miR-146a and Krüppel-like factor 4 form a feedback loop to participate in vascular smooth muscle cell proliferation
miR-146a and KLF4 negatively regulate each other's expression and miR-146a thereby promotes vascular smooth muscle cell proliferation. Depletion of miR-146a in injured rat carotid arteries decreases hyperplasia and this might have therapeutic implications for proliferative vascular diseases.
Keywords: Krüppel-like factor 4, microRNA, miR-146a, proliferation, vascular smooth muscle cells
Abstract
MicroRNAs are phenotypic regulators of vascular smooth muscle cells (VSMCs). In this paper, we demonstrate that miR-146a targets the Krüppel-like factor 4 (KLF4) 3′-untranslated region and has an important role in promoting VSMC proliferation in vitro and vascular neointimal hyperplasia in vivo. Silencing of miR-146a in VSMCs increases KLF4 expression, whereas overexpression of miR-146a decreases KLF4 levels. Furthermore, we demonstrate that KLF4 competes with Krüppel-like factor 5 (KLF5) to bind to and regulate the miR-146a promoter, and that KLF4 and KLF5 exert opposing effects on the miR-146a promoter. Overexpression of KLF4 in VSMCs decreases miR-146a transcription levels. By using both gain-of-function and loss-of-function approaches, we found that miR-146a promotes VSMC proliferation in vitro. Transfection of antisense miR-146a oligonucleotide into balloon-injured rat carotid arteries markedly decreased neointimal hyperplasia. These findings suggest that miR-146a and KLF4 form a feedback loop to regulate each other's expression and VSMC proliferation.
Introduction
MicroRNAs (miRNAs) are being identified as a class of novel endogenous regulators of gene expression at the post-transcriptional level. They have essential roles in regulating many cellular events, including cell proliferation, differentiation and apoptosis. Therefore, miRNAs might be important for normal development and physiology. Deregulation of miRNAs might cause a variety of diseases, including tumours and cardiovascular disease (Ji et al, 2007; Cordes et al, 2009).
Vascular smooth muscle cells (VSMCs) oscillate between proliferated and differentiated states in response to local environmental cues. VSMC phenotypic modulation and proliferation are the pathological bases of proliferative vascular diseases, including atherosclerosis and restenosis. Earlier studies have shown that VSMC phenotypic modulation is controlled by complex interactions between general and SMC-specific proteins (Kumar & Owens, 2003; Kawai-Kowase & Owens, 2007). Recent studies have shown that miRNAs participate in the phenotypic control of VSMCs (Cordes et al, 2009; Davis et al, 2009). More importantly, miR-146a was upregulated in rat balloon-injured arteries (Ji et al, 2007) and in oxidized low-density lipoprotein (oxLDL)-stimulated human monocytes/macrophages (Chen et al, 2009). However, little is known about the role of miR-146a and the mechanism for its action in VSMC phenotypic modulation. Here, we demonstrate that miR-146a targets the Krüppel-like factor 4 (KLF4) 3′-untranslated region (UTR) and has an important role in promoting VSMC proliferation in vitro and vascular neointimal hyperplasia in vivo. We found that miR-146a transcription is regulated by KLF4 and Krüppel-like factor 5 (KLF5), and that miR-146a and KLF4 form a feedback loop to regulate each other's expression and VSMC proliferation.
