plays an important role in sustaining cellular viability and that its dysfunction is associated with human inherited diseases. It still remains unclear as to whether the requirement for Hsp60 is neuron specific or whether the protein is more generally essential for mammalian development and postnatal survival. In the present study, we describe the generation and characteristics of a mutant mouse line that harbors an inactivating gene-trap insertion in the Hspd1 gene encoding Hsp60. We show that homozygosity for the null allele causes early embryonic lethality, while heterozygosity for the inactivated allele permits embryonic development and postnatal survival.
Materials and methods
Generation of mice heterozygous for an inactivating insertion in the Hspd1 gene
Mice that were heterozygous for an inactivating insertion in the Hspd1 gene were obtained from Lexicon Pharmaceuticals, Inc. (The Woodlands, TX). They were produced using mouse embryonic stem cells (129/SvEvBrd) that contained a gene-trap vector insertion in intron 2 in one allele of the Hspd1 gene (cell clone OST171441). The gene-trap vector (OmniBank VICTR48 comprising 5,174 base pairs), which contains a promoter-less neomycin resistance gene and a triple polyadenylation sequence, surrounded by splice acceptor and donor sequences (Fig. 1a), leads to the production of a mature Hspd1 exon 1–2/neomycin fusion transcript as first determined by 5′ rapid amplification of complementary DNA (cDNA) ends (RACE) PCR performed by Lexicon Pharmaceuticals, Inc. The OST171441 embryonic stem cells were expanded and injected into host mouse blastocysts. The blastocysts were implanted into pseudopregnant females and resulted in four male chimeras, which were then mated to C57BL/6J albino females. Two of the chimeras bred successfully, and one had germline transmission of the gene-trap insertion as indicated by the presence of the inactivated allele in the offspring produced. All investigations were carried out in F1 offspring from heterozygous intercrossings unless otherwise indicated.
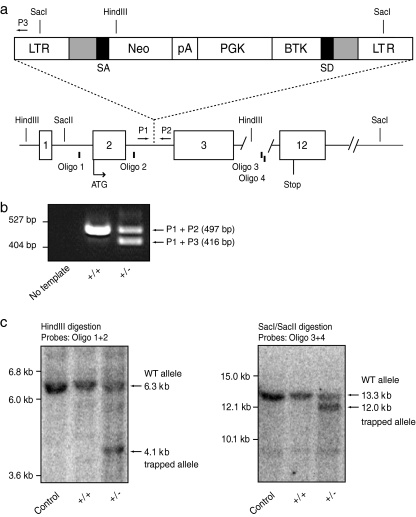
Fig. 1.
Retroviral insertion and disruption of the murine Hspd1 gene. a Schematic representation of the VICTR48 gene-trap vector (top) and the modified region of the mouse Hspd1 gene (bottom). The retroviral construct, which contains long terminal repeats (LTRs) for genomic insertion, is located between exon 2 and exon 3 of the Hspd1 gene. The modified allele is expected to prevent normal splicing of exons 2 and 3 by the production of a mature Hspd1 exon1–2/neomycin fusion transcript. Only relevant restriction endonuclease sites are shown. Schematic, not drawn to scale. b PCR-based identification of Hspd1+/+ (+/+) and Hspd1+/− (+/−) mice. Routine multiplex PCR with Hspd1-specific (P1 and P2) and LTR-specific (P3) primers produces two independent products of 497 base pairs (P1-P2) and 416 base pairs (P1-P3) for, respectively, the Hspd1 WT allele and the trapped allele. No PCR product spanning the entire gene-trap insertion was produced from the trapped allele by the P1 and P2 primers under the present amplification conditions. PCR primer locations are shown in a (arrows) and described under Materials and Methods. c Southern blotting analysis of the Hspd1 locus using genomic DNA isolated from Hspd1+/+ (+/+) and Hspd1+/− (+/−) liver tissue. Samples were digested with either HindIII (left panel) or SacI/SacII (right panel), and fragments were detected with oligonucleotide probe mixtures distributed in the 5′ (oligo 1 + 2, left panel) or 3′ (oligo 3 + 4, right panel) flanking regions of the insertion point. Control genomic DNA isolated from mouse strain 129 embryonic stem cells. Oligonucleotide probe locations are shown in a (bars) and described under Materials and Methods. Neo promoter-less neomycin resistance gene, pA triple polyadenylation sequence, PGK phosphoglycerate kinase promoter, BTK Bruton's tyrosine kinase exon 1, SD splice donor site, SA splice acceptor site, WT wild type
Genotyping
Oligonucleotide primers (P1: Hspd1 upper, 5′-TAAGACAGCATTTCTCCGGTAG-3′; P2: Hspd1 lower, 5′-CTGAGTGTTGGGATTATGCAG-3′; and P3: LTR lower, 5′-GCCAGTCCTCCGATTGAC-3′) were used concomitantly in a multiplex PCR to amplify corresponding Hspd1 alleles on mouse chromosome 1. The expected sizes of the PCR products from the wild-type and trapped alleles are 497 and 416 base pairs, respectively. Purified mouse tail genomic DNA (10–500 ng) was used as a template for the PCR in a 25-μL reaction volume. Cycling conditions were 95°C for 30 s, 54°C for 30 s, and 72°C for 30 s (40 cycles). Amplified products were separated on 1.5% agarose gels (Fig. 1b). The identity of the individual bands was confirmed by bidirectional direct sequencing using BigDye Terminator v1.1 Cycle Sequencing Kit and ABI PRISM® 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, CA).
Southern blotting
Total genomic DNA was isolated from liver tissue using the Puregene DNA Purification Kit (Gentra Systems, Minneapolis, MN) adding proteinase K during cell lysis for maximum yield. RNase treatment, protein precipitation, DNA precipitation, and DNA hydration were performed exactly according to the manufacturer’s protocol. Only DNA samples having an A260/A280 OD ratio between 1.8 and 2.0 were used. Restriction endonuclease digestion was performed with standard procedures, using 10-µg genomic DNA. The resulting DNA fragments were separated by electrophoresis in a 0.7% agarose Tris–borate–ethylenediaminetetraacetic acid (EDTA) gel for 357 Vh and blotted to a Zeta Probe nylon membrane (Bio-Rad Laboratories, Hercules, CA) by capillary blotting. For sensitive and specific detection of DNA fragments of the Hspd1 gene, Starfire oligonucleotide probes with 3′ hexamer extension were selected and produced (Integrated DNA Technologies, Coralville, IA) to fulfill the following criteria: (1) no presence of repetitive sequences, (2) no presence of exonic sequences, (3) Tm > 85°C, (4) guanine–cytosine content > 50%, (5) 50–55 base pairs long, and (6) >95% full length (high-performance liquid chromatography purified). Sequences of the four probes selected (oligo 1–4) are available upon request. Labeling of the probes was achieved by annealing polyT to the 3′ extension of the template oligonucleotides and subsequent addition of 60 µCi [α-32P]dATP (6,000 Ci/mmol) per 0.5-pmol oligonucleotide by DNA polymerase extension using 5U Exo− Klenow DNA polymerase (Stratagene, La Jolla, CA) and subsequent purification on ProbeQuant G25 microcolumns (GE Health Care, Uppsala, Sweden) according to the manufacturer's instructions. Hybridization of probe mixtures (oligo 1 + 2 or oligo 3 + 4) to the membrane was performed at 60°C using standard laboratory procedures. Visualization was performed using a STORM 840 PhosphorImager (Molecular Dynamics, Sunnyvale, CA) and the ImageQuant 5.0 software (Molecular Dynamics). The expected sizes of the fragments produced by HindIII and SacI/SacII digestion are, respectively, 6.6 and 13.3 kb for the wild-type allele and 4.1 and 12.0 kb for the trapped allele.
