Abstract
Oxygen is required for aerobic energy production but its levels have to be tightly regulated to avoid deleterious effects. Thus, animals have evolved mechanisms to monitor and respond to fluctuations in oxygen availability. Here, the evolution of the HIF system is discussed in light of a report that reveals its presence in the simplest animal.
EMBO Rep (2010) advance online publication. doi: 10.1038/embor.2010.170
All animal life requires molecular oxygen as the terminal electron acceptor in aerobic energy production. A lack of oxygen can reduce the rate of energy production, whereas an excess of oxygen leads to the accumulation of toxic reactive oxygen species. Hence, animals have evolved sophisticated mechanisms with which to monitor and respond to fluctuations in oxygen availability, in order to maintain cellular homeostasis. In all animal taxa examined so far, the maintenance of physiological oxygen homeostasis is mediated by the oxygen-dependent post-translational hydroxylation of a heterodimeric transcription factor, termed hypoxia-inducible factor (HIF; Kaelin & Ratcliffe, 2008). The hydroxylation reaction is catalysed by prolyl hydroxylase (PHD) enzymes, which are direct sensors of cellular oxygen tension. Under normoxia, HIFα is hydroxylated in a PHD-dependent manner, which leads to its ubiquitination by the von Hippel–Lindau protein (VHL) and proteasomal degradation. Under hypoxia, the hydroxylase activity of PHD enzymes is inhibited, thereby allowing the stable formation of the heterodimeric HIF transcription factor and its activation. HIF is then translocated to the nucleus where it activates the transcription of numerous target genes involved in processes that enhance oxygen delivery—such as erythropoiesis and angiogenesis—or improve prospects for survival under hypoxia, by altering energy metabolism.
The evolutionary origins of this central physiological regulatory system have been unclear, as the regulatory interactions of the constituent HIF and PHD genes have not been experimentally characterized in non-bilaterian animals. In this issue of EMBO reports, the Schofield lab demonstrate that the HIF system has a regulatory oxygen-sensing function in the simplest known animal, the placozoan Trichoplax adhaerens (Loenarz et al, 2010). The ancestors of T. adhaerens seem to have diverged from the lineage leading to bilaterian animals more than 550 million years ago, in the Precambrian.
Loenarz and colleagues conducted a comparative genomic analysis revealing that the three main components of the HIF system—HIF, PHD and VHL—are present in all the metazoans, including T. adhaerens, but are not found in non-metazoan taxa such as choanoflagellates or other protists. The authors then demonstrated that T. adhaerens HIFα has an oxygen-dependent degradation (ODD) domain containing the critical proline residue that is hydroxylated by PHD. This proline-containing ODD domain sets HIFα apart from other metazoan bHLH–PAS transcription factors. Purified T. adhaerens PHD can hydroxylate ODD peptide fragments of HIFα from both T. adhaerens and human HIFα in cell culture, and the expression of T. adhaerens PHD rescues the effects of silencing PHD2 in human cells. Furthermore, ODD peptides from other distantly related animal taxa are hydroxylated by human PHD2, which demonstrates that the core PHD–HIF interaction has been conserved over more than half a billion years of evolution.
…a HIF-mediated switch from aerobic mitochondrial energy production to anaerobic energy production is conserved in all metazoan taxa
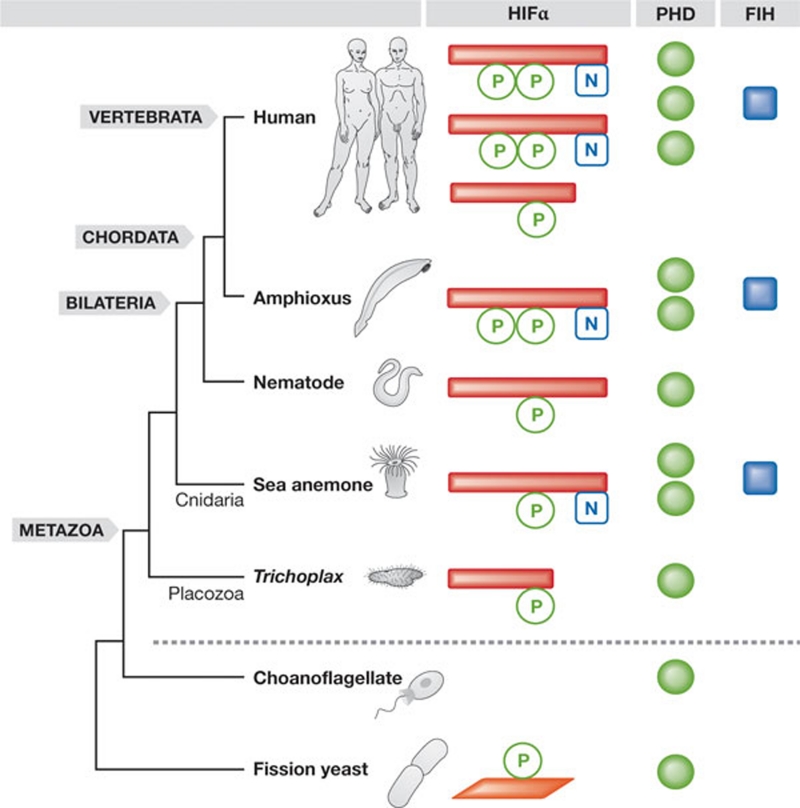
Interestingly, T. adhaerens seems to have many levels of feedback regulation that function to avoid an excessive HIF response. First, the authors observed hypoxic upregulation of PHD. Second, an alternatively spliced HIFα without ODD was described, the relative abundance of which decreased under hypoxia. The oxygen-dependent splicing of HIFα has previously been demonstrated in protostomes such as Drosophila, as well as deuterostomes such as humans. Although T. adhaerens is arguably the simplest living animal—it superficially resembles a multicellular amoeba—this organism has a HIF system with complex feedback regulation. This suggests that the core components of the HIF system originated in the metazoan common ancestor. Over the course of animal evolution, the HIF system has been further elaborated and refined in each of the main descendant lineages (Fig 1). In fact, the genomic locations of PHD and HIFα-gene duplicates in vertebrates suggest that two rounds of whole-genome duplication in the stem lineage of vertebrates might have had a crucial role in the functional diversification of the HIF system.