Results
miR-146a inhibits KLF4 expression by targeting its 3′-UTR
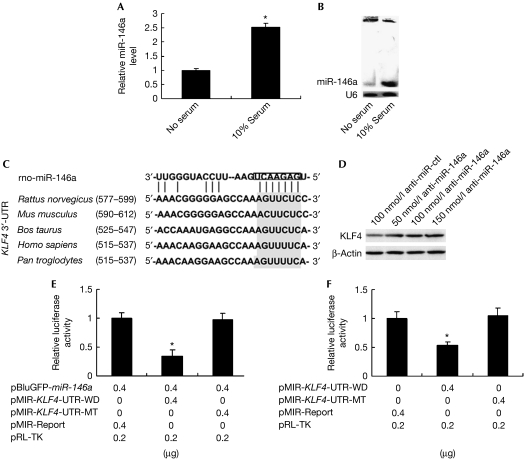
To identify the potential role of miR-146a in VSMC biology and pathobiology, we first examined the expression signature of miR-146a in quiescent and proliferative VSMCs by using quantitative real-time (qRT)–PCR and northern blotting analyses. The expression level of miR-146a in 10% serum-induced proliferative VSMCs was significantly higher than that in serum-starved quiescent VSMCs (Fig 1A,B). The higher expression signature of miR-146a in proliferative VSMCs implies that miR-146a might depress the expression of some antiproliferative genes. Studies have shown that KLF4 is an antiproliferative regulator in VSMCs. To test whether KLF4 is a direct target of miR-146a, we used a bioinformatics approach to search for the potential matching site of miR-146a in the KLF4 3′-UTR. There was an miR-146a-binding site at nucleotides 577–599 of the rat KLF4 3′-UTR—which is highly conserved in vertebrates—suggesting it is important in regulating gene expression (Fig 1C). To evaluate the effect of miR-146a on KLF4 expression, we synthesized antisense miR-146a oligonucleotide (anti-miR-146a) with modification by locked nucleic acid and transfected VSMCs. Anti-miR-146a, but not the control oligonucleotide (anti-miR-ctl), could markedly decrease 10% serum-induced miR-146a expression (supplementary Fig S1 online). We transfected anti-miR-146a and anti-miR-ctl into VSMCs grown in 2% fetal bovine serum (FBS), and found that anti-miR-146a increased KLF4 protein level in a dose-dependent manner (Fig 1D). To further confirm whether miR-146a directly targets the KLF4 3′-UTR, we constructed the wild-type pMIR-KLF4-UTR-WD and its mutant pMIR-KLF4-UTR-MT and co-transfected 293A cells with pBluGFP-miR-146a and pMIR-KLF4-UTR-WD or pMIR-KLF4-UTR-MT. As shown in Fig 1E, mutation of the miR-146a-binding site within the 3′-UTR of KLF4 abrogated the inhibitory effect of miR-146a on reporter gene expression. As expected, endogenous miR-146a induced by 10% FBS in VSMCs also decreased pMIR-KLF4-UTR-WD-mediated luciferase activity compared with the control (Fig 1F). Taken together, these data suggest that miR-146a inhibits KLF4 expression through a special miR-146a-binding site in the KLF4 3′-UTR.
Figure 1.
miR-146a inhibits KLF4 expression. (A) VSMCs were serum starved or 10% serum induced for 24 h, and then miR-146a expression was detected by qRT–PCR. *P<0.05 compared with no serum control. (B) miR-146a expression in serum-starved or 10% serum-induced VSMCs for 24 h was detected by northern blotting. (C) The miR-146a-binding site resides at nucleotides 577–599 of the rat KLF4 mRNA 3′-UTR, and is highly conserved in vertebrates. (D) Total protein was harvested from VSMCs transfected with anti-miR-ctl (100 nmol/l) or anti-miR-146a (50, 100 and 150 nmol/l), and analysed by western blotting for KLF4. (E) 293A cells were co-transfected with pBluGFP-miR-146a and the wild-type pMIR-KLF4-UTR-WD or with its mutant pMIR-KLF4-UTR-MT. After 24 h, luciferase activities were measured. *P<0.05 compared with pMIR-Report control. (F) 10% serum-induced VSMCs were transfected with pMIR-KLF4-UTR-WD or pMIR-KLF4-UTR-MT. After 24 h, luciferase activities were measured. *P<0.05 compared with pMIR-Report control. All experiments were repeated three times. KLF, Krüppel-like factor; qRT–PCR, quantitative real-time PCR; UTR, untranslated region; VSMC, vascular smooth muscle cell.
