Macrophages recognize streptococci through bacterial single-stranded RNA
Recognition of streptococci and other Gram-positive bacteria by macrophages and monocytes is shown here to rely on bacterial ssRNA. SsRNA interacts with a signaling complex, which comprises the TLR adapters MyD88 and UNC-93B, but not the established MyD88-dependent ssRNA sensors.
Keywords: bacterial infection, RNA, phagocytosis, signal transduction
Abstract
Group B streptococcus (GBS) is a leading cause of both neonatal sepsis and meningitis, two diseases that are characterized by inflammation. However, the manner in which GBS organisms are recognized by monocytes and macrophages is poorly understood. In this study, we report that the recognition of GBS and other Gram-positive bacteria by macrophages and monocytes relies on bacterial single-stranded RNA (ssRNA). ssRNA interacts with a signalling complex, which comprises the Toll-like receptor adaptors MyD88 and UNC-93B, but not the established MyD88-dependent ssRNA sensors. The role of ssRNA in the recognition of Gram-positive bacteria—leading to the induction of inflammatory cytokines—has potential implications for sepsis pathogenesis, diagnosis and treatment.
Introduction
The concept of pathogen sensing by germline-encoded pattern recognition receptors and the more recent discovery of Toll-like receptors (TLRs) have transformed our understanding of innate immunity to bacteria (Medzhitov et al, 1997). TLRs expressed by host immune cells such as macrophages, detect conserved products of microbial biosynthetic pathways, resulting in the formation of inflammatory cytokines. The ligand interaction sites of TLRs face either the extracellular space or the luminal side of phagosomes. In accordance with a crucial role of the TLR system in the recognition of group B streptococcus (GBS), MyD88—the intracellular signalling adaptor of all TLRs, except TLR3 (Biondo et al, 2005)—is essential for the instruction of the cytokine response in macrophages when they are exposed to these bacteria (Henneke & Golenbock, 2002). However, the specific TLR, or combination of TLRs, which interacts with the GBS particle and MyD88, has not been identified in macrophages. Furthermore, activation of the inflammasome does not account for activation of cytokines by GBS (Kenzel et al, 2009). Next to the cognate receptor, the microbial substructure, which accounts for the bulk of the cytokine-inducing properties of GBS particles or other streptococci and staphylococci, is unclear. Many candidate surface molecules—such as capsular polysaccharide, peptidoglycan, lipoteichoic acid and lipoproteins—are not essential in this context (Henneke & Golenbock, 2001; Henneke et al, 2005, 2008; Kenzel et al, 2009). Therefore, both the macrophage and microbial contributions to the receptor–ligand interface for bacterial particles—which are potent inducers of inflammatory cytokines—have not been defined for GBS or other non-flagellated Gram-positive bacteria (Valenti-Weigand et al, 1996; Lembo et al, 2003). By contrast, extracellular GBS lipoproteins are known to interact with TLR2/6 and thereby contribute to GBS sepsis pathogenesis (Henneke et al, 2005, 2008; Gratz et al, 2008). Furthermore, GBS DNA induces type I interferons through an unknown receptor, but this mechanism does not account for the induction of type I inflammatory cytokines—such as tumour necrosis factor (TNF)—in response to GBS (Charrel-Dennis et al, 2008). Knowledge of discrete molecular events—which determine the response of the single-tissue macrophage-encountering invading GBS—is important for the understanding of the infection-fighting abilities of the host.
Here, we report on a new and conserved mechanism through which Gram-positive bacteria are recognized by macrophages. It involves bacterial single-stranded RNA (ssRNA) and the host molecules MyD88 and UNC-93B, but it occurs independently of known nucleotide-sensing TLRs.
