Elegant post-translational regulation is achieved by ‘microProteins', which form homotypic dimers with their targets and act through the dominant–negative suppression of protein complex function. The recent identification of new microProteins suggests their role is general and has evolved in both the plant and animal kingdoms.
Keywords: homotypic interactions, Id-like proteins, protein–protein interaction, transcription factors
Abstract
Many proteins achieve their function by acting as part of multi-protein complexes. The formation of these complexes is highly regulated and mediated through domains of protein–protein interaction. Disruption of a complex or of the ability of the proteins to form homodimers, heterodimers or multimers can have severe consequences for cellular function. In this context, the formation of dimers and multimers can be perturbed by proteins referred to here as ‘microProteins'. These disruptive protein species contain the protein-interaction domains of bona fide interaction partners, but lack the functional domains required for the activation of, for example, transcription or DNA binding. MicroProteins thus behave as post-translational regulators by forming homotypic dimers with their targets, and act through the dominant–negative suppression of protein complex function. Although the first microProtein was identified more than two decades ago, the recent discovery and characterization of three further small protein species in plants emphasizes their importance. The studies discussed in this review demonstrate that the action of microProteins is general and that it has evolved in both the animal and the plant kingdoms.
See Glossary for abbreviations used in this article.
Glossary.
- bHLH
basic helix-loop-helix
- BLAST
Basic Local Alignment Search Tool
- BMAL
brain and muscle aryl hydrocarbon receptor nuclear translocator-like
- BR
plant Brassinosteroid hormones
- BRI1
Brassinosteroid insensitive 1, BR-receptor kinase
- CRY
CRYPTOCHROME
- HFR1
long hypocotyl in far-red, atypical bHLH
- IBH1
ILI1 binding bHLH
- ILI1
increased leaf–lamina inclination1; rice HLH protein
- KDR
KIDARI, PRE-like HLH protein
- KNATM
MEINOX-domain protein
- NMD
non-sense-mediated mRNA decay
- PAR
phytochrome rapidly upregulated, atypical bHLH
- PER1
PERIOD 1
- PRE
paclobutrazol resistance, HLH protein family
- RACE
rapid amplification of cDNA ends
- SAW1
SAWTOOTH 1
- ZPR
LITTLE ZIPPER, small leucine zipper proteins
Introduction
Herein, we discuss a group of proteins that perturb the formation of functional protein dimers by forming non-functional, homotypic protein complexes with their targets, which they regulate in a dominant-negative manner (Fig 1). We refer to these protein species as ‘microProteins' (miPs)—although some are not small—because the results of their actions are analogous to microRNAs (miRNAs), which are negative regulators of mRNAs.
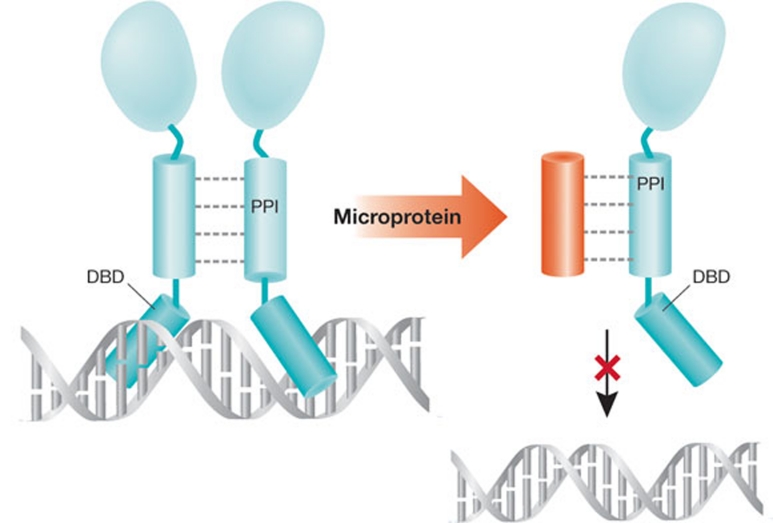
Figure 1.
Model of microProtein interference. Targets of microProteins are often transcriptional regulators that bind to DNA as active homodimers. microProteins interfere with their targets by forming non-functional heterodimeric complexes that cannot bind to DNA. DBD, DNA-binding domain; PPI, protein–protein interaction domain.
miPs can potentially act in the context of any protein that needs to form functional dimers in order to perform its function. This review considers several such cases, many of which are transcription factors. In eukaryotes, protein-encoding genes are transcribed by a protein complex containing the enzyme RNA polymerase II. In conjunction with the basal transcription machinery, the rate of transcription is fine-tuned with the help of several transcriptional regulators—also called transcription factors—which act as DNA-binding factors that can either enhance or repress the transcription of a gene. A common feature of transcriptional regulators—which is often a prerequisite for DNA binding—is that they form functional protein homodimers.