KLF4 competes with KLF5 for the miR-146a promoter
To evaluate the mechanisms by which miR-146a might be regulated in a physiological setting, we cloned 2 kb of the 5′ regulatory sequence of the rat miR-146a gene into the pGL3-luc reporter to construct pGL3-miR-146a-luc, and transfected 293A cells with it. Luciferase activity assay showed that the 2-kb 5′-flanking region was sufficient for miR-146a expression (Fig 2A). To identify potential regulatory elements in the 2-kb miR-146a promoter, we analysed this region using the TESS computational program. Interestingly, we found one typical KLF4/KLF5-binding motif (CACCC) and its two reverse orientation sequences (GGGTG) in this region (Fig 2B). To investigate whether KLF4 and KLF5 interact with the miR-146a promoter in vivo, we performed a chromatin immunoprecipitation assay. Both KLF4 and KLF5 were recruited to the miR-146a promoter (Fig 2C). Electrophoretic mobility shift assay (EMSA) showed that KLF4 and KLF5 indeed bind to the miR-146a promoter directly in vitro (Fig 2D). As three KLF4/KLF5-binding motifs are located immediately adjacent to each other, we synthesized double-stranded oligonucleotides containing these three motifs and their mutants at different motifs, and defined the motif to which KLF4 and KLF5 were bound. Oligo pull-down assay showed that KLF4 could bind to each of the three motifs, whereas KLF5 mainly bound to motifs 1 and 2 (Fig 2E).
Figure 2.
KLF4 competes with KLF5 to bind to and regulate the miR-146a promoter. (A) 293A cells were transfected with pGL3-miR-146a-luc or pGL3-Basic. After 24 h, luciferase activities were measured. **P<0.01 compared with pGL3-Basic control. (B) Partial sequence of the miR-146a promoter and KLF4/KLF5-binding motifs. The underlined sequences are primers for ChIP assay. (C) Proteins and DNA in VSMCs were crosslinked. ChIP assays were performed with KLF4 or KLF5 antibodies and the miR-146a promoter region containing KLF4/KLF5-binding sites was amplified by PCR. (D) Recombinant KLF4 or KLF5 was incubated with a biotin-labelled probe containing KLF4/KLF5-binding sites in the miR-146a promoter. Protein–DNA complexes were separated by non-denaturing polyacrylamide gel electrophoresis and then visualized by chemiluminescence. Super-shift assays were performed by adding KLF4 or KLF5 antibodies. (E) Oligonucleotide pull-down assay was performed with biotin-labelled double-stranded oligonucleotides for the KLF4/KLF5-binding sites and their mutants as probes. The DNA-bound proteins were collected with streptavidin-agarose beads and analysed by western blotting with KLF4 or KLF5 antibodies. (F) 293A cells were co-transfected with pGL3-miR-146a-luc and KLF4 or KLF5 expression plasmid. After 24 h, luciferase activities were measured. *P<0.05 compared with empty vector pEGFP-N1 control. (G) 293A cells were co-transfected with a constant amount of PMT-KLF5 and with increasing amounts of pEGFP-KLF4, along with pGL3-miR-146a-luc. After 24 h, luciferase activities were measured. *P<0.05 compared with PMT-KLF5 alone. (H) 293A cells were co-transfected with a constant amount of pEGFP-KLF4 and with increasing amounts of PMT-KLF5, along with pGL3-miR-146a-luc. After 24 h, luciferase activities were measured. *P<0.05 compared with pEGFP-KLF4 alone. (I) VSMCs were transduced with Ad-KLF4, Ad-KLF5 or Ad-GFP. After 48 h, KLF4, KLF5 and miR-146a expression levels were detected by using western blotting and qRT–PCR. *P<0.05 compared with Ad-GFP control. All experiments were repeated three times. Ab, antibody; ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility shift assay; GFP, green fluorescent protein; KLF, Krüppel-like factor; qRT–PCR, quantitative real-time PCR; VSMC, vascular smooth muscle cell.