Results And Discussion
GBS recognition by macrophages depends on bacterial ssRNA
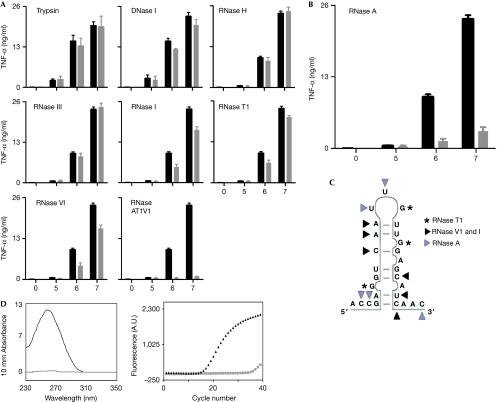
The molecules in streptococcal and staphylococcal particles, which induce inflammatory cytokines in macrophages and monocytes are largely unknown. We investigated this by using systematic enzymatic digestion of GBS, followed by a loss-of-cytokine-induction model. We found that GBS treatment with RNase A, which degrades ssRNA at 3′ cytosine and uracil residues, reduced TNF formation by around 80% (Fig 1B). This was in contrast to the depletion of GBS double-stranded RNA (dsRNA; with RNase III), RNA–DNA hybrids (with RNase H) and DNA (with DNase I), none of which reduced the ability of GBS to induce TNF (Fig 1A). Furthermore, trypsin digestion of GBS proteins did not impair the TNF induction by GBS (Fig 1A). These data indicate that bacterial ssRNA is more effective than bacterial DNA in inducing TNF in macrophages. Furthermore, the effect of ssRNA was sequence specific; as RNase I, which degrades ssRNA at all dinucleotide pairs, RNase T1, which degrades ssRNA at the 3′ end of guanine residues and RNase V1, which degrades ssRNA at all base-paired nucleotides, had only a minor effect on the TNF response (Fig 1A). The stimulating ssRNA was non-ribosomal, as RNase III—which cleaves dsRNA present in ribosomal RNA (rRNA; 16S rRNA and 23S rRNA) from the polycistronic RNA operon in prokaryotes (D'Alessio & Riordan, 1997)—failed to show loss of TNF induction. For the induction of TNF, 3′ unpaired cytosine and uracil residues of GBS ssRNA are most important, as shown by the cleavage at these sites with RNase A. RNA is an attractive target for the sampling of prokaryotes by macrophages, as RNA quantitatively exceeds DNA by approximately tenfold and a high concentration of bacterial RNA is present in the lumen of bacteria containing phagosomes (Mancuso et al, 2009).
Figure 1.
GBS ssRNA with 3′ of single-stranded cytosine and uracil residues is essential for transcriptional activation of inflammatory cytokines. BMDM from wild-type mice were stimulated with GBS (105, 106 and 107 per ml), which had been depleted for specific macromolecules by enzymatic treatment (grey bars). (A) Trypsin (digestion of proteins), DNase I (degradation of DNA), RNase H (degradation of RNA strands in RNA–DNA hybrids), RNase III (degradation of dsRNA), RNase I (degradation of ssRNA at all dinucleotide pairs), RNase T1 (degradation of ssRNA at 3′ of all single-stranded guanine residues), RNase V1 (degradation of ssRNA at all base-paired nucleotides), combination of RNases A, T1 and V1 (maximal degradation of ssRNAs). (B) RNase A (degradation of ssRNA at 3′ of single-stranded cytosine and uracil residues). (C) Model for sequence and structural specificities of ssRNA-degrading enzyme RNase A (grey triangle), RNase V1 and I (black triangle), RNase T1(asterisk). (D) Absorbance spectrum of RNA and real-time PCR amplification plot of complementary DNA (GBS caf gene). RNA isolated from RNase A-treated (grey) and -untreated (black) bacteria. Data shown are mean values±s.d. (n=3), representative of at least three independent experiments. BMDM, bone-marrow-derived macrophages; dsRNA, double-stranded RNA; GBS, group B streptococci; ssRNA, single-stranded RNA; TNF, tumour necrosis factor; WT, wild-type.