Quantitative reverse transcription PCR
Tissues were dissected from 5-month-old animals and stored at −80°C in RNAlater RNA Stabilization Reagent according to the manufacturer’s instructions (Qiagen Sciences, Germantown, MD). Total RNA was isolated from frozen sections using the SV Total RNA Isolation System (Promega, Madison, WI) with a DNase I treatment included. Only RNA samples that showed no apparent degradation when subjected to denaturing gel electrophoresis and had an A260/A280 OD ratio above 1.8 were analyzed. cDNA was synthesized from 1-μg total RNA in a 20-μL reaction with 100-pmol “anchored” oligo(dT) primers (18T + N), using the Advantage RT-for-PCR Kit (Clontech, Mountain View, CA) according to the manufacturer’s instructions.
Relative quantification of cDNA by PCR was performed with the ABI7000 real-time sequence detection system and TaqMan probe chemistry (Applied Biosystems, Nærum, Denmark) in a manner similar to that described previously (Hansen et al. 2008). Probes for Hspd1 and Hspe1 transcripts were labeled with 5-carboxyfluorescein in the 5′ end and a nonfluorescent quencher group in the 3′ end. Primers and probes were designed using Primer Express software (Applied Biosystems) and sequences are available upon request. Probes were designed to span exon boundaries to avoid detection of potential contaminating genomic DNA. PCR was carried out in a 25-μL reaction mixture containing 2-μL cDNA, 250-nM probe, 900-nM of each primer, and universal PCR master mix (Applied Biosystems). The PCR conditions were 95°C for 10 min and 40 reaction cycles at 95°C for 15 s and 60°C for 1 min. Each cDNA sample was analyzed in triplicate.
Relative gene expression was calculated by the “standard curve method” (Heid et al. 1996; Winer et al. 1999), and the expression of the gene of interest was normalized to the expression of the endogenous beta-actin control gene, Actb (Assay Mm00607939_s1, Applied Biosystems).
Western blotting
Frozen tissue samples dissected from 5-month-old animals (stored at −80°C) were homogenized in a lysis buffer: 50-mM Tris–HCl (pH 7.7), containing 5-mM EDTA, 1% Triton X-100, 1-mM dithiothreitol, 0.5-μg/mL leupeptin, 10-μg/mL aprotinin, and one Complete Protease Inhibitor Cocktail Tablet (Roche, Basel, Switzerland) per 10 mL of lysis buffer, followed by three freeze–thaw cycles and a 30-s burst in a cooled water bath sonicator. The resulting extract was centrifuged at 4°C for 30 min at 15,000×g, and the protein concentration of the supernatant was determined by Bradford protein assay (Bio-Rad Laboratories). Sample aliquots, representing identical total amounts of protein, were prepared in a standard sodium dodecyl sulfate sample buffer, boiled for 5 min, and separated on either 4–15% gradient or 15% polyacrylamide gel electrophoresis gels for the detection of, respectively, Hsp60 and Hsp10. Proteins were blotted onto polyvinylidene fluoride membranes (Millipore, Copenhagen, Denmark) and developed using horseradish peroxidase-conjugated secondary antibodies (DAKO, Copenhagen, Denmark) and the enhanced chemiluminescence ECL+ Western Blotting Detection System (GE Healthcare, Hillerød, Denmark) as recommended by the manufacturer. The following primary antibodies were used to detect the Hsp60 and Hsp10 chaperonins, respectively: mouse monoclonal anti-Hsp60 antibody (H-3524, Sigma-Aldrich, St. Louis, MO) diluted 1:25,000 and rabbit polyclonal anti-Hsp10 antibody (SPA-110, Stressgen, Assay Designs, Ann Arbor, MI) diluted 1:10,000. The following primary antibodies were used as, respectively, cytosolic and mitochondrial loading controls: mouse monoclonal anti-tubulin-beta (MS-582-P, NeoMarkers, Union City, CA) diluted 1:10,000 and mouse monoclonal anti-VDAC-1/Porin antibody (ab14734, Abcam, Cambridge, UK) diluted 1:5,000. Images were obtained with the ChemiDoc-It Imaging System (UVP, LLC, Upland, CA), and protein levels were estimated using the analysis software VisionWorksLS Image Acquisition and Analysis Software (UVP).
Histological analysis of embryos
Upon timed mating, female mice were killed by cervical dislocation at the days indicated. Embryos were fixed in the uterus with 4% freshly dissolved paraformaldehyde in phosphate-buffered saline (PBS; without Ca2+ and Mg2+) overnight. After washing for 1 h in PBS, embryos were dehydrated and embedded in the following dilutions of isopropanol: 30% for 3 h, 50% for 2 h, 75% for 3 h, 90% for 6 h, 100% for two times 4 h, 1:1 isopropanol–paraffin for 12 h at 60°C, and 100% paraffin for 8 h at 60°C. Paraffin blocks were cut into 5-μm sections and mounted on glass slides. Sections were rehydrated and stained with hematoxylin and eosin (HE) before light microscopy analysis.
Statistical analysis
Group means were compared by the Student’s two-sample t test using the Microsoft Office Excel 2007 software (Microsoft Corporation). Observed genotype distributions were compared to the expected distributions by the Pearson’s χ2 test, using the Vassar Stats Web Site for Statistical Computation (http://faculty.vassar.edu/lowry/VassarStats.html). P values < 0.05 were regarded as statistically significant.
Animals and housing
Breeding and experiments were conducted according to institutional and national guidelines for the care and use of laboratory animals and were approved by the Danish Experimental Animal Inspectorate, Ministry of Justice. Animals were kept in standard plastic cages on wood-chip bedding with nest material, wooden chewing blocks, and metal houses for environmental enrichment. They were kept in a 12-h light/dark cycle with ad libitum access to tap water and standard laboratory diet.