The first miP identified that is able to disrupt a functional transcription factor complex—inhibitor of DNA binding (Id)—was isolated about two decades ago in a search for genes encoding basic helix–loop–helix (bHLH) transcription factors from a murine erythroleukaemia cell cDNA library (Benezra et al, 1990). The Id protein contains a regular HLH domain, but lacks the adjacent basic domain that comprises canonical bHLH transcription factors and is required for transcriptional activation (Benezra et al, 1990). As such, although Id can interact with the bHLH transcriptional regulators MyoD, E12 and E47, it prevents MyoD, for example, from binding to DNA. As MyoD is a tissue-specific transcription factor that is required for muscle differentiation, the researchers assumed that Id acts in a negative manner to fine-tune muscle development. However, later studies showed that Id has a higher affinity for the more ubiquitously expressed E-type bHLH transcriptional regulators, suggesting that Id probably regulates the abundance of MyoD/E-type heterodimers and thus allows the MyoD homodimers to exert their action in a tissue-specific manner (Ohtani et al, 2001; Yates et al, 1999; Fig 2).
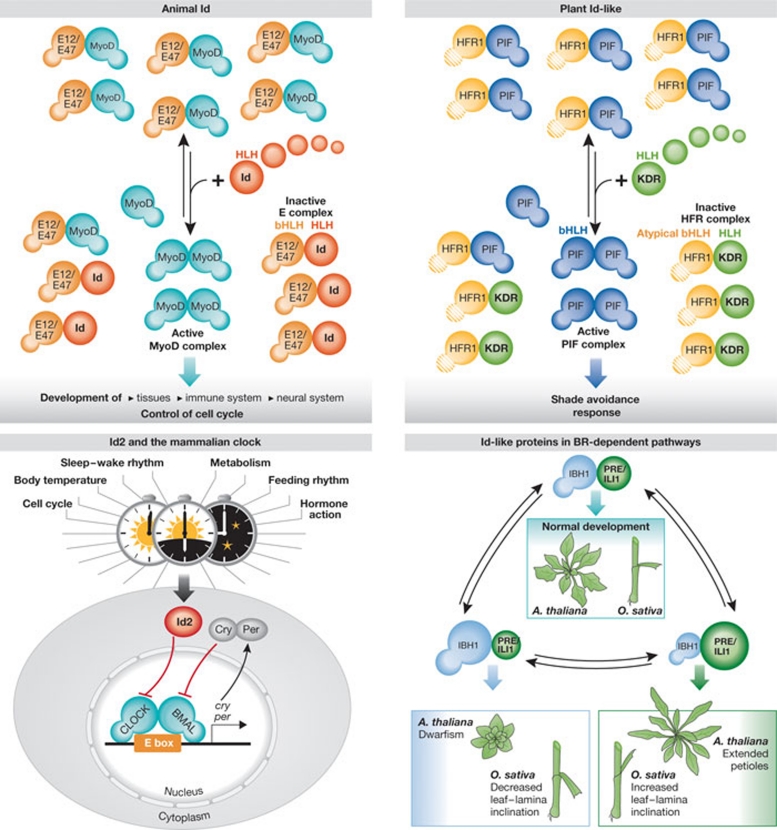
Figure 2.
Animal and plant Id-like proteins. Left: in animals, MyoD forms heterodimers with the more common E12/E47 bHLH proteins. If the HLH protein Id is expressed, it interacts with E12/E47 and sequesters them into nonfunctional heterodimers, allowing MyoD to bind to DNA as a homodimer. Id2 influences the circadian clock of flies, probably by sequestering the bHLH transcription factors BMAL and CLOCK. Right: in plants, PIF-type bHLH proteins activate downstream target genes to regulate light responses. The atypical bHLH protein HFR1 suppresses PIF activity by forming non-DNA-binding complexes with PIFs. The plant Id-like KDR protein sequesters HFR1, shifting the equilibrium to PIFs and allowing them to form active homodimers. Plant Id-like proteins regulate light-dependent growth and BR-dependent growth. Overexpression of the bHLH target (IBH1) causes decreased leaf–lamina inclination in rice and dwarfism in Arabidopsis, and overexpression of the Id-like proteins PRE or ILI1 increases the leaf–lamina inclination in rice (Oryza sativa) and induces petiole elongation in Arabidopsis. bHLH, basic helix–loop–helix; BR, brassinosteroid hormone; HLH, helix–loop–helix; IBH1, ILI1 binding bHLH; Id, inhibitor of DNA binding; KDR, KIDARI; PIF, phytochrome-interacting factor.
Id is small and contains a protein-interaction domain that allows the formation of non-functional heterodimeric complexes; two characteristics of miPs. Thus, miPs have the potential to interfere with both positive and negative regulators of cellular machinery, influencing the fine-tuned feedback loops that are important to regulatory networks, for example in circadian clocks or the cell cycle. Artificial miPs could therefore be useful biotechnological tools in vivo, for example to release active regulatory proteins from inactive complexes, thus allowing the control of protein activity in a dose-dependent manner. With respect to the potential of generating large ultrasensitive responses by protein sequestration, miPs might help us to understand protein regulatory networks, as these responses have been shown to be tunable (Buchler & Cross, 2009).
Animal microProteins and the circadian clock
A common characteristic of the family of Id proteins in mammals is their regulation of cell-fate choices. Id proteins operate in different tissues and cells including myoblasts (Atherton et al, 1996), the neural system (Kondo & Raff, 2000) and the immune system (Hacker et al, 2003; Morrow et al, 1999). In addition to acting as modulators of the transcription machinery and influencing developmental cell fates, Id proteins also interfere with cell-cycle control and are overexpressed in a variety of human tumours (Ruzinova & Benezra, 2003). The molecular function of Id proteins in developmental processes has been well-studied, but little is known about the function of Id proteins in adults.