To investigate the effect of KLF4 and KLF5 on miR-146a promoter activity, we co-transfected 293A cells with pGL3-miR-146a-luc and expression plasmids for KLF4 (pEGFP-KLF4) or KLF5 (PMT-KLF5). Western blot analysis showed that pEGFP-KLF4 or PMT-KLF5 was expressed successfully in 293A cells (supplementary Fig S2A online). Luciferase activity assay showed that KLF4 decreased the miR-146a promoter activity to about 20% of empty vector control level. By contrast, KLF5 exerted an opposite effect on the miR-146a promoter, increasing the reporter activity by about 60% compared with the control level (Fig 2F). One site mutation of the three KLF4/KLF5-binding motifs in the miR-146a promoter did not significantly affect the regulation of KLF4/KLF5 on the miR-146a promoter, whereas deletion of the region containing all three motifs abolished the effect of KLF4/KLF5 on the miR-146a promoter activity, compared with wild-type miR-146a promoter (supplementary Fig S3B,C online).
We co-transfected 293A cells with a constant amount of PMT-KLF5 and increasing amounts of pEGFP-KLF4, along with pGL3-miR-146a-luc. Western blot analysis showed that pEGFP-KLF4 or PMT-KLF5 was expressed successfully in 293A cells (supplementary Fig S2B,C online). As shown in Fig 2G, the stimulatory effect of KLF5 on the miR-146a promoter activity was gradually abolished with increases in the amount of pEGFP-KLF4. By contrast, when 293A cells were co-transfected with a constant amount of pEGFP-KLF4 and increasing amounts of PMT-KLF5, KLF5 abrogated the inhibitory effect of KLF4 on the miR-146a promoter in a concentration-dependent manner (Fig 2H). Taken together, these results suggest that KLF4 competes with KLF5 to bind to and regulate the miR-146a promoter as a pair of positive and negative regulators.
To investigate the effect of KLF4 and KLF5 on miR-146a expression in VSMCs, we transfected VSMCs grown in 10% FBS with Ad-KLF4 or Ad-KLF5. The results showed that overexpression of KLF4 decreased the expression level of miR-146a by 40%, whereas overexpressing KLF5 in cells increased miR-146a expression activity by 35%, compared with those transfected with Ad-GFP (Fig 2I).
miR-146a promotes VSMC proliferation in vitro
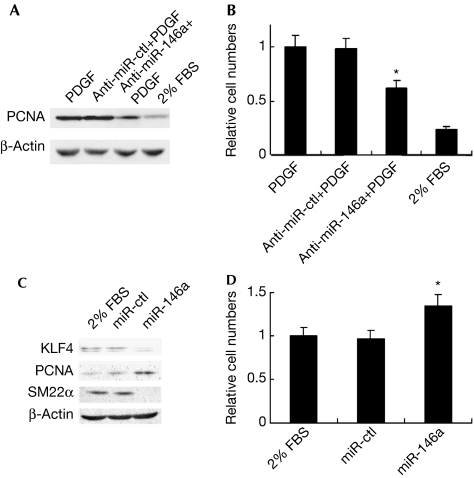
To identify the potential role of miR-146a in VSMC proliferation, 5% serum-induced VSMCs were transfected with anti-miR-146a or anti-miR-ctl and then stimulated with platelet-derived growth factor-BB. As expected, transfection of anti-miR-146a, but not anti-miR-ctl, significantly attenuated platelet-derived growth factor-BB-induced expression of proliferating cell nuclear antigen (PCNA), a proliferation marker (Fig 3A). Cell counting obtained a similar result to that shown by PCNA expression (Fig 3B). By contrast, transfection of miR-146a mimics, but not miR-ctl, into quiescent VSMCs significantly decreased the expression of KLF4 and SM22α (a differentiation marker), and increased PCNA expression (Fig 3C). Cell counting showed that quiescent VSMCs transfected with miR-146a mimics had a higher proliferative ability than those transfected with miR-ctl (Fig 3D). These data suggest that miR-146a exerts a proliferative effect on VSMCs in vitro.