Results
Generation of heterozygous mutant mice
A mouse embryonic stem cell clone carrying a gene-trap insertion in intron 2 of the Hspd1 gene (Fig. 1a) as determined by 5′ RACE PCR was used to generate heterozygous Hspd1+/Gt(OST171441)Lex (Hspd+/−) mice by blastocyst injection and breeding of transgenic offspring.
A BLAST analysis of the Hspd1 cDNA sequence against the mouse genome revealed the presence of a set of matching regions on several different chromosomes. These regions included at least nine processed Hspd1 pseudogenes (derived by retrotransposition of messenger RNA [mRNA]) and six nonprocessed Hspd1 pseudogenes (derived by gene duplication) as determined by searching the mouse pseudogene database (Zhang et al. 2004). In order to confirm the presence of the gene-trap insertion in the Hspd1 gene, DNA was isolated from liver tissue from heterozygous Hspd1+/− mice and their Hspd1+/+ wild-type littermates and subjected to Southern blot analysis (Fig. 1c). Bands of expected sizes were obtained using two different combinations of restriction enzymes (HindIII and SacI/SacII), in combination with oligonucleotide probes complementary to intron regions either upstream (oligo 1 + 2) or downstream (oligo 3 + 4) of the gene-trap insertion. This confirmed the integration of the gene-trap insertion in the genuine Hspd1 gene as neither the intron-less processed Hspd1 pseudogenes nor the mutated non-processed Hspd1 pseudogenes would be detected by the present panel of highly specific intronic oligonucleotide probes.
The exact integration site of the gene-trap vector in intron 2 of the Hspd1 gene, 967 base pairs downstream of exon 2, was determined by PCR and subsequent DNA sequence analysis (data not shown). The gene-trap insertion is predicted to eliminate completely the normal Hspd1 gene product by preventing normal splicing of exons 2 and 3 and results in the production of an Hspd1 exon 1–2/neomycin fusion transcript. The presence of the phosphoglycerate kinase promoter (PGK) and a modified Bruton's tyrosine kinase exon 1 (BTK) with stop codons in all three reading frames upstream of the gene-trap splice donor site is predicted to result in a fusion transcript between BTK and exons 3–12 of the Hspd1 gene. This transcript will be a substrate for nonsense-mediated mRNA decay due to the presence of premature stop codons and further prevent proper splicing of exons 2 and 3 of the Hspd1 gene as well as the translation of downstream exons (Zambrowicz et al. 2003).
The effect of the gene-trap insertion on Hspd1 mRNA levels
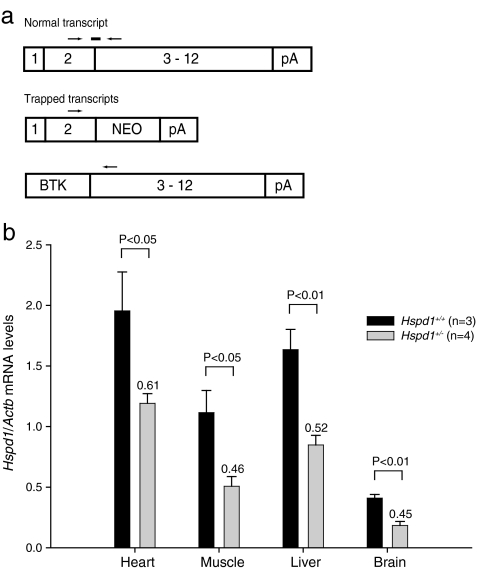
In order to determine whether the gene trap insertion is partially circumvented by direct splicing from exon 2 to exon 3 of the Hspd1 gene, we performed quantitative RT-PCR analysis on RNA extracted from various tissues using a TaqMan probe-based assay with a probe spanning the Hspd1 exon 2–exon 3 boundary (Fig. 2a). By this method, we found that the levels of correctly spliced Hspd1 mRNA, normalized to beta-actin (Actb) mRNA as an internal control, were significantly decreased in heterozygous Hspd1+/− mice (n = 4) compared to their Hspd1+/+ wild-type littermates (n = 3) in all tissues examined (Fig. 2b). The ratios between the levels of correctly spliced Hspd1 mRNA within the two groups (Hspd1+/− compared to Hspd1+/+) were 0.61 ± 0.19, 0.46 ± 0.18, 0.52 ± 0.13, and 0.45 ± 0.15 (mean ± SD) for the heart, muscle, the liver, and the brain, respectively. These values are not statistically different from the 50% reduction expected in heterozygous animals if the gene-trap insertion leads to complete transcriptional truncation of the trapped allele of the Hspd1 gene without compensatory transcriptional up-regulation of the wild-type allele.
Fig. 2.
Quantitative RT-PCR analysis of the Hspd1 gene. a The location of the TaqMan probe (solid bar) and primers (arrows) used for quantitative RT-PCR analysis of the Hspd1 gene in schematic representations of the normal and trapped transcripts. The probe was designed to span the Hspd1 exon 2–exon 3 boundary to allow specific detection of only the Hspd1 mRNA produced by normal splicing of exons 2 and 3. Neo, promoter-less neomycin resistance gene; pA, poly(A) tail; BTK, Bruton's tyrosine kinase exon 1. Schematic, not drawn to scale. b Quantitative RT-PCR analysis of the Hspd1 gene on RNA extracted from various tissues. The levels of correctly spliced Hspd1 mRNA normalized to Actb mRNA as an internal control were significantly decreased in heterozygous Hspd1+/− mice (n = 4) compared to their Hspd1+/+ wild-type littermates (n = 3) in all tissues examined. Values plotted are mean ± SEM. The ratios between the levels within the two groups (Hspd1+/− compared to Hspd1+/+) are indicated above the bars
The effect of the gene-trap insertion on Hsp60 protein levels
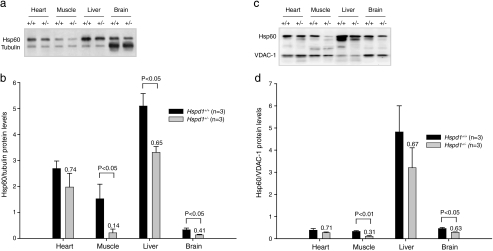
To investigate whether or not the reduced levels of correctly spliced Hspd1 mRNA were accompanied by decreased Hsp60 protein levels, we performed a semiquantitative Western blot analysis using an anti-Hsp60 antibody on protein extracted from the same panel of tissues and animals as analyzed with quantitative RT-PCR. As a loading control, the Western blots were analyzed concomitantly using an anti-tubulin antibody. Protein bands with the expected apparent molecular weights (Hsp60 60 kDa and tubulin 55 kDa) were detected in all tissues (Fig. 3a). Based upon densitometric analysis, we found that the levels of immunoreactive Hsp60 relative to cytosolic tubulin were significantly decreased in heterozygous Hspd1+/− mice, compared to Hspd1+/+ wild-type littermates, in all tissues examined (Fig. 3b), except in the heart. The ratios between the levels within the two groups (Hspd1+/− compared to Hspd1+/+) were 0.74 ± 0.30, 0.14 ± 0.19, 0.65 ± 0.13, and 0.41 ± 0.18 (mean ± SD) for the heart, muscle, the liver, and the brain, respectively.