GBS ssRNA induces cytokines via MyD88 and UNC-93B
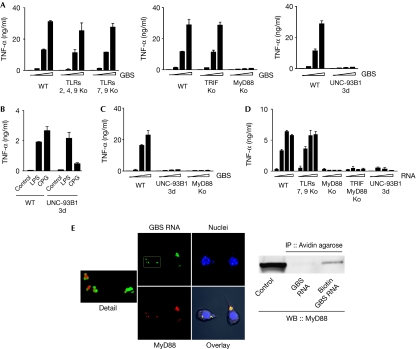
Previously, we had found that MyD88 is a key adaptor in the recognition of GBS organisms and that phagosomal processing of the bacteria is a prerequisite for cytokine induction. Hence, we wondered whether endosomal nucleotide sensing mediated the response to bacterial ssRNA. To address this question experimentally, we analysed macrophages from mice with a point mutation in UNC-93B, resulting in a H412R amino-acid exchange (Tabeta et al, 2006). UNC-93B is a multispanning transmembrane endoplasmic reticulum protein, which enables localization and processing of endosomal TLRs (Kim et al, 2008). We found that UNC-93B was required for efficient cytokine induction by both fixed and live bacteria (Fig 2A,C). Mutation of UNC-93B residue 412 abrogates the interaction with the endosomal nucleotide sensors TLRs 3, 7 and 9 (Brinkmann et al, 2007). TLR7 was an attractive candidate receptor for streptococcal ssRNA, as it was recently reported to mediate the induction of type I interferon by GBS in conventional dendritic cells (cDCs; Mancuso et al, 2009). We therefore analysed macrophages from mice deficient in both TLR7 and TLR9. However, in contrast to our expectations, we found that TNF induction by GBS occurred independently of these receptors and of TLRs 2 and 4 (Fig 2A). Moreover, TRIF (TIR domain-containing adaptor-inducing interferon β)—the essential adaptor of the dsRNA receptor TLR3—had a redundant role in cytokine induction by GBS organisms (Fig 2A). Most experimental evidence suggests that TLR8, which recognizes ssRNA in humans, is not functional in mice (Heil et al, 2004; Bauer et al, 2010; Martinez et al, 2010). Moreover, we determined TLR8 transcript levels in mouse macrophages by using quantitative PCR and found marginal to no expression (data not shown), consistent with previous studies (Fukui et al, 2009). Accordingly, the single TLRs—which have been previously shown to sense endosomal nucleic acids—were not involved in the MyD88- and UNC-93B-dependent recognition of GBS. Finally, the retinoic acid inducible gene I (RIG-I)-like helicases RIG-I and MDA5, which interact with ssRNA from viruses, were not essential in this context, as indicated by the normal cytokine response of both RIG-I- and MDA5-deficient mouse macrophages (data not shown).
Figure 2.
GBS and GBS RNA induce inflammatory cytokines in a MyD88-dependent manner—independently of TLRs. BMDM lacking the indicated TLRs or TLR adaptors or UNC-93B were stimulated with (A) GBS (105, 106 and 107 per millilitre) or (D) GBS RNA (0, 1, 5 and 10 μg/ml). After 24 h, the TNF level in the supernatants was measured by ELISA. (B) TNF production by wild-type and mutant macrophages stimulated with LPS or CPG. (C) BMDM lacking MyD88 or UNC-93B were stimulated with live GBS (5, 10 and 100 MOI) and the TNF level in the supernatants was measured by ELISA. (E) RNA from ethyluridine-incorporated GBS was labelled with the Click-iT RNA labelling kit. MyD88-YFP-expressing RAW264.7 macrophages were stimulated with labelled GBS RNA and observed with CLSM. ‘Detail' shows a magnified version of the area indicated by the rectangle in GBS RNA, representing FITC and YFP two-channel overlay after spectral unmixing. ‘GBS RNA', ‘MyD88' and ‘Nuclei' represent merged confocal planes, ‘Overlay' is a single plane simultaneous acquisition in all three and PMT channel. Thus, not all events of the merged images are visible in ‘Overlay'. MyD88-YFP-expressing RAW264.7 macrophages were stimulated with biotinylated GBS RNA (1 μg/ml) for 2 h. Lysates of these cells were subjected to IP with Avidin agarose and WB analysis with MyD88-specific antibodies. Data shown are mean values±s.d. (n=3), representative of at least three independent experiments. BMDM, bone-marrow-derived macrophages; CPG, Cpg-DNA; CLSM, confocal laser scanning microscope; ELISA, enzyme-linked immunosorbent assay; FITC, fluorescein isothiocyanate; GBS, group B streptococci; IP, immunoprecipitation; LPS, lipopolysaccharide; MOI, multiplicity of infection; TLR, Toll-like receptor; TNF, tumour necrosis factor; WB, western blotting; WT, wild type; YFP, yellow fluorescent protein.