Duffield and colleagues have recently shown that the miP Id2 has a role in the mammalian circadian clock (Duffield et al, 2009). The main mammalian circadian clock is located in the suprachiasmatic nucleus—a structure within the hypothalamus of the brain—and consists of interlocking feedback loops at the molecular level that involve the bHLH transcriptional regulators BMAL and CLOCK. BMAL and CLOCK regulate the expression of the mouse proteins PER1 and CRY, which in turn downregulate BMAL/CLOCK in a negative feedback loop (Ko & Takahashi, 2006; Reppert & Weaver, 2001, 2002). Id2 is able to repress BMAL/CLOCK function in in vitro transactivation assays in a dose-dependent manner. Id2 loss-of-function mutant mice resemble PER mutant mice; they show faster photonic entrainment (recovery from jetlag) than wild-type mice (Duffield et al, 2009), supporting the role of Id2 as a negative inhibitor of BMAL and CLOCK function. These findings demonstrate that components of basic developmental programmes—in this case, Id-like miPs—can also function in the circadian system, perhaps connecting the clock with the regulation of development and, potentially, disease.
The core circadian clock of both plants and animals consists of interlocking transcriptional–translational feedback loops of transcription factors that generate a rhythmic output. Studies in mice have revealed that the core components BMAL and CLOCK—which are bHLH transcription factors—are targeted by the miP Id2 (Duffield et al, 2009). In plants, bHLH transcription factors have not been shown to act in the core circadian clock. Interestingly, the phytochrome-interacting factor (PIF)-type bHLH transcription factors—known to be involved in plant responses to light—have recently been shown to act in the clock output pathway, mediating rhythmic growth responses in Arabidopsis (Nozue et al, 2007). These PIFs are regulated by the atypical bHLH protein HFR1, which is regulated by the HLH miP KDR. These findings demonstrate mechanistic similarities in the regulation of protein activity of circadian-clock-associated proteins by miPs.
Plant microProteins and light
Unlike animals, most plants are sessile organisms and therefore have to cope with the environmental conditions in which they live. The availability of light is crucial for plant growth and development, as plants harvest light to produce energy-rich sugars. Plants often grow in close proximity to each other and therefore struggle to access optimal light conditions. They have evolved refined mechanisms to sense changes in light quality, for example to avoid living beneath the canopy of taller plants or trees. In response to shade, plants act to find better light for themselves and their offspring: the main stem becomes elongated, leaf blades become larger, and seed germination and flowering are accelerated. The bHLH PIF transcription factors acting downstream from the photoreceptor systems—as well as other influencing factors—regulate shade-avoidance responses (Lorrain et al, 2008).
PIF bHLH transcription factors are positive regulators of shade-avoidance responses, whereas HFR1—an atypical bHLH—suppresses excessive shade responses (Sessa et al, 2005; Fig 2). HFR1 functions by trapping the PIF factors PIF4 and PIF5 into non-functional complexes that cannot bind to DNA, resulting in the loss of PIF activity (Hornitschek et al, 2009). In addition to interacting with PIFs, HFR1 also interacts with an Id-HLH protein called KIDARI (KDR; Hyun & Lee, 2006). Transgenic plants overexpressing KDR develop elongated hypocotyls that resemble those of hfr1 mutant plants. Overexpression of KDR and HFR1 together results in transgenic plants with elongated hypocotyls, suggesting that KDR suppresses HFR1 activity. As HFR1 itself sequesters PIF factors into non-functional dimers, the function of KDR might be to influence the ratio of functional to non-functional PIF dimers, by trapping the negative regulator HFR1 (Fig 2). Whether KDR has other targets and how KDR activity is regulated remains unknown. Further research is necessary to determine the conditions under which different protein dimers are formed, in order to elucidate how inhibitory miPs function in the light-signalling pathways.
Plant microProteins and hormone signalling
Other Id-like miPs have recently been identified in plants (Wang et al, 2009; Zhang et al, 2009) and also interact with bHLH transcription factors to regulate developmental programmes—for example, by interfering with signalling in response to brassinosteroid hormones (BRs).
BRs are plant steroidal hormones that are involved in regulating many physiological responses such as growth, cell elongation, differentiation of transport elements and stress tolerance. In plants, BR acts in the tissue where it is synthesized and does not undergo long distance transport (Bishop et al, 1996; Shimada et al, 2003; Symons & Reid, 2004), which is not the case in animals. Thus, negative regulation of BR synthesis is required to prevent overactivation of the membrane-bound BR receptor complex (He et al, 2000; Symons & Reid, 2004; Wang et al, 2001).
Arabidopsis mutants that are insensitive to BR or deficient in BR have dark green leaves and show dwarfism, among other phenotypes. The function of the Arabidopsis Id-like family of miPs, PRE1–6, has been studied in this context. The overexpression of any of the PRE family of miPs suppresses the dwarf phenotype of the weak BR-receptor mutant bri1 (Wang et al, 2009).