Fig. 3.
Semi-quantitative Western blot analysis using an anti-Hsp60 antibody on protein extracted from various tissues. a A representative blot demonstrating that protein bands of the expected molecular weights (Hsp60 60 kDa and tubulin 55 kDa) were detected in all tissues analyzed. b Densitometric analysis of protein amounts revealed that the relative levels of immunoreactive Hsp60 to cytosolic tubulin were decreased in heterozygous Hspd1+/− mice (n = 3) compared to Hspd1+/+ wild-type littermates (n = 3) in all tissues examined, however, not to a statistically significant level in heart. Values plotted are mean ± SEM. The ratios between the levels within the two groups (Hspd1+/− compared to Hspd1+/+) are indicated above the bars. c A representative blot demonstrating that protein bands of the expected molecular weights (Hsp60 60 kDa and VDAC-1 39 kDa) were detected in all tissues analyzed. d Densitometric analysis of protein amounts revealed that the relative levels of immunoreactive Hsp60 to mitochondrial VDAC-1 were decreased in heterozygous Hspd1+/− mice (n = 3) compared to Hspd1+/+ wild-type littermates (n = 3) in all tissues examined, however, not to a statistically significant level in the heart and the liver. Values plotted are mean ± SEM. The ratios between the levels within the two groups (Hspd1+/− compared to Hspd1+/+) are indicated above the bars
In order to test whether or not decreased levels of Hsp60 protein were accountable to decreased amount of mitochondrial protein in the tissues examined, we performed a similar analysis substituting the antibody against cytosolic tubulin with an antibody directed against the mitochondria-specific protein, VDAC-1 as a loading control. Protein bands with the expected apparent molecular weights (Hsp60 60 kDa and VDAC-1 39 kDa) were detected in all tissues (Fig. 3c). The levels of immunoreactive Hsp60 relative to mitochondrial VDAC-1 were decreased in heterozygous Hspd1+/− mice, compared to Hspd1+/+ wild-type littermates, in all tissues examined (Fig. 3d), however, not to a statistically significant level in the heart and the liver. The ratios between the levels within the two groups (Hspd1+/− compared to Hspd1+/+) were 0.71 ± 0.29, 0.31 ± 0.20, 0.67 ± 0.43, and 0.63 ± 0.12 (mean ± SD) for the heart, muscle, the liver, and the brain, respectively. Thus, the levels of Hsp60 are decreased both in relation to cytosolic tubulin and mitochondrial VDAC-1 arguing that the total amount of mitochondrial protein is not reduced in the heterozygous Hspd1+/− mice.
Generation of homozygous mutant mice
Heterozygous Hspd1+/− mice were bred in order to obtain homozygous Hspd−/− mice, and weaned pups were genotyped by PCR analysis (Table 1). Genotype distribution among 114 weaned animals from this breeding diverged significantly from Mendelian expectations (p < 0.0001) in that no homozygous Hspd1−/− mice were detected. Wild-type Hspd1+/+ and heterozygous Hspd1+/− mice were born in the expected Mendelian ratio, i.e., 1:2 (p = 0.923, i.e., the probability for a 1:2:0 genotype distribution given the observed numbers, see Table 1).
Table 1.
Genotypes of offspring derived from crosses between Hspd1+/− mice
| No. of mice with indicated genotype among 114 offspring | Mendelian expectation 1:2:1 (P value) | Mendelian expectation 1:2:0 (P value) | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| 40 | 74 | 0 | <0.0001a | 0.923a |
aProbability of the indicated Mendelian outcome as determined by a χ2 test
The effects of Hsp60 deficiency during embryonic development
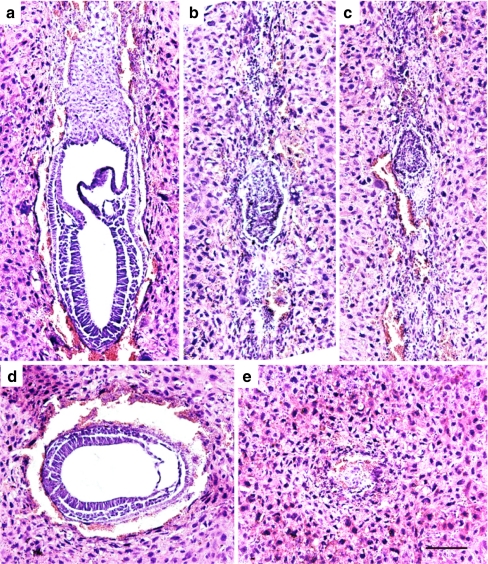
In order to determine at what time point homozygous Hspd1−/− embryos were lost, we made histological examinations of embryos derived from heterozygous Hspd1+/− intercrosses at various developmental stages ranging from 4.5 dpc to live born mice (Table 2 and Fig. 4). Starting at day 6 of gestation, we observed embryos that appeared smaller and progressively retarded. After 7.5 dpc, there was a significant number of degenerated embryos. In the 12 litters analyzed between 6 and 9.75 dpc, 34 of 98 embryos (35%) were either smaller than normal, retarded or degenerated, which fits well with the expected 25% homozygous embryos. We conclude from these observations that homozygous Hspd1−/− embryos are able to implant but stop developing at Theiler stage 8 to 9 when the egg cylinder differentiates.
Table 2.
Synopsis of the histological examination of 18 litters derived from crosses between Hspd1+/− mice
| Litter | Age (dpc)a | Normal | Small | Retarded | Degenerated |
|---|---|---|---|---|---|
| 1 | 5.25 | 6 | |||
| 2 | 5.25 | 10 | |||
| 3 | 5.50 | 12 | |||
| 4 | 5.75 | 8 | |||
| 5 | 5.75 | 13 | |||
| 6 | 5.75 | 10 | |||
| 7 | 6.00 | 6 | 4 | ||
| 8 | 6.00 | 5 | 2 | ||
| 9 | 6.50 | 5 | 3 | ||
| 10 | 6.75 | 3 | 3 | ||
| 11 | 6.75 | 8 | 2 | ||
| 12 | 6.75 | 4 | 1 | ||
| 13 | 7.00 | 7 | 1 | 2 | |
| 14 | 7.50 | 5 | 1 | 4 | |
| 15 | 7.50 | 8 | 1 | ||
| 16 | 8.50 | 3 | 2 | ||
| 17 | 8.50 | 6 | 4 | ||
| 18 | 9.75 | 4 | 4 |
aThe age of the litters was determined by the developmental stage of normal-appearing embryos
Fig. 4.