Next, we wondered whether RNA from GBS was not only a crucial and potent inflammatory stimulus in GBS organisms, but whether it also induced type I cytokines as a purified structure. We found that purified GBS RNA engaged the same signalling pathways as whole GBS organisms; that is, MyD88 and UNC-93B were required for cytokine induction, whereas TLRs 7 and 9, RIG-I and MDA5 were not (Fig 2D; data not shown). Thus, the response to GBS RNA was similar to that of GBS organisms.
With our understanding of the role of MyD88 in sensing GBS RNA, we assessed whether these structures localized to each other in macrophages. First, we monitored the spatial distribution of GBS RNA—which was labelled with green fluorescent 5-ethynyl uridine—and MyD88. At 60 min after the addition of GBS RNA to macrophages, we found that it colocalized with MyD88-yellow fluorescent protein in distinct perinuclear spots (Fig 2E). Second, colocalization of GBS RNA and MyD88 in one complex was confirmed by coimmunoprecipitation, experiments in which MyD88 was pulled down together with biotinylated GBS RNA (Fig 2E).
ssRNA is a specific pattern of Gram-positive bacteria
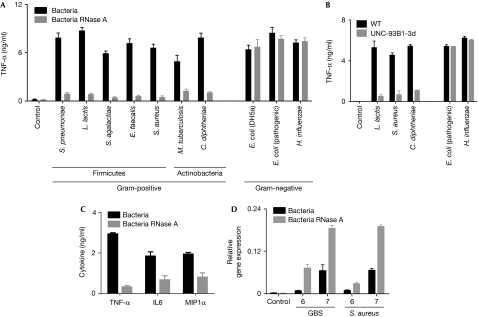
Next, we investigated whether the essential role of ssRNA is a particular feature of GBS-induced cytokine formation, or whether other bacteria are recognized through a similar mechanism. We found that Streptococcus pneumoniae, Lactococcus lactis, Enterococcus faecalis and Staphylococcus aureus—which belong to the Firmicutes group—lost 70–90% of their TNF-inducing capacity when depleted of ssRNA using RNase A (Fig 3A). Similarly, TNF induction by the Actinobacteria Corynebacterium diphtheriae and Mycobacterium tuberculosis was abrogated on ssRNA digestion. By contrast, the TNF-inducing properties of the Gram-negative rods Escherichia coli and Haemophilus influenzae were preserved after depletion of ssRNA (Fig 3A) and in UNC-93B-deficient cells (Fig 3B). In accordance with this finding, RNase A digestion of purified nucleic acids from E. coli did not significantly reduce their cytokine-inducing ability (data not shown). These findings suggest that bacterial ssRNA is needed for the macrophage response to Gram-positive, but not Gram-negative bacteria.
Figure 3.
ssRNA from Gram-positive but not Gram-negative bacteria essentially contributes to cytokine formation; ssRNA recognition is evolutionarily conserved. (A) BMDM from wild-type mice were stimulated with the indicated species of Gram-positive or Gram-negative bacteria (black bars) or with similar bacterial preparations depleted of ssRNA (grey bars, each 106 per millilitre). After 24 h, TNF level was determined by ELISA. (B) BMDM from wild-type mice (black bars) and UNC-93B mice (grey bars) were stimulated with the indicated species of bacteria (each 106 per millilitre), and after 24 h TNF level was determined by ELISA. (C) Human peripheral blood mononuclear cells were stimulated with GBS and RNase A-treated GBS (concentrations 105, 106 and 107 per millilitre) for 24 h. Cytokine level was determined by ELISA. (D) Fully expanded leaves of Arabidopsis thaliana were infiltrated with GBS or Staphylococcus aureus (106 and 107 per millilitre) with and without RNase A treatment, and PR1b gene expression was measured after 24 h by quantitative PCR. Data shown are mean values±s.d. (n=3), representative of at least three independent experiments. BMDM, bone-marrow-derived macrophages; ELISA, enzyme-linked immunosorbent assay; GBS, group B streptococci; IL6, interleukin-6; PR1b, pathogenesis-related gene 1; ssRNA, single-stranded RNA; TNF, tumour necrosis factor; WT, wild type.