Further evidence of a link between BR signalling and miP function comes from rice. Zhang and colleagues have identified the rice mutant ili1-D, which overexpresses ILI1—the rice homologue of the PRE1 miP in Arabidopsis. These rice plants show an increased leaf–lamina inclination (an erect leaf) phenotype resembling that of rice plants treated with BR (Zhang et al, 2009). Conversely, the reduction of ILI1 expression by RNA interference reduces lamina inclination. Thus, in both Arabidopsis and rice, the phenotype of plants that overexpress Id-like miPs is similar to the phenotype of plants overexposed to BR, and the reverse is also true.
ILI1 interacts with the bHLH transcription factor IBH1, which is suppressed by BR (Zhang et al, 2009). Overexpression of rice IBH1 in Arabidopsis results in a dwarf phenotype, which is suppressed by concomitant overexpression of rice ILI1, suggesting that the proteins interact and form homotypic bHLH/HLH heterodimers. It seems that ILI1/PRE1 fine-tunes the transcriptional activity of IBH1 in response to BR signalling events. Generally, it seems that a regulatory module of HLH/bHLH protein dimers controls hormone signalling in monocotyledonous species such as rice and dicotyledonous species such as Arabidopsis, analogously to Id/MyoD in animals.
Plant leucine-zipper-like microProteins
Homeodomain transcription factors regulate many basal developmental programmes in both animals and plants. Recently, miPs have been identified in plants that target homeodomain proteins from different classes, some of which we discuss here.
Homeodomain–leucine-zipper (HD-ZIP) transcription factors are specific to plants and can be subdivided into four classes on the basis of their protein domain structure and organization. Class III HD-ZIP proteins (HD-ZIPIII) evolved early in land plants (Prigge & Clark, 2006) and are found in all recently sequenced plants, indicating that they have a function in all green organisms. HD-ZIPIII proteins act as master regulators of shoot apical meristem (the tissue in the shoot tip containing the plant stem cells) development, vascular differentiation and leaf development. REVOLUTA (REV) is one of five Arabidopsis HD-ZIPIII genes and is involved in shoot and leaf development (Bowman & Floyd, 2008). Analysis of the REV target genes using microarray analyses identified four small genes encoding leucine-zipper miPs, together called the LITTLE ZIPPER (ZPR) proteins (Wenkel et al, 2007). HD-ZIPIII proteins transcriptionally activate ZPR expression and ZPR proteins interact with HD-ZIPIII proteins through the leucine-zipper domain, thereby attenuating HD-ZIPIII activity (Kim et al, 2008; Wenkel et al, 2007; Fig 3). Transgenic plants overexpressing ZPR resemble HD-ZIPIII loss-of-function mutant plants, whereas ZPR mutants show defects similar to HD-ZIPIII gain-of-function plants (Kim et al, 2008; Wenkel et al, 2007).
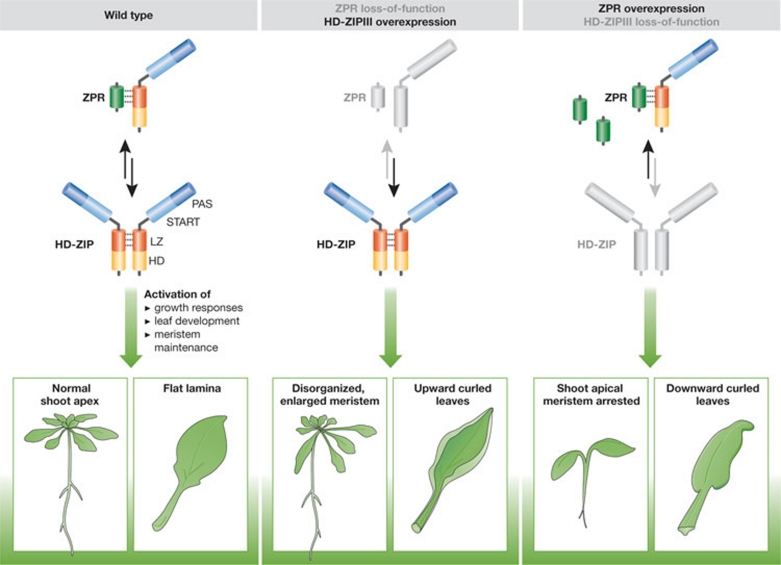
Figure 3.
LITTLE ZIPPER-type miProteins in plants. Left: in wild-type plants, HD-ZIPIII transcription factors bind to DNA as homodimers to control developmental processes including meristem and leaf development. Middle: In hd-zipIII gain-of-function mutant plants or multiple zpr loss-of-function mutant plants, the meristem becomes larger and disorganized and leaf development can be disturbed. Right: If HD-ZIPIII activity is reduced or lost, or if ZPR proteins are over-expressed, the shoot meristem can terminate and leaves show downward curling. HD-ZIPIII, class III homeodomain-leucine zipper; ZPR, LITTLE ZIPPER.