Figure 3.
miR-146a promotes VSMC proliferation in vitro. (A) Western blotting for PCNA expression in VSMCs transfected with anti-miR-146a (100 nmol/l) or anti-miR-ctl (100 nmol/l), and then treated with PDGF-BB (10 ng/ml). (B) Cell count of VSMCs transfected with anti-miR-146a or anti-miR-ctl and then treated with PDGF-BB. *P<0.05 compared with PDGF control. (C) Western blotting analysis for KLF4, PCNA and SM22α expression in VSMCs transfected with miR-146a mimics (20 nmol/l) or miR-ctl (20 nmol/l). (D) Cell count of VSMCs transfected with miR-146a mimics or miR-ctl. *P<0.05 compared with 2% FBS control. All experiments were repeated three times. FBS, fetal bovine serum; KLF, Krüppel-like factor; PCNA, proliferating cell nuclear antigen; PDGF-BB, platelet-derived growth factor-BB; VSMC, vascular smooth muscle cell.
miR-146a promotes vascular neointimal hyperplasia in vivo
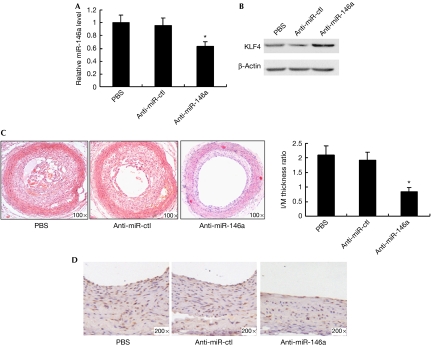
When balloon-injured rat carotid arteries were treated with anti-miR-146a or anti-miR-ctl, anti-miR-146a, but not anti-miR-ctl, decreased miR-146a expression and increased KLF4 expression in injured vascular wall—shown by qRT–PCR and western blotting analyses (Fig 4A,B). Anti-miR-146a significantly decreased neointimal hyperplasia and the number of PCNA-positive cells in injured vascular wall—shown by haematoxylin–eosin staining and immunohistochemistry staining for PCNA (Figs 4C,D). By contrast, anti-miR-ctl had no effect on VSMC proliferation and neointima formation induced by balloon injury. In addition, anti-miR-146a treatment also increased SM22α expression (supplementary Fig S4A online) and decreased SMEMB—a differentiation marker—expression (supplementary Fig S4B online), as shown by immunohistochemistry staining. These results suggest that miR-146a promotes vascular neointimal hyperplasia in vivo.
Figure 4.
miR-146a promotes vascular neointimal hyperplasia in vivo. (A) miR-146a expression in carotid arteries from PBS-, anti-miR-ctl (33 μg)- and anti-miR-146a (33 μg)-treated rats at 14 days after balloon injury, as detected by qRT–PCR. *P<0.05 compared with PBS and anti-miR-ctl control. (B) Western blotting for KLF4 expression in carotid arteries from PBS-, anti-miR-ctl- and anti-miR-146a-treated rats at 14 days after balloon injury. (C) Haematoxylin–eosin staining of carotid arteries from PBS-, anti-miR-ctl- and anti-miR-146a-treated rats at 14 days after balloon injury. Neointimal formation was calculated as the I/M ratio. *P<0.05 compared with PBS and anti-miR-ctl control. (D) PCNA immunohistochemical staining of carotid arteries from PBS-, anti-miR-ctl- and anti-miR-146a-treated rats at 14 days after balloon injury. I/M, intima/media; KLF, Krüppel-like factor; PBS, phosphate-buffered saline; PCNA, proliferating cell nuclear antigen; qRT–PCR, quantitative real-time PCR.