HE staining of paraffin sections of five embryos from the same 7.5 dpc litter. a–c Longitudinal sections; d, e cross section. Each picture was taken at the level where the respective embryo had its largest width. a, d Normal-appearing embryos. b, c, and e Degenerating embryos in different stages of resorption. Scale bar = 100 μm
After 8.5 dpc when embryos can be prepared without considerable maternal contamination, all surviving embryos were genotypically either wild-type or heterozygous Hspd1+/− (Table 3).
Table 3.
Appearance of embryos derived from crosses between Hspd1+/− mice between 8.5 and 10.5 dpc
| No. of mice with indicated phenotype among 28 embryos | P valuea | ||
|---|---|---|---|
| Normal | Degenerated | ||
| Observed (expected) | 19 (21) | 9 (7) | 0.512 |
aProbability of a Mendelian distribution of 3:1 (expected number of Hspd1+/+ and Hspd1+/− compared to Hspd1−/−) determined by a χ2 test
The effect of reduced expression of the Hspd1 gene on reproductive performance
To investigate whether or not the reproductive performance of the Hspd1+/− mice was affected by other means, we determined litter sizes and the sex ratios of the offspring in crosses between Hspd1+/− mice. The average litter size was 5.7 ± 2.2 (mean ± SD, n = 20), which is similar to the average litter size of 12 different inbred strains, namely, 5.7 ± 0.9, as calculated from data available on the Mouse Genome Informatics homepage (http://www.informatics.jax.org). The overall percentage of males born was 58.7%, which is significantly higher than the average percentage of male offspring produced by the same 12 inbred strains, namely, 50.7% ± 3.1. To further substantiate this observation, we evaluated offspring sex ratios according to the genotype of the parents in independent crosses between wild-type Hspd1+/+ and heterozygous Hspd1+/− mice (Table 4). We found that a significant excess of males (p = 0.039), namely, 63.4% ± 4.0, was born in crosses in which the genotype of the male parent was Hspd1+/−, compared to crosses in which the male parent was wild-type Hspd1+/+, namely, 45.3% ± 5.4. The genotype distribution between wild-type Hspd1+/+ and heterozygous Hspd1+/− mice among the offspring was 1:1 as expected. The 1:1 distribution was also present if males and females were recorded separately, arguing that it is the genotype of the spermatogonia and not the genotype of the derived offspring that is responsible for the changed sex ratio.
Table 4.
Offspring sex ratio enumerated according to the genotype of the parents
| Genotype male parent | Genotype female parent | % males |
|---|---|---|
| +/+ | +/− | 45.3 ± 5.4a |
| +/− | +/+ | 63.4 ± 4.0b |
aNumbers indicated are mean ± SD, n = 3 (independent breeding pairs)
bNumbers indicated are mean ± SD, n = 4 (independent breeding pairs)
The effect of reduced expression of the Hspd1 gene on Hspe1 expression
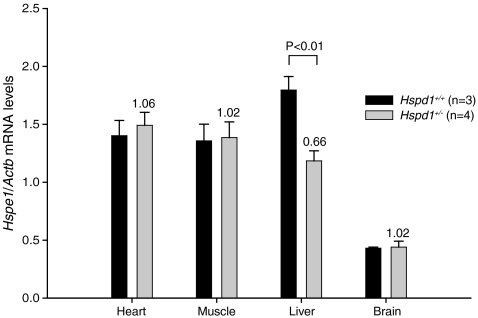
In order to investigate whether or not the reduced expression of the Hspd1 gene in heterozygous Hspd1+/− mice affects the expression of the Hspe1 gene that encodes the Hsp10 co-chaperonin transcribed from a common bidirectional promoter (Hansen et al. 2003), we performed a quantitative RT-PCR analysis on the same RNA that was analyzed above. The Hspe1 mRNA was quantified using a TaqMan probe that spanned the exon 2–exon 3 boundary of the Hspe1gene. The levels of Hspe1 mRNA relative to Actb mRNA were similar in heterozygous Hspd1+/− mice and wild-type mice in all tissues examined except the liver, where the level was significantly decreased by a factor of 0.66 ± 0.12 (mean ± SD) in heterozygous Hspd1+/− animals (Fig. 5). Thus, at the transcriptional level, liver tissue seems to down-regulate the Hspe1 gene either as a result of a distant liver-specific control element that is affected by the insertion of the gene-trap in the Hspd1 gene or less likely, in response to decreased Hspd1 mRNA and/or Hsp60 protein by a liver-specific feedback loop.
Fig. 5.
Quantitative RT-PCR analysis of the Hspe1 gene on RNA extracted from various tissues. The relative levels of Hspe1 mRNA normalized to Actb mRNA as an internal control were similar in heterozygous Hspd1+/− mice (n = 4) and Hspd1+/+ wild-type mice (n = 3) in all tissues examined except the liver where the level was significantly decreased by a factor 0.66 ± 0.12 (mean ± SD) in heterozygous Hspd1+/− mice. Values plotted are mean ± SEM. The ratios between the levels within the two groups (Hspd1+/− compared to Hspd1+/+) are indicated above the bars
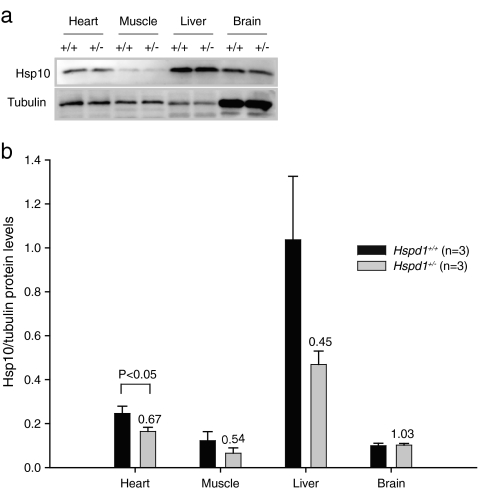
In order to investigate whether or not the similarities and differences in the levels of Hspe1 mRNA in the various tissues analyzed by quantitative RT-PCR were accompanied by similar changes in Hsp10 protein levels, we performed a semiquantitative Western blot analysis using an anti-Hsp10 antibody on protein extracted from the same tissues. As a loading control, the samples were analyzed concomitantly with an anti-tubulin antibody. Protein bands with the expected apparent molecular weights (Hsp10 10 kDa and tubulin 55 kDa) were detected in all tissues (Fig. 6a). We further found that the levels of immunoreactive Hsp10 relative to cytosolic tubulin were less in heterozygous Hspd1+/− mice than in wild-type animals in all tissues examined except the brain (Fig. 6b). The ratios between the levels within the two groups (Hspd1+/− compared to Hspd1+/+) were 0.67 ± 0.20, 0.54 ± 0.46, 0.45 ± 0.24, and 1.03 ± 0.24 (mean ± SD) for the heart, muscle, the liver, and the brain, respectively. Thus, the levels of Hsp10 protein seem to be adjusted to the Hsp60 protein level, either at the transcriptional level (liver) or further down-stream, e.g., by reduced protein stability due to missing interaction partners (heart and muscle). Only in brain tissue does the amount of Hsp60 and Hsp10 seem to be disproportionate.