HD-ZIPIII proteins are involved in the establishment of the shoot apical meristem during early embryonic development. In the adult plant they contribute to the maintenance of the apical stem-cells and specify adaxial leaf development in the early leaf primordia (Bowman & Floyd, 2008). Loss of HD-ZIPIII activity can lead to meristem arrest and consumption of the apical stem cells, whereas overexpression results in enlarged meristems. Conversely, overexpression of ZPR proteins can cause downward curling of the leaf blade (abaxialization) and, in severe cases, meristem arrest. Loss of ZPR function—as seen in little zipper3;little zipper4 double mutants—causes an enlargement of the shoot apical meristem (Kim et al, 2008) that is probably due to the uncoupling of miP function (Fig 3). These results suggest that, in wild-type plants, a dynamic balance between active HD-ZIPIII protein homodimers and inactive HD-ZIPIII/ZPR protein complexes regulates meristem and leaf development. HD-ZIPIII proteins have a putative steroid–lipid binding domain, the function of which could be to regulate the ratio of inactive to active protein dimers, through ligand binding.
Other plant microProteins
Homeodomain transcription factors belonging to the three-amino-acid loop-extension (TALE) family of proteins are also targets of miPs in plants. TALE homeodomain transcriptional regulators are characterized by an insertion of three amino acids between α-helices 1 and 2 of the homeodomain. TALE homeodomain proteins are evolutionarily conserved and exist in animals, fungi and plants, indicating that they evolved in a common ancestor. In plants and animals, TALE proteins regulate many developmental processes.
In addition to the TALE homeodomain, the KNOTTED-related homeobox (KNOX) domain is conserved in plant KNOX genes. Animal myeloid ectopic viral integration site (MEIS) proteins also have a KNOX domain and a TALE homeodomain, which was subsequently called MEINOX (Bürglin, 1998). Another common feature in MEINOX proteins is that they interact physically with other classes of non-MEINOX TALE-homeodomain transcription factors (Berthelsen et al, 1998; Modrusan et al, 1994).
In Arabidopsis, two classes of KNOX genes exist, each of which includes four genes encoding MEINOX-TALE-homeodomain proteins, here referred to as KNOX proteins. Plant KNOX proteins interact physically with BEL-like homeodomain proteins (BELLs) and mutant analyses suggest that this interaction is functional (Bhatt et al, 2004; Smith & Hake, 2003). It is assumed that the formation of KNOX/BELL dimers is specific to each family member and that it is a prerequisite for the regulation of downstream target genes.
A bioinformatics-based approach using the plant MEINOX domain as the search term in BLAST identified KNATM, a MEINOX miP that lacks the TALE homeodomain (Magnani & Hake, 2008).
KNATM interacts physically with BELL proteins. Overexpression of KNATM resulted in transgenic plants with elongated petioles (leaf stalks), whereas overexpression of a BELL protein (in this case SAW1) resulted in transgenic plants with shortened petioles (Magnani & Hake, 2008; Fig 4). Co-expression of BELL and KNATM proteins in double transgenic plants resulted in plants that were phenotypically similar to the wild type, indicating that KNATM inhibits BELL function and KNATM is trapped by BELL into non-functional complexes, suppressing the KNATM overexpression phenotype (Magnani & Hake, 2008). A KNATM protein called PETROSELINUM (PTS) was simultaneously identified in tomato. It has been shown that PTS functions similarly to KNATM and regulates leaf shape in tomato (Kimura et al, 2008). The example of KNATM shows that miPs interfere not only with target proteins that form homodimers, but also with proteins that require heterodimerization to be active.
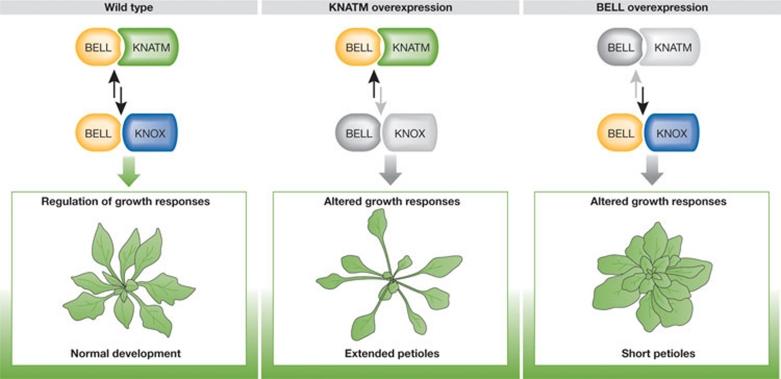
Figure 4.
KNATM microProtein function. Left: In wild-type plants, an equilibrium of KNATM/BELL and KNOX/BELL proteins regulate normal development. Middle: If KNATM is overexpressed, development is altered and transgenic plants have elongated leaf stalks (petioles). Right: overexpression of BELL causes short petioles. BELL, BEL-like homeodomain protein; KNOX, KNOTTED-related homeobox protein.