Discussion
KLF4 and KLF5 are closely related members of the KLF family of transcription factors; a pair of positive and negative regulators of cellular proliferation. They bind to a similar DNA sequence that has either a CACCC homology or is rich in GC content. KLF4 expression is regulated directly by interaction of KLF4 and KLF5 with the same GC-rich elements on the KLF4 promoter (Dang et al, 2002). Here, we demonstrate a new regulatory mechanism whereby KLF4 expression is regulated indirectly by KLF4 and KLF5 through miR-146a-mediated post-transcriptional inhibition. In this regulatory network, miR-146a inhibits KLF4 expression by targeting its 3′-UTR (Fig 1), and KLF4 inhibits miR-146a transcription by binding to CACCC (or GGGTG) elements on the miR-146a promoter (Fig 2). miR-146a and KLF4 form a feedback loop to regulate each other's expression. Our results provide a new insight into the direct relationship between miR-146a and KLF4. In addition, KLF5 exhibits an opposing effect on miR-146a transcription as a KLF4 competitor. KLF5 expression was suppressed by anti-miR-146a (supplementary Fig S2D online), which might be due to the anti-miR-146a-mediated KLF4 upregulation.
Cell-specific physiological effects are one of the defining characteristics of miRNAs. For example, miR-21 has an anti-apoptotic effect in VSMCs (Ji et al, 2007) and glioblastoma cells (Chan et al, 2005); however, it does not have the same effect in HeLa cells (Cheng et al, 2005). It has been reported that miR-146a has important roles in innate immune responses and cancer metastasis (Williams et al, 2008). We found that miR-146a promotes VSMC proliferation and vascular neointimal hyperplasia, and that KLF4 is involved in a miR-146a-mediated proliferative effect on VSMCs. How does KLF4 work in regulating VSMC proliferation? We—as well as other groups—have demonstrated that KLF4 has an antiproliferative effect on VSMCs by upregulating p21 (Wassmann et al, 2007) and VSMC differentiation genes SM22α and α-smooth muscle actin, and by downregulating the dedifferentiation gene Smemb (Wang et al, 2008). This effect depends on the platelet-derived growth factor-BB receptor-mediated phosphoinositide 3-kinase signalling pathway (Zheng et al, 2009).
In conclusion, the data presented here, in conjunction with previous work, show that miR-146a and KLF4 form a feedback loop to regulate VSMC proliferation and vascular neointimal hyperplasia (supplementary Fig S5 online). miR-146a might represent a new therapeutic target for proliferative vascular diseases.
Methods
See the supplementary information online for synthetic oligonucleotides, cell culture and transfection, construction of recombinant vectors, oligonucleotide pull-down assay, site-directed mutagenesis, northern blotting, western blotting, balloon injury model and oligonucleotide treatment, morphology analysis and immunohistochemistry.
RNA preparation and qRT–PCR. Small RNAs from cultured VSMCs were extracted using a mirVana miRNA Isolation Kit (Ambion) according to the manufacturer's recommendations. qRT–PCR of miR-146a was performed using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems) with the mirVana qRT–PCR miRNA Detection Kit (Ambion). As an internal control, U6 primers were used for RNA template normalization. All PCRs were performed in triplicate. The relative expression level was calculated using the following equation: relative gene expression=2−(ΔCtsample–ΔCtcontrol).
Reporter gene assay. 293A cells were grown to 70% confluence and were transfected in triplicate with different constructs along with pRL-TK. The cells were harvested after 24 h and the activities of both firefly and Renilla luciferases were measured in the LB 955 Luminometer system using the dual luciferase reporter system (Promega), according to the manufacturer's recommendations. The activity of firefly luciferase was normalized to that of Renilla luciferase.
Chromatin immunoprecipitation assay. Briefly, VSMCs were treated with 1% formaldehyde for 15 min to crosslink proteins with DNA. The crosslinked chromatin was then prepared and sonicated to an average size of 0.4–0.5 kb. The samples were precleared with protein A-agarose/salmon sperm DNA (Sigma) for 30 min at 4°C, followed by an overnight incubation at 4°C with KLF4 or KLF5 antibodies (1:400 dilution, Santa Cruz). DNA was extracted after the reversal of crosslinking at 65°C for 4 h. The genomic region of miR-146a promoter containing all three potential KLF4/KLF5-binding sites was amplified by PCR with primers 5′-TCGGGGTACCGAGAATGCCTGTGAGGGGACTAT-3′ and 5′-GAAAGCTACTGGTCACAGGACAGC-3′.