The recognition of streptococci by their ssRNA was not only a key event in mouse macrophages but also in human monocytes (Fig 3C) and monocyte-derived macrophages (data not shown). Furthermore GBS ssRNA was crucial for the induction of interleukin-6, type I interferons and chemokine macrophage inflammatory protein 1α (MIP1α), supporting the importance of nucleotide recognition in antibacterial inflammation (Fig 3C; data not shown).
Outside the mammalian class, in plants Gram-positive bacteria induce various pathogenesis-related (PR) genes such as PR1B and plant defensin PDF1.2 (Prithiviraj et al, 2005; Gust et al, 2007). Although some components of the immune systems are evolutionarily conserved in plants and mammals, the specific response to a given microbial stimulus can differ substantially (Prithiviraj et al, 2005; Boller & He, 2009). Here, we determined the contribution of Gram-positive bacterial ssRNA to the activation of PR genes, which are known to be upregulated in bacterial infection (Prithiviraj et al, 2005). We found that the infiltration of Arabidopsis with GBS or S. aureus enhanced the transcription of PR1b and PDF1.2. However, in contrast to the cytokine response in mammalian phagocytes, depletion of ssRNA from GBS resulted in enhanced expression of PR1b and PDF1.2 (Fig 3D; data not shown). It seems that recognition of bacterial ssRNA is conserved in all kingdoms, although during the course of evolution the specific response has apparently been adapted for cell-specific functions. In accordance with this model, the Pseudomonas syringae effector HopU1 modifies several Arabidopsis RNA-binding proteins (Boller & He, 2009). The exact mechanisms by which bacterial ssRNA is recognized by plants remain to be elucidated.
Our results concur with other studies that have found endosomal processing of extracellular bacteria and transcriptional activation of cytokines to be closely interlinked in macrophages. However, our study challenges the current paradigm, which assigns bacterial DNA and lipidated proteins exceptional roles in pattern recognition of Gram-positive bacteria by macrophages (Talati et al, 2008). Neither of these microbial structures or their respective cognate receptors (TLRs 2 and 9) were shown to be crucial for initiating a potent macrophage response to whole bacterial organisms (Fig 2; data not shown). By contrast, recognition of bacterial ssRNA was required for the cytokine responses and MyD88 and UNC-93B were essential in this process. MyD88 and UNC-93B have complementary adaptor functions in the context of TLRs 3, 7, 8 and 9; however, neither of these endosomal receptors is essential for the recognition of GBS. The interaction between UNC-93B and MyD88 is poorly understood, and several issues therefore remain to be clarified to understand fully this new mechanism of bacterial recognition. First, it is unclear whether MyD88 partly exerts its effect by propagation of phagosomal processing and cleavage of TLRs, which seems to be a prerequisite for the sensing of endosomal nucleic acids (Blander & Medzhitov, 2004; Ewald et al, 2008). Second, the polytopic membrane protein UNC-93B delivers the nucleotide-sensing receptors TLR7 and TLR9 from the endoplasmic reticulum to endolysosomes and does not seem to be involved directly in signal transduction by these receptors (Kim et al, 2008). However, foreign nucleotides are sensed through the cytoplasmic RIGI-like helicase system and it is unclear whether UNC-93B is involved in this pathway.
One important finding of this study is that ssRNA is involved in the recognition of Gram-positive bacteria, whereas Gram-negative bacteria seem to be detected through other molecular structures. Moreover, recognition of bacterial ssRNA occurs in a cell-lineage-specific manner. In contrast to macrophages, cDCs recognize GBS ssRNA by means of TLR7 (Mancuso et al, 2009). Whether differences in the recognition of microbial substructures relate to compartmentalization of bacterial classes to colonizing sites (predominantly urogenital and gastrointestinal compared with respiratory and cutaneous) remains to be determined.