Fig. 6.
Semiquantitative Western blot analysis using an anti-Hsp10 antibody on protein extracted from various tissues. a A representative blot demonstrating that protein bands of the expected molecular weights (Hsp10 10 kDa and tubulin 55 kDa) were detected in all tissues analyzed. b Densitometric analysis of protein amounts revealed that the relative levels of immunoreactive Hsp10 to cytosolic tubulin were decreased in heterozygous Hspd1+/− mice (n = 3) compared to Hspd1+/+ wild-type littermates (n = 3) in all tissues examined except the brain. Values plotted are mean ± SEM. The ratios between the levels within the two groups (Hspd1+/− compared to Hspd1+/+) are indicated above the bars
Discussion
Our data support the hypothesis that loss of the Hspd1 gene is lethal. They further indicate that a ≈50% reduction of the amount of the encoded chaperonin Hsp60 is compatible with overall embryonic development and prenatal viability. However, reduced amounts of mitochondrial Hsp60 seem to be associated with, on the one hand, a sperm phenotype resulting in the production of a disproportionately large number of male offspring and tissue-dependent differences in the regulation of the expression of the Hsp10 co-chaperonin.
In the present study, we achieved inactivation of one allele of the Hspd1 gene in the mouse genome by random integration of a gene-trap vector in intron 2 of the Hspd1 gene (OST171441, Lexicon Pharmaceuticals, Inc.; Fig. 1a). We confirmed the integration of the gene-trap vector in intron 2 of the Hspd1 gene, 967 base pair downstream of exon 2 by Southern blotting analysis (Fig. 1c) as well as by PCR and direct sequencing (data not shown).
Using quantitative RT-PCR, we found a ≈50% reduction of correctly spliced Hspd1 mRNA in all tissues examined (Fig. 2), which indicates that there is no feedback regulation of the Hspd1 gene at the transcriptional level. At the protein level, we exclusively identified Hsp60 protein of the expected molecular weight of 60 kDa. We further demonstrated a reduction in the amount of Hsp60 protein compared to the amount of cytosolic tubulin in all tissues (Fig. 3b). The reduction was statistically significant in the majority of tissues; however, in the heart, the reduction was less pronounced. The decreased levels of Hsp60 protein were probably not accountable to decreased amount of total mitochondrial protein, as similar results were obtained when the amount of Hsp60 protein was compared to the amount of mitochondrial VDAC-1 protein (Fig. 3d). As the transcription of the Hspd1 gene is reduced to approximately 50%, i.e., no apparent product feedback loop, a less than 50% reduction in the Hsp60 protein level could reflect a reduced protein turnover.
A prenatal loss of homozygous Hspd−/− mice was initially indicated by the genotype distribution among 114 weaned animals from heterozygous Hspd+/− intercrosses in that no homozygous Hspd1−/− mice were identified (Table 1). By comprehensive and systematic histological examination of embryos at various embryonic stages, derived from timed pregnancies (Table 2 and Fig. 4), we found the first evidence of retarded growth at 6 dpc and embryo degeneration at 7.5 dpc, indicating that embryonic lethality due to decreased levels of Hsp60 occurs at an early time point after implantation. After 8.5 dpc, no homozygous Hspd1−/− embryos could be detected.
The mechanisms that allow Hspd1−/− embryos to survive during the very early stages of embryogenesis are not clear. It is well documented that proper mitochondrial function is critical for embryonic development. Its dysfunction may not only compromise developmental processes but also trigger apoptosis (reviewed by Dumollard et al. 2009). It is further generally accepted that limited or no mitochondrial biogenesis occurs during early embryogenesis (Dumollard et al. 2006; Jansen 2000; Poulton and Marchington 2002). One possible mechanism for homozygous Hspd1−/− embryos to survive during the very early stages of embryogenesis could be the presence of an adequate amount of Hsp60 activity in the mitochondria that has descended through the oocyte from their heterozygous Hspd1+/− mothers. This mechanism requires the maternally encoded Hsp60 protein to be quite stable during early development. Remarkable stability like this has been reported for several other mitochondrial proteins (pyruvate dehydrogenase, cytochrome c, and dihydrolipoamide dehydrogenase), all of which are ultimately required for oxidative phosphorylation in that, murine embryos lacking any of those survive until implantation (Johnson et al. 1997, 2001; Li et al. 2000). Based upon these observations, we suggest that Hspd1−/− embryos most likely die due to mitochondrial dysfunction that arises soon after the developmental time point at which mitochondrial biogenesis resumes, Hsp60 derived from the mother is depleted, and no additional Hsp60 is produced due to the inactivation of both alleles of the Hspd1 gene in the nucleus of the cells of the embryo.
Our finding that wild-type Hspd1+/+ and heterozygous Hspd1+/− mice were born in the expected Mendelian ratio, i.e., 1:2, supports the notion that a ≈50% reduction in the amount of Hsp60 is permissive with regard to overall embryonic development and prenatal viability (Table 1).
A role of Hsp60 in proper mitochondrial function during spermatogenesis is indicated by the observation that Hsp60, among a number of other mitochondrial proteins, is differentially expressed during distinct phases of spermatogenesis (Meinhardt et al. 1999). In the present study, we did not find that reduced amounts of Hsp60 in heterozygous Hspd1+/− males affect their overall fertility. However, in crosses in which the genotype of the male parent was Hspd1+/−, we did find that significantly, more males (≈63%) were born than in crosses in which the male parent was wild-type (≈45%; Table 4). It is tempting to speculate that this phenomenon is a consequence of X chromosome-bearing sperm cells being more sensitive to the reduced levels of Hsp60 than Y chromosome-bearing sperm cells. It has previously been reported that Y chromosome-bearing spermatozoa are smaller in size and exhibit a greater downward swimming velocity than X chromosome-bearing spermatozoa (Ericsson et al. 1973). Whether or not this difference in motility is affected by reduced amounts of Hsp60 is not known. Sperm cell motility is dependent on ATP production (for review, see Ramalho-Santos et al. 2009), and although controversial, it is possible that oxidative phosphorylation that occurs in the midpiece mitochondria of the sperm cell is an important source of ATP (Narisawa et al. 2002). This phenomenon is of great interest for future studies. Our observation that the sex ratios were disproportionate among both heterozygous Hspd1+/− and wild-type Hspd1+/+ offspring further indicates that Hsp60 is produced at the stage when the spermatogonia are still connected; otherwise, the sex ratios would have been disproportionate only among heterozygous Hspd1+/− offspring.