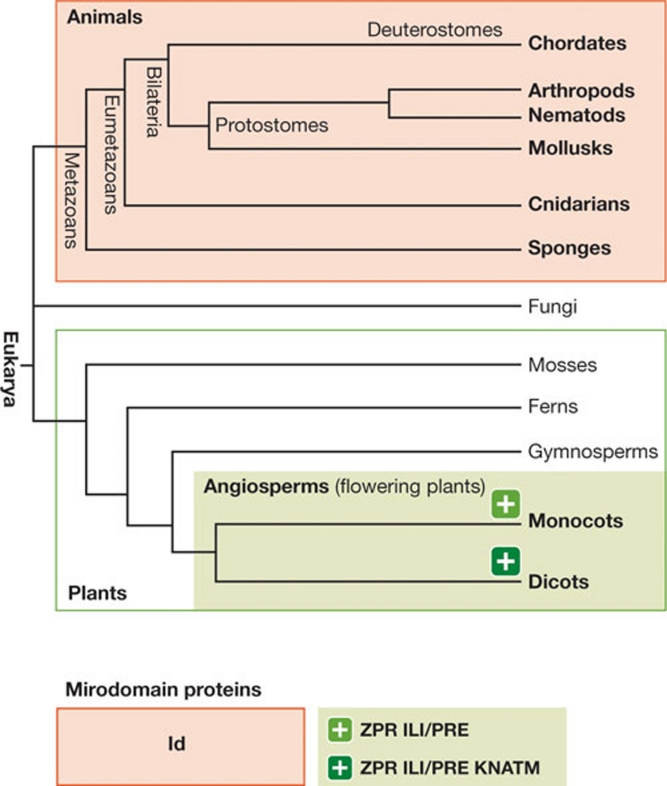
The results of database searches indicate that plant miPs have evolved recently, as the proteins identified so far can only be found in higher plant species (Fig 5). Seed plants are subdivided into Gymnospermae—including conifers, cycads and Gingko—and Angiospermae including flowering plants. Database searches of sequenced plants suggest that both the plant Id-like miPs and the ZPR-like miPs seem to be found in angiosperms, but not in gymnosperms, indicating that they evolved before the split of the monocotyledonous and dicotyledonous lineages of flowering plants. KNATM miPs evolved in the dicotyledonous lineage of flowering plants (Magnani & Hake, 2008) and are therefore more recent. On the basis of database searches, no plant Id-like miP is found in lower plants (mosses or ferns), even though the protein exists in the entire animal lineage. This suggests that HLH proteins evolved independently in both the animal and plant lineages, probably through gene duplication and subsequent domain loss.
Figure 5.
Evolution of microProteins. The simplified cladogram shows that Id-like proteins are found in all animals. In plants, the microProteins identified so far only exist in higher plants. Id, inhibitor of DNA binding; ILI, increased leaf–lamina inclination; PRE, paclobutrazol resistance, HLH family; ZPR, LITTLE ZIPPER.
Alternative splicing as a source for microProteins
Splicing is the process of removing intronic sequences from pre-mRNA molecules. Alternatively spliced mRNAs often produce protein isoforms that differ in structure and sometimes function. In the case of transcription factors that function as dimers, alternative splicing has the potential to generate miPs that contain only the protein-interaction domain and can thus inhibit the function of the full-length target proteins.
The animal MEINOX-TALE-homeodomain MEIS2 transcription factor is expressed in five splice variants (MEIS2A–E). It regulates developmental processes such as neural development in a cooperative manner with other homeodomain proteins and has a ubiquitous expression pattern. Nevertheless, during brain development MEIS2 specifically activates expression of the D1A dopamine receptor in the striatum (Yang et al, 2000). In this context, MEIS2A–D activate D1A and the MEIS2E splice variant acts as a dominant-negative factor of MEIS2A–D function. MEIS2E does not contain the homeodomain required for DNA binding and thus traps MEIS2A–D into non-functional complexes (Yang et al, 2000). MEIS2E has an expression pattern that depends on the tissue and stage of development, suggesting that MEIS2E functions in a developmentally controlled manner (Yang et al, 2000). As MEIS2E is structurally related to KNATM, it is interesting to note that a protein existing as a unique miP in plants—KNATM—is generated in animals by alternative splicing.
Another example of miPs arising from alternatively spliced transcripts is the animal ETS1 transcription factor, which regulates growth responses in animals. The ETS1 protein contains an ETS domain that is required for binding to the cis-acting ETS-binding site (EBS) of ETS1 target genes. In addition to the ETS domain, ETS1 contains a Pointed domain and a transactivation domain, both of which are required for the activation of target genes. Surrounding the ETS DNA-binding domain are auto-inhibitory domains that prevent ETS1 from binding to DNA (Lee et al, 2005). In order to bind to DNA, ETS1 requires protein partners to counteract this auto-inhibition (Pufall & Graves, 2002). Recently, a splice variant of ETS1 was identified that encodes a 27-kDa protein (ETS1 27p) lacking the Pointed and the transactivation domains (Laitem et al, 2009). ETS1 p27 is able to interact physically with ETS1, but blocks the ETS1-mediated induction of target genes, resulting in dominant-negative suppression of ETS1 function. In addition, ETS1 27p induces the translocation of the ETS1/ETS1 p27 complex from the nucleus to the cytoplasm, adding a second layer of negative regulation (Laitem et al, 2009). ETS1 27p mRNA shows a tissue- and stage-specific expression pattern and is strongly expressed in various tumours (Laitem et al, 2009), suggesting a physiological role for the ETS1 27p miP in human disease.
A third example of miP generation by alternative splicing is the human homologue of the Drosophila seven in absentia (SINA) gene—Siah1—which encodes a RING-finger E3 ubiquitin ligase that mediates the degradation of target proteins including β-catenin. SIAH1 contains an amino-terminal RING domain that enables it to interact with E2 proteins, and a carboxy-terminal substrate recognition domain. A splice variant, SIAH1S, was recently identified that acts in a dominant-negative manner (Mei et al, 2007). SIAH1S can interact with SIAH1 to form a heterodimeric complex and the binding of SIAH1S to SIAH1 attenuates SIAH1 function so that target substrate proteins cannot be bound. This is a prerequisite for the induction of protein degradation. Similarly to ETS1 27p, SIAH1S shows a spatiotemporal expression pattern and is induced in tumour tissue, suggesting a biological role for these miPs.