EMSA. EMSA was performed using the LightShift Chemiluminscent EMSA Kit (Pierce), according to the manufacturer's recommendations. Briefly, a biotin-labelled probe containing three KLF4/KLF5-binding sites for the miR-146a promoter was synthesized by Sangon. The sequence is as follows: 5′-AAGGAGGGTGGCACCCTCCCCTGGGTGGTGTC-3′. A total of 10 ng of purified recombinant KLF4 or KLF5 was incubated with the biotin-labelled probe for 30 min at 25°C, loaded onto a 6% non-denaturating polyacrylamide gel and run at 100 V for 40 min, then electrotransferred to a nylon membrane. The probe was crosslinked to the membrane by exposure to 254 nm ultraviolet radiation for 15 min. Finally, the biotin-labelled probe was detected by chemiluminescence, according to the manufacturer's instructions. To specifically identify KLF4 or KLF5 protein in binding complexes, 2 μg of KLF4 or KLF5 antibody (Santa Cruz Biotechnology) was added to the binding reaction mix and was incubated for 30 min at 25°C before adding the probe.
Statistical analysis. Data presented as bar graphs are the means±s.e.m. of at least three independent experiments. Statistical analysis was performed using Student's t-test.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of the People's Republic of China (No. 30971457, 90919035, 31071003, 30871272), the Natural Science Foundation of Hebei Province of China (No. C2009001095) and the Office of Education Foundation of Hebei Province of China (No. 2009150).
Footnotes
The authors declare that they have no conflict of interest.
References
- Chan JA, Krichevsky AM, Kosik KS (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65: 6029–6033 [DOI] [PubMed] [Google Scholar]
- Chen T, Huang ZQ, Wang LS, Wang Y, Wu FZ, Meng S, Wang CQ (2009) MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res 83: 131–139 [DOI] [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, Shelton J, Ford LP (2005) Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 33: 1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D (2009) miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460: 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A (2009) Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem 284: 3728–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang DT, Zhao W, Mahatan CS, Geiman DE, Yang VW (2002) Opposing effects of Krüppel-like factor 4 (gut-enriched Krüppel-like factor) and Krüppel-like factor 5 (intestinal-enriched Krüppel-like factor) on the promoter of the Krüppel-like factor 4 gene. Nucleic Acids Res 30: 2736–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C (2007) MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res 100: 1579–1588 [DOI] [PubMed] [Google Scholar]
- Kawai-Kowase K, Owens GK (2007) Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. Am J Physiol Cell Physiol 292: C59–C69 [DOI] [PubMed] [Google Scholar]
- Kumar MS, Owens GK (2003) Combinatorial control of smooth muscle-specific gene expression. Arterioscler Thromb Vasc Biol 23: 737–747 [DOI] [PubMed] [Google Scholar]
- Wang C, Han M, Zhao XM, Wen JK (2008) Krüppel-like factor 4 is required for the expression of vascular smooth muscle cell differentiation marker genes induced by all-trans retinoic acid. J Biochem 144: 313–321 [DOI] [PubMed] [Google Scholar]
- Wassmann S, Wassmann K, Jung A, Velten M, Knuefermann P, Petoumenos V, Becher U, Werner C, Mueller C, Nickenig G (2007) Induction of p53 by GKLF is essential for inhibition of proliferation of vascular smooth muscle cells. J Mol Cell Cardiol 43: 301–307 [DOI] [PubMed] [Google Scholar]
- Williams AE, Perry MM, Moschos SA, Larner-Svensson HM, Lindsay MA (2008) Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem Soc Trans 36: 1211–1215 [DOI] [PubMed] [Google Scholar]
- Zheng B, Bernier M, Zhang XH, Meng F, Miao SB, He M, Zhao XM, Wen JK (2009) Krüppel-like factor 4 inhibits proliferation by platelet-derived growth factor receptor beta-mediated, not by retinoic acid receptor alpha-mediated, phosphatidylinositol 3-kinase and ERK signaling in vascular smooth muscle cells. J Biol Chem 284: 22773–22785 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.