Figure 1.
Hypoxia-inducible factor pathway evolution. The core components of the HIF pathway were established in the metazoan common ancestor, and the pathway has been subject to further refinements and elaborations in each of the descendant lineages. HIFα homologues are only present in metazoans, whereas PHD homologues are also found in other eukaryotes. FIH homologues are not as widespread in metazoans as PHD enzymes. Two rounds of whole-genome duplication in the stem lineage of vertebrates could be responsible for the many HIFα and PHD paralogues that are found in the human genome. In fission yeast, a PHD homologue interacts with Sre1. FIH, factor inhibiting HIF; HIF, hypoxia-inducible factor; PHD, prolyl hydroxylase; Sre1, sterol regulatory element binding protein.
An additional mechanism of oxygen-dependent regulation of the HIF pathway is provided by a second hydroxylase enzyme called factor inhibiting HIF (FIH). This enzyme regulates transcriptional activity by hydroxylating an asparaginyl residue in the carboxyl terminus of HIFα. Loenarz and colleagues found that the FIH homologues and asparaginyl hydroxylation site of HIFα are absent in T. adhaerens, but present in a non-bilaterian cnidarian, Nematostella vectensis. They also demonstrated that a peptide including the N. vectensis FIH asparaginyl hydroxylation site was hydroxylated by human FIH, suggesting that the regulatory role of FIH was already established in the common ancestor of Cnidaria and Bilateria. However, compared with FIH, it seems that the PHD enzymes have a higher degree of regulatory control in the HIF pathway.
Regarding the HIF target genes, computational analyses by Loenarz and colleagues provide evidence that hypoxia response elements are enriched in the promoters of metazoan genes, compared with those of non-metazoans. When T. adhaerens was exposed to hypoxia, glycolytic enzymes such as pyruvate dehydrogenase kinase were upregulated. Thus, a HIF-mediated switch from aerobic mitochondrial energy production to anaerobic energy production is conserved in all metazoan taxa. In a broader sense, this checkpoint of energy homeostasis could date back to the origins of mitochondria in eukaryotic cells, when endosymbiosis was established between an anaerobic bacterium and an aerobic proto-eukaryote.
Early in animal evolution, increases in body size and mobility presented new challenges as to how to increase the rate of oxidative metabolism while minimizing the production of reactive oxygen species. A direct mechanistic link was recently found for the crosstalk between the HIF pathway and mitochondria. Barth and colleagues (2009) reported that a specific PHD domain, MYND, anchors PHD2 to mitochondrial or endoplasmic reticulum membranes through the protein FKBP38. The authors suggested that membrane-bound PHD2 is degraded, whereas cytosolic PHD2 is stable (Barth et al, 2009). Additionally, the anchor component FKBP38 contains a calcium-binding domain. The evolutionary preservation of the PHD MYND domain noted by the Schofield group suggests that oxygen regulation associated with mitochondrial membrane-bound proteins could have ancient origins.
A recent cross-kingdom comparison of transcriptional responses to hypoxia highlighted that the switch from aerobic to anaerobic energy production is common to bacteria, fungi, plants and animals, but has different signalling and transcription factor components (Mustroph et al, 2010). HIFα belongs to a family of bHLH–PAS transcription factors. The PAS domain responsible for HIF dimerization has ancient origins, as there are prokaryote homologues involved in environmental sensing (Taylor & Zhulin, 1999). However, genome comparisons have shown that the association of PAS domains with a bHLH DNA-binding domain was probably a metazoan invention.
Are there any non-metazoan systems that share characteristics with the PHD–HIF system? Yes, but there is only one documented example of a prolyl hydroxylase and transcription factor dyad in non-metazoan eukaryotes. In fission yeast (Schizosaccharomyces pombe), the predicted prolyl hydroxylase family member Ofd1 regulates a transcription factor called Sre1 (sterol regulatory element binding protein) in an oxygen-dependent manner (Hughes & Espenshade, 2008). However, in budding yeast (Saccharomyces cerevisiae) oxygen sensing is mediated through haem or sterols without the involvement of prolyl hydroxylases (Grahl & Cramer, 2010). Other organisms, such as amoeba (West et al, 2007) and plants (Mustroph et al, 2010), have oxygen-sensing prolyl hydroxylases, but there are no reports connecting these to transcription factors. Future studies in basal metazoans, including sponges, and other eukaryotes could shed light on how the HIF pathway or other prolyl hydroxylase/transcription factor dyads evolved through the co-option of pre-existing proteins for novel oxygen-sensing functions.
References
- Barth S et al. (2009) J Biol Chem 284: 23046–23058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahl N, Cramer RA (2010) Med Mycol 48: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes BT, Espenshade PJ (2008) EMBO j 27: 1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Ratcliffe PJ (2008) Mol Cell 30: 393–402 [DOI] [PubMed] [Google Scholar]
- Loenarz et al. (2010) EMBO Rep doi:10.1038/embor.2010.170 [Google Scholar]
- Mustroph A et al. (2010) Plant Physiol 152: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB (1999) Microbiol Mol Biol Rev 63: 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West CM et al. (2007) Development 134: 3349–3358 [DOI] [PubMed] [Google Scholar]