It is believed that Hsp60 needs to combine with its co-chaperonin Hsp10 into a barrel-like structure that comprises two seven-mer Hsp60 ring structures and a seven-mer Hsp10 lid, before it can assume its proper function in the mitochondrial matrix (Horwich et al. 2007). As in humans (Hansen et al. 2003), the murine Hspd1 gene encoding Hsp60 is organized in a head-to-head orientation with the Hspe1gene encoding Hsp10. Both genes are transcribed from a common bidirectional promoter, which among other transcription factor binding sites, comprises heat shock elements and a CHOP binding site involved in mitochondrial stress response (Zhao et al. 2002). We investigated whether or not the reduced expression of the Hspd1 gene in heterozygous Hspd1+/− mice affects the expression of the nearby Hspe1 gene both at the transcriptional level (Fig. 5) and at the protein level (Fig. 6). At the transcriptional level, we found a marked reduction (≈1/3) in the relative expression of the Hspe1 gene in the liver. However, there were no changes in any of the other tissues examined. This reduction in Hspe1 expression could be the result of the actions of a distant liver-specific control element that is affected by the insertion of the gene-trap in the Hspd1 gene or less likely, the result of a liver-specific feedback loop activated by decreased Hspd1 mRNA and/or Hsp60 protein. At the protein level, we further found that the levels of immunoreactive Hsp10 relative to cytosolic tubulin had a tendency to be reduced in heterozygous Hspd1+/− mice compared to wild-type animals in all tissues examined except the brain. Although these observations should be interpreted with caution since the densitometric analysis of such small proteins (10 kDa) has some uncertainty, they do indicate that Hsp10 stability depends on interaction with Hsp60.
In conclusion, our results show that Hspd1 is an essential gene for mammalian survival, while the haploid amount of the gene product results in only mild phenotypic changes in mice. Future studies are needed to investigate whether or not the heterozygous Hspd1+/− mice will develop symptoms of hereditary spastic paraplegia by age. Even though mice are both smaller and have a shorter life-span than humans, they have proven to be able to develop mitochondrial dysfunction and late-onset degeneration of the longest motor neurons in the spinal cord due to genetic inactivation of hereditary spastic paraplegia genes (Ferreirinha et al. 2004; Kasher et al. 2009; Tarrade et al. 2006). Besides, if heterozygous Hspd1+/− mice exhibit a discreet underlying mitochondrial stress phenotype before the onset of more pronounced disease symptoms, they will have the potential to serve as a modifying genetic background for mouse models of various diseases in which mitochondrial dysfunction is a potential common denominator (e.g., Parkinson’s disease, amyotrophic lateral sclerosis, Alzheimer’s disease, Huntington’s disease, and multiple sclerosis; Dutta et al. 2006; Kwong et al. 2006). Investigation of the molecular and cellular effects of reduced levels of Hsp60 in various tissues in our mouse model could further provide a better understanding of the mechanisms by which various mammalian cell types compensate for unbalanced and/or deficient mitochondrial protein quality control under normal and stress conditions.
Acknowledgments
The authors thank Birgitte Grann for her excellent technical assistance and Helle Christiansen for checking mice vaginal plugs each morning during the analysis of timed pregnancies. The work was supported by grants from the Ludvig and Sara Elsass Foundation, the Lundbeck Foundation, the EU 6th Framework Program, the Novo Nordisk Foundation, the Augustinus Foundation, “Elvira og Rasmus Riisforts Almenvelgørende Fond” [the Elvira and Rasmus Riisfort’s Common Charitable Foundation], and “Grosserer A.V. Lykfeldt og Hustrus Legat” [the Merchant A.V. Lykfeldt and Wife’s Grant], and Aarhus University.
References
- Arya R, Mallik M, Lakhotia SC. Heat shock genes—integrating cell survival and death. J Biosci. 2007;32:595–610. doi: 10.1007/s12038-007-0059-3. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/S0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Chandra D, Choy G, Tang DG. Cytosolic accumulation of HSP60 during apoptosis with or without apparent mitochondrial release: evidence that its pro-apoptotic or pro-survival functions involve differential interactions with caspase-3. J Biol Chem. 2007;282:31289–31301. doi: 10.1074/jbc.M702777200. [DOI] [PubMed] [Google Scholar]
- Cheng MY, Hartl FU, Martin J, Pollock RA, Kalousek F, Neupert W, Hallberg EM, Hallberg RL, Horwich AL. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 1989;337:620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- Deluca GC, Ebers GC, Esiri MM. The extent of axonal loss in the long tracts in hereditary spastic paraplegia. Neuropathol Appl Neurobiol. 2004;30:576–584. doi: 10.1111/j.1365-2990.2004.00587.x. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Duchen M, Sardet C. Calcium signals and mitochondria at fertilisation. Semin Cell Dev Biol. 2006;17:314–323. doi: 10.1016/j.semcdb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Carroll J, Duchen MR, Campbell K, Swann K. Mitochondrial function and redox state in mammalian embryos. Semin Cell Dev Biol. 2009;20:346–353. doi: 10.1016/j.semcdb.2008.12.013. [DOI] [PubMed] [Google Scholar]
- Dutta R, McDonough J, Yin X, Peterson J, Chang A, Torres T, Gudz T, Macklin WB, Lewis DA, Fox RJ, Rudick R, Mirnics K, Trapp BD. Mitochondrial dysfunction as a cause of axonal degeneration in multiple sclerosis patients. Ann Neurol. 2006;59:478–489. doi: 10.1002/ana.20736. [DOI] [PubMed] [Google Scholar]
- Ericsson RJ, Langevin CN, Nishino M. Isolation of fractions rich in human Y sperm. Nature. 1973;246:421–424. doi: 10.1038/246421a0. [DOI] [PubMed] [Google Scholar]