Taken together, the data presented here and elsewhere, indicate that the recognition of ssRNA is important for both the type I macrophage response and the type I interferon cDC response to Gram-positive bacteria. Sensing of streptococcal ssRNA probably contributes to invasive disease by some of the most dangerous infectious agents worldwide.
Methods
Animals and cell lines. Mice lacking MyD88 and those lacking TLRs, along with respective wild-type mice (Adachi et al, 1998; Takeuchi et al, 1999), were kindly provided by Dr S. Akira, Department of Host Defence, Osaka University (Osaka, Japan). Mice with an H412R mutation of UNC-93B were kindly provided by Bruce Beutler (La Jolla, CA, USA). Immortalization of MyD88-TRIF-deficient mice was performed as described previously (Charrel-Dennis et al, 2008). RIG-I- and MDA5-deficient BMDMs were kindly provided by Kathrin Fitzgerald, Department of Infectious Diseases and Immunology, University of Massachusetts (Worcester, MA, USA).
Preparation of bacteria and digestion of bacterial RNA/nucleic acids. The GBS type III strain COH1—which was originally isolated from a newborn infant with sepsis—was cultured to exponential growth phase in a chemically defined medium and heat fixed, as described previously (Kenzel et al, 2009). For depletion of nucleic acids from GBS, 109 heat-inactivated GBS was incubated with the indicated nucleases according to the manufacturer's instructions (MBI-Fermentas). Depletion of nucleic acids was confirmed using spectrophotometric observation (A260/280), followed by real-time PCR (Fig 1D). Functional and structural integrity of nucleic-acid-depleted bacteria was confirmed by phagocytosis, fluorescence and electron microscopy.
Macrophage infection. Macrophage differentiation and culturing were carried out as described previously (Charrel-Dennis et al, 2008). Before infection, non-adherent cells were removed by washing with phosphate-buffered saline, and the medium was replaced by a medium supplemented with 10% FBS, with or without GBS. Samples were analysed at different time points after infection, up to 48 h. Live bacterial infections were performed as described previously (Mancuso et al, 2009), with some changes: GBS were grown in chemically defined medium to exponential growth phase, followed by extensive washing with phosphate-buffered saline. Bacteria were added to monolayers at various multiplicity of infection (bacteria/cell). Monolayers were washed extensively after 30 min at 37 °C and incubated for various time intervals in the presence of antibiotics. The bacterial count was determined by colony counting on blood agar, according to standard procedures.
Determination of TNF, interleukin-6 and MIP1α levels. Cytokine levels in the macrophage culture medium were quantified by using enzyme-linked immunosorbent assay kits, according to the manufacturer's instructions (R&D Systems).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We are grateful to Patrick Tieu-Cuot and Sarah Dubrac (Paris) for helping us to label bacterial RNA and Taras Pasternak (Freiburg) for helping with Arabidopsis thaliana. We are indebted to L. Fuchs, S.-H. Seibel, A. Leibinger, S. Sellner, A. Imm and M. Häffner for outstanding technical assistance. This work was supported by grants from the Bundesministerium für Bildung und Forschung (BMBF 01 EO 0803), the Deutsche Forschungsgemeinschaft (He 3127/2-3 and 3-1) and the National Institutes of Health (ROI AI052455-06A1, to D.T.G.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagawi M, Nakanishi K, Akira S (1998) Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9: 143–150 [DOI] [PubMed] [Google Scholar]
- Bauer S et al. (2010) A major role for TLR8 in the recognition of vaccinia viral DNA by murine pDC? Proc Natl Acad Sci USA 107: E139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondo C, Midiri A, Messina L, Tomasello F, Garufi G, Catania MR, Bombaci M, Beninati C, Teti G, Mancuso G (2005) MyD88 and TLR2, but not TLR4, are required for host defense against Cryptococcus neoformans. Eur J Immunol 35: 870–878 [DOI] [PubMed] [Google Scholar]
- Blander JM, Medzhitov R (2004) Regulation of phagosome maturation by signals from Toll-like receptors. Science 304: 1014–1018 [DOI] [PubMed] [Google Scholar]
- Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM (2007) The interaction between the ER membrane protein UNC-93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol 177: 265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel-Dennis M, Latz E, Halmen KA, Trieu-Cuot P, Fitzgerald KA, Kasper DL, Golenbock DT (2008) TLR-independent type I interferon induction in response to an extracellular bacterial pathogen via intracellular recognition of its DNA. Cell Host Microbe 4: 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessio G, Riordan JF (1997) Ribonucleases: Structure and Function. New York and London: Academic [Google Scholar]
- Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM (2008) The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456: 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui R, Saitoh S, Matsumoto F, Kozuka-Hata H, Oyama M, Tabeta K, Beutler B, Miyake K (2009) UNC-93B biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA- but against RNA-sensing. J Exp Med 206: 1339–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz N et al. (2008) Group A streptococcus activates type I interferon production and MyD88-dependent signaling without involvement of TLR2, TLR4, and TLR9. J Biol Chem 283: 19879–19887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust AA et al. (2007) Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J Biol Chem 282: 32338–32348 [DOI] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S (2004) Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303: 1526–1529 [DOI] [PubMed] [Google Scholar]
- Henneke P et al. (2008) Lipoproteins are critical TLR2 activating toxins in group B streptococcal sepsis. J Immunol 180: 6149–6158 [DOI] [PubMed] [Google Scholar]
- Henneke P, Golenbock DT (2001) TIRAP: how Toll receptors fraternize. Nat Immunol 2: 828–830 [DOI] [PubMed] [Google Scholar]
- Henneke P, Golenbock DT (2002) Innate immune recognition of lipopolysaccharide by endothelial cells. Crit Care Med 30: S207–S213 [DOI] [PubMed] [Google Scholar]
- Henneke P et al. (2005) Role of lipoteichoic acid in the phagocyte response to group B streptococcus. J Immunol 174: 6449–6455 [DOI] [PubMed] [Google Scholar]
- Kenzel S, Santos-Sierra S, Deshmukh SD, Moeller I, Ergin B, Fitzgerald KA, Lien E, Akira S, Golenbock DT, Henneke P (2009) Role of p38 and early growth response factor 1 in the macrophage response to group B streptococcus. Infect Immun 77: 2474–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Brinkmann MM, Paquet ME, Ploegh HL (2008) UNC-93B delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 452: 234–238 [DOI] [PubMed] [Google Scholar]
- Lembo A, Kalis C, Kirschning CJ, Mitolo V, Jirillo E, Wagner H, Galanos C, Freudenberg MA (2003) Differential contribution of Toll-like receptors 4 and 2 to the cytokine response to Salmonella enterica serovar Typhimurium and Staphylococcus aureus in mice. Infect Immun 71: 6058–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, Akira S, Teti G, Beninati C (2009) Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat Immunol 10: 587–594 [DOI] [PubMed] [Google Scholar]
- Martinez J, Huang X, Yang Y (2010) Toll-like receptor 8-mediated activation of murine plasmacytoid dendritic cells by vaccinia viral DNA. Proc Natl Acad Sci USA 107: 6442–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Janeway CA Jr (1997) A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388: 394–397 [DOI] [PubMed] [Google Scholar]
- Prithiviraj B, Bais HP, Jha AK, Vivanco JM (2005) Staphylococcus aureus pathogenicity on Arabidopsis thaliana is mediated either by a direct effect of salicylic acid on the pathogen or by SA-dependent, NPR1-independent host responses. Plant J 42: 417–432 [DOI] [PubMed] [Google Scholar]
- Tabeta K et al. (2006) The UNC-93B mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol 7: 156–164 [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Hashino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S (1999) Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11: 443–451 [DOI] [PubMed] [Google Scholar]
- Talati AJ, Kim HJ, Kim YI, Yi AK, English BK (2008) Role of bacterial DNA in macrophage activation by group B streptococci. Microbes Infect 10: 1106–1113 [DOI] [PubMed] [Google Scholar]
- Valenti-Weigand P, Benkel P, Rohde M, Chhatwal GS (1996) Entry and intracellular survival of group B streptococci in J774 macrophages. Infect Immun 64: 2467–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.