Outlook and perspectives
As some miPs are small, it is difficult to identify them in forward genetic screens. As shown for the SIAH1 ubiquitin ligase, it is not only transcription factors that can be the targets of miPs, but also proteins with other cellular functions, increasing the physiological range of miP action. The thorough characterization of protein-interaction domains in proteins that function as dimers or even multimers should allow the identification of new miPs that encode only the protein-interaction domain. A common characteristic of all miPs discussed in this review is their dominant-negative mode of action.
On the basis of domain homology, miPs can be found by position-specific iterative BLAST searches (PSI-BLAST). The KNATM microdomain was identified by PSI-BLAST searches with the KNOX MEINOX domain (Magnani & Hake, 2008). The LITTLE ZIPPER proteins were first identified in microarray experiments as target genes of the HD-ZIPIII transcription factor REVOLUTA, but their original annotation—‘expressed protein'—did not suggest that they had a biological function (Wenkel et al, 2007). PSI-BLAST searches with the ZPR protein sequences showed sequence homology within the leucine-zipper domain of HD-ZIPIII and ZPR proteins. By using well-characterized protein-interaction motifs as a query in PSI-BLAST searches, it is possible to identify new miPs and study their function using reverse genetic approaches. In addition, splice variants of proteins that are known to function as dimers and contain a protein-interaction motif can also be identified by screening available expressed sequence tag databases. Alternatively, genes encoding proteins of interest can be experimentally tested by 3′ and 5′ RACE, to identify alternatively spliced variants.
The function of miPs—whether they exist in the genome or are produced by alternative splicing—can be tested by using overexpression approaches. As they are predicted to function as dominant-negative inhibitors, overexpression should produce phenotypes similar to the loss-of-function mutants of their predicted targets. Conversely, loss of miP function should result in gain-of-function phenotypes, due to uncoupling of inactive dimer formation.
Similarly to miRNAs, miPs act as negative regulators. Artificial miRNAs are valuable tools for the disruption of target mRNAs in a cell-type-specific or inducible manner. miRNAs can be designed that affect a single mRNA or a whole family of targets (Alvarez et al, 2006; Ossowski et al, 2008; Schwab et al, 2006), and miRNAs function by base-pairing with target mRNAs. One advantage of working with miRNAs is therefore that potential targets can be predicted bioinformatically and artificial miRNAs can be designed. Unlike miRNAs, miPs and their targets cannot be easily identified bioinformatically and have to be experimentally tested. Protein composition and the cellular environment can both influence protein–protein interactions. Nevertheless, understanding the processes that led to the evolution of miPs and studying the way in which they modulate the activity of their target proteins is of biotechnological importance.
In addition to their potential inhibitory effect on protein function, artificial miPs could be attached to other functional domains and used as ‘molecular Velcro' to transiently modify or trap target proteins in defined subcellular spaces, making them a versatile tool with which to analyse the function of target proteins. By designing artificial miPs with different binding characteristics, it might also be possible to maintain target proteins in an attenuated—that is, non-functional—state. The release of the target proteins from the miP could then be induced by a second miP species that would target the first and thereby shift the equilibrium of miP/target to an inhibited miP complex.
As miPs function post-translationally, it is interesting to speculate whether they could act as rapid relays in response to endogenous or environmental signals. In the case of an equilibrium between active homodimers and inactivated miP complexes, the balance could shift instantaneously in response to changing conditions—for example, redox state or phosphorylation—as this would not rely on the transcription of new protein, but only on an alteration of the binding properties of the partners, as might occur in HD-ZIPIII–ZPR interactions.
Finally, within the framework of synthetic biology, miPs have enormous potential. By combining different promoters—tissue-specific or responsive to various cues—with protein-interaction domains of varying strengths, novel regulatory circuits with defined properties can be established (Sidebar A).
Sidebar A | In need of answers.
What controls the formation of non-functional protein dimers?
What happens to the non-functional protein dimer after it has formed?
Do miPs homodimerize themselves?
Can two miPs compete for one target?
Do miPs act non-cell-autonomously?
Do miPs act mainly as direct negative regulators or do they keep active regulators in check?
How is the equilibrium between microP complexes regulated?
MiPs act as dominant-negative inhibitors of protein function by interfering with protein-complex formation. They are found in animals and plants and regulate a range of physiological processes from basic patterning to adaptive development. MiPs can sequester target proteins that function as homodimers, but—as seen for KNATM in Arabidopsis—they can also inhibit the formation of functional heterodimers. Further research is necessary to understand the equilibrium between miPs and their targets, and how other factors can influence this to elicit physiological responses.
Acknowledgments
We are grateful to Sabine Mueller, Sascha Laubinger, Kristina Ile, Gabriel Schaaf and Ulrike Zentgraf for critical reading of the manuscript. We also thank two anonymous reviewers for their helpful suggestions.