- Ferreirinha F, Quattrini A, Pirozzi M, Valsecchi V, Dina G, Broccoli V, Auricchio A, Piemonte F, Tozzi G, Gaeta L, Casari G, Ballabio A, Rugarli EI. Axonal degeneration in paraplegin-deficient mice is associated with abnormal mitochondria and impairment of axonal transport. J Clin Invest. 2004;113:231–242. doi: 10.1172/JCI20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habich C, Burkart V. Heat shock protein 60: regulatory role on innate immune cells. Cell Mol Life Sci. 2007;64:742–751. doi: 10.1007/s00018-007-6413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JJ, Durr A, Cournu-Rebeix I, Georgopoulos C, Ang D, Nielsen MN, Davoine CS, Brice A, Fontaine B, Gregersen N, Bross P. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet. 2002;70:1328–1332. doi: 10.1086/339935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JJ, Bross P, Westergaard M, Nielsen MN, Eiberg H, Borglum AD, Mogensen J, Kristiansen K, Bolund L, Gregersen N. Genomic structure of the human mitochondrial chaperonin genes: HSP60 and HSP10 are localised head to head on chromosome 2 separated by a bidirectional promoter. Hum Genet. 2003;112:71–77. doi: 10.1007/s00439-002-0837-9. [DOI] [PubMed] [Google Scholar]
- Hansen J, Svenstrup K, Ang D, Nielsen MN, Christensen JH, Gregersen N, Nielsen JE, Georgopoulos C, Bross P. A novel non-synonymous variation in the HSPD1 gene associated with hereditary spastic paraplegia. J Neurol. 2007;254:897–900. doi: 10.1007/s00415-006-0470-y. [DOI] [PubMed] [Google Scholar]
- Hansen J, Corydon TJ, Palmfeldt J, Durr A, Fontaine B, Nielsen MN, Christensen JH, Gregersen N, Bross P. Decreased expression of the mitochondrial matrix proteases Lon and ClpP in cells from a patient with hereditary spastic paraplegia (SPG13) Neuroscience. 2008;153:474–482. doi: 10.1016/j.neuroscience.2008.01.070. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Hemmingsen SM, Woolford C, Vies SM, Tilly K, Dennis DT, Georgopoulos CP, Hendrix RW, Ellis RJ. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: physiology and mechanism. Annu Rev Cell Dev Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- Jansen RP. Germline passage of mitochondria: quantitative considerations and possible embryological sequelae. Hum Reprod. 2000;15(Suppl 2):112–128. doi: 10.1093/humrep/15.suppl_2.112. [DOI] [PubMed] [Google Scholar]
- Johnson MT, Yang HS, Magnuson T, Patel MS. Targeted disruption of the murine dihydrolipoamide dehydrogenase gene (Dld) results in perigastrulation lethality. Proc Natl Acad Sci U S A. 1997;94:14512–14517. doi: 10.1073/pnas.94.26.14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MT, Mahmood S, Hyatt SL, Yang HS, Soloway PD, Hanson RW, Patel MS. Inactivation of the murine pyruvate dehydrogenase (Pdha1) gene and its effect on early embryonic development. Mol Genet Metab. 2001;74:293–302. doi: 10.1006/mgme.2001.3249. [DOI] [PubMed] [Google Scholar]
- Kasher PR, Vos KJ, Wharton SB, Manser C, Bennett EJ, Bingley M, Wood JD, Milner R, McDermott CJ, Miller CC, Shaw PJ, Grierson AJ. Direct evidence for axonal transport defects in a novel mouse model of mutant spastin-induced hereditary spastic paraplegia (HSP) and human HSP patients. J Neurochem. 2009;110:34–44. doi: 10.1111/j.1471-4159.2009.06104.x. [DOI] [PubMed] [Google Scholar]
- Kwong JQ, Beal MF, Manfredi G. The role of mitochondria in inherited neurodegenerative diseases. J Neurochem. 2006;97:1659–1675. doi: 10.1111/j.1471-4159.2006.03990.x. [DOI] [PubMed] [Google Scholar]
- Li K, Li Y, Shelton JM, Richardson JA, Spencer E, Chen ZJ, Wang X, Williams RS. Cytochrome c deficiency causes embryonic lethality and attenuates stress-induced apoptosis. Cell. 2000;101:389–399. doi: 10.1016/S0092-8674(00)80849-1. [DOI] [PubMed] [Google Scholar]
- Magen D, Georgopoulos C, Bross P, Ang D, Segev Y, Goldsher D, Nemirovski A, Shahar E, Ravid S, Luder A, Heno B, Gershoni-Baruch R, Skorecki K, Mandel H. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am J Hum Genet. 2008;83:30–42. doi: 10.1016/j.ajhg.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt A, Wilhelm B, Seitz J. Expression of mitochondrial marker proteins during spermatogenesis. Hum Reprod Updat. 1999;5:108–119. doi: 10.1093/humupd/5.2.108. [DOI] [PubMed] [Google Scholar]
- Narisawa S, Hecht NB, Goldberg E, Boatright KM, Reed JC, Millan JL. Testis-specific cytochrome c-null mice produce functional sperm but undergo early testicular atrophy. Mol Cell Biol. 2002;22:5554–5562. doi: 10.1128/MCB.22.15.5554-5562.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perezgasga L, Segovia L, Zurita M. Molecular characterization of the 5′ control region and of two lethal alleles affecting the hsp60 gene in Drosophila melanogaster. FEBS Lett. 1999;456:269–273. doi: 10.1016/S0014-5793(99)00963-1. [DOI] [PubMed] [Google Scholar]
- Pockley AG, Muthana M, Calderwood SK. The dual immunoregulatory roles of stress proteins. Trends Biochem Sci. 2008;33:71–79. doi: 10.1016/j.tibs.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Poulton J, Marchington DR. Segregation of mitochondrial DNA (mtDNA) in human oocytes and in animal models of mtDNA disease: clinical implications. Reproduction. 2002;123:751–755. doi: 10.1530/rep.0.1230751. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos J, Varum S, Amaral S, Mota PC, Sousa AP, Amaral A. Mitochondrial functionality in reproduction: from gonads and gametes to embryos and embryonic stem cells. Hum Reprod Updat. 2009;15:553–572. doi: 10.1093/humupd/dmp016. [DOI] [PubMed] [Google Scholar]
- Salinas S, Proukakis C, Crosby A, Warner TT. Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7:1127–1138. doi: 10.1016/S1474-4422(08)70258-8. [DOI] [PubMed] [Google Scholar]
- Svenstrup K, Bross P, Koefoed P, Hjermind LE, Eiberg H, Born AP, Vissing J, Gyllenborg J, Norremolle A, Hasholt L, Nielsen JE. Sequence variants in SPAST, SPG3A and HSPD1 in hereditary spastic paraplegia. J Neurol Sci. 2009;284:90–95. doi: 10.1016/j.jns.2009.04.024. [DOI] [PubMed] [Google Scholar]
- Tarrade A, Fassier C, Courageot S, Charvin D, Vitte J, Peris L, Thorel A, Mouisel E, Fonknechten N, Roblot N, Seilhean D, Dierich A, Hauw JJ, Melki J. A mutation of spastin is responsible for swellings and impairment of transport in a region of axon characterized by changes in microtubule composition. Hum Mol Genet. 2006;15:3544–3558. doi: 10.1093/hmg/ddl431. [DOI] [PubMed] [Google Scholar]
- Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- Zambrowicz BP, Abuin A, Ramirez-Solis R, Richter LJ, Piggott J, BeltrandelRio H, Buxton EC, Edwards J, Finch RA, Friddle CJ, Gupta A, Hansen G, Hu Y, Huang W, Jaing C, Key BW, Jr, Kipp P, Kohlhauff B, Ma ZQ, Markesich D, Payne R, Potter DG, Qian N, Shaw J, Schrick J, Shi ZZ, Sparks MJ, Van SI, Vogel P, Walke W, Xu N, Zhu Q, Person C, Sands AT. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci U S A. 2003;100:14109–14114. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carriero N, Gerstein M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 2004;20:62–67. doi: 10.1016/j.tig.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]