Footnotes
The authors declare that they have no conflict of interest
References
- Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton G, Travers H, Deed R, Norton J (1996) Regulation of cell differentiation in C2C12 myoblasts by the Id3 helix–loop–helix protein. Cell Growth Differ 7: 1059–1066 [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H (1990) The protein Id: a negative regulator of helix–loop–helix DNA binding proteins. Cell 61: 49–59 [DOI] [PubMed] [Google Scholar]
- Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F (1998) The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J 17: 1434–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AM, Etchells JP, Canales C, Lagodienko A, Dickinson H (2004) VAAMANA—a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328: 103–111 [DOI] [PubMed] [Google Scholar]
- Bishop GJ, Harrison K, Jones J (1996) The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell 8: 959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK (2008) Patterning and polarity in seed plant shoots. Annu Rev Plant Biol 59: 67–88 [DOI] [PubMed] [Google Scholar]
- Buchler NE, Cross FR (2009) Protein sequestration generates a flexible ultrasensitive response in a genetic network. Mol Syst Biol 5: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR (1998) The PBC domain contains a MEINOX domain: coevolution of Hox and TALE homeobox genes? Dev Genes Evol 208: 113–116 [DOI] [PubMed] [Google Scholar]
- Duffield GE, Watson NP, Mantani A, Peirson SN, Robles-Murguia M, Loros JJ, Israel MA, Dunlap JC (2009) A role for Id2 in regulating photic entrainment of the mammalian circadian system. Curr Biol 19: 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker C et al. (2003) Transcriptional profiling identifies Id2 function in dendritic cell development. Nat Immunol 4: 380–386 [DOI] [PubMed] [Google Scholar]
- He Z, Wang Z-Y, Li J, Zhu Q, Lamb C, Ronald P, Chory J (2000) Perception of Brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288: 2360–2363 [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Lee I (2006) KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol Biol 61: 283–296 [DOI] [PubMed] [Google Scholar]
- Kim Y-S et al. (2008) HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20: 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Koenig D, Kang J, Yoong FY, Sinha N (2008) Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr Biol 18: 672–677 [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS (2006) Molecular components of the mammalian circadian clock. Hum Mol Genet 15: R271–R277 [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M (2000) The Id4 HLH protein and the timing of oligodendrocyte differentiation. EMBO J 19: 1998–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitem C, Leprivier G, Choul-Li S, Begue A, Monte D, Larsimont D, Dumont P, Duterque-Coquillaud M, Aumercier M (2009) Ets-1 p27: a novel Ets-1 isoform with dominant-negative effects on the transcriptional properties and the subcellular localization of Ets-1 p51. Oncogene 28: 2087–2099 [DOI] [PubMed] [Google Scholar]
- Lee GM, Donaldson LW, Pufall MA, Kang H-S, Pot I, Graves BJ, McIntosh LP (2005) The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J Biol Chem 280: 7088–7099 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Magnani E, Hake S (2008) KNOX Lost the OX: The Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20: 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Xie C, Xie W, Wu Z, Wu M (2007) Siah-1S, a novel splice variant of Siah-1 (seven in absentia homolog), counteracts Siah-1-mediated downregulation of β-catenin. Oncogene 26: 6319–6331 [DOI] [PubMed] [Google Scholar]
- Modrusan Z, Reiser L, Feldmann KA, Fischer RL, Haughn GW (1994) Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 6: 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow MA, Mayer EW, Perez CA, Adlam M, Siu G (1999) Overexpression of the helix–loop–helix protein Id2 blocks T cell development at multiple stages. Mol Immunol 36: 491–503 [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJG, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E (2001) Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409: 1067–1070 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Clark SE (2006) Evolution of the class III HD-Zip gene family in land plants. Evol Dev 8: 350–361 [DOI] [PubMed] [Google Scholar]
- Pufall MA, Graves BJ (2002) Autoinhibitory domains: modular effectors of cellular regulation. Annu Rev Cell Dev Biol 18: 421–462 [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR (2001) Molecular analysis of mammalian circadian rhythmns. Annu Rev Physiol 63: 647–676 [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR (2002) Coordination of circadian timing in mammals. Nature 418: 935–941 [DOI] [PubMed] [Google Scholar]
- Ruzinova MB, Benezra R (2003) Id proteins in development, cell cycle and cancer. Trends Cell Biol 13: 410–418 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I (2005) A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S (2003) Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HMS, Hake S (2003) The Interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15: 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons GM, Reid JB (2004) Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol 135: 2196–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhu Y, Fujioka S, Asami T, Li J, Li J (2009) Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix–loop–helix proteins. Plant Cell 21: 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Wenkel S, Emery J, Hou B-H, Evans MMS, Barton MK (2007) A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 19: 3379–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hwang CK, D'Souza UM, Lee S-H, Junn E, Mouradian MM (2000) Three-amino acid extension loop homeodomain proteins Meis2 and TGIF differentially regulate transcription. J Biol Chem 275: 20734–20741 [DOI] [PubMed] [Google Scholar]
- Yates PR, Atherton GT, Deed RW, Norton JD, Sharrocks AD (1999) Id helix-loop-helix proteins inhibit nucleoprotein complex formation by the TCF ETS-domain transcription factors. EMBO J 18: 968–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L-Y et al. (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]