Abstract
The dissociation chemistry of somatostatin-14 was examined using various tandem mass spectrometry techniques including low energy beam type and ion trap CID of protonated and deprotonated forms of the peptide, CID of peptide-gold complexes, and ETD of cations. Most of the sequence of somatostatin-14 is present within a loop defined by the disulfide linkage between Cys-3 and Cys-14. The generation of readily interpretable sequence-related ions from within the loop requires the cleavage of at least one of the bonds of the disulfide linkage and the cleavage of one polypeptide backbone bond. CID of the protonated forms of somatostatin did not appear to give rise to an appreciable degree of dissociation of the disulfide linkage. Sequential fragmentation via multiple alternative pathways tended to generate very complex spectra. CID of the anions proceeded through CH2-S cleavages extensively but relatively few structurally diagnostic ions were generated. The incorporation of Au(I) into the molecule via ion/ion reactions followed by CID gave rise to many structurally relevant dissociation products, particularly for the [M+Au+H]2+ species. The products were generated by a combination of S-S bond cleavage and amide bond cleavage. Electron transfer dissociation of the [M+3H]3+ ion generated rich sequence information, as did CID of the electron transfer products that did not fragment directly upon electron transfer. The electron transfer results suggest that both the S-S bond and an N-Cα bond can be cleaved following a single electron transfer reaction.
Keywords: somatostatin-14, electron transfer dissociation, ion/ion reactions, CID, gold(I) cationization
Introduction
Tandem mass spectrometry is commonly applied to the characterization of the primary structures of proteins and peptides via dissociation reactions of gaseous ions derived from the species of interest. The dissociation chemistry of protein and peptide ions is influenced by factors associated with the ion itself, which include ion polarity,1 electronic structure (e.g., radical or even-electron species),2 three dimensional structure,3 molecular size,4 and the identity of any agents used to impart an excess charge (e.g., protons, metal ions, anions, etc.).5,6,7,8,9,10 The conditions under which an ion derived from a peptide or protein is activated or otherwise probed can also be particularly important in determining the extent and quality of information.11 Protein and peptide ions are most frequently generated via electrospray ionization (ESI) or matrix assisted laser desorption ionization (MALDI) as species with one or more excess protons. Protonated molecules are usually fragmented using collision-induced dissociation (CID), which results predominantly in backbone amide bond cleavages.11 Alternatively, capture of an electron, either from an anion or a free electron, by multiply-charged protein and peptide cations can result in N-Cα bond cleavages. These processes are referred to as electron transfer dissociation (ETD)12 and electron capture dissociation (ECD),13,14 respectively, and they usually provide complementary information to CID.
Disulfide linkages are post-translational modifications that play key roles in stabilizing the native structures of many proteins and the identification and localization of such linkages is often an important objective in primary structure characterization.15 The presence of disulfide linkages, however, can play a major role in the extent to which peptide or protein sequence information can be obtained. Various methods involving the gas-phase dissociation of ions with disulfide linkages have been employed, which include, for example, CID,16,17 electron-induced dissociation (EID),18 post-source decay associated with MALDI,19 infrared multiphoton dissociation (IRMPD),20 ECD,21,22 ETD,23,24,25 electron detachment dissociation (EDD),20 and UV-photodissociation.26 However, the structural characterization of disulfide bond-containing polypeptides via gas-phase ion dissociation is not universally successful with every dissociation approach. Under normal CID conditions, for example, backbone amide bond cleavages are dominant dissociation channels for multiply protonated peptides and proteins whereas disulfide bond cleavage is often uncompetitive. Furthermore, it has been noted that a disulfide bond stabilizes the region of the ion that falls within the loop formed by the disulfide linkage, thereby affecting the extent of primary structural information that can be obtained.27,28 However, when proton mobility of an ion is limited, disulfide bond cleavages are often the main fragmentation channels in the CID of cations,29 and ordinarily dominant in the CID of multiply charged anions.16
Incorporating metal ions is an alternative strategy for disulfide bond cleavage in proteins and peptides. However, there is only a limited number of reports directed to this objective. The first study reporting transition metal ions as disulfide bond cleavage reagents in organic molecules appeared in 1989.30 In a more recent study, a gas phase ion/ion reaction was reported involving the reaction of Fe+ with multiply charged insulin anions. In that study, two iron cations were possibly involved in the cleavage of two disulfide bonds between the two chains of insulin.31 Since then, disulfide bond cleavage in proteins and peptides associated with different transition metal ions has been examined employing ECD32 and CID.33,34 The use of cationization by sodium and various alkaline earth metals in conjunction with low energy CID has also been demonstrated to give rise to disulfide bond cleavage in polypeptide systems.35,36 Gold-cationization through an ion-ion reaction method has been suggested as a means for selective cleavage of disulfide linkages in peptides37. The incorporation of gold cations into peptides in the gas phase via ion/ion reaction avoids complications associated with directly adding a metal salt to the peptide solution, which can introduce variability in the number of metal adducts, variability in charge state, and possible matrix effects.
Intramolecular disulfide linkages can be particularly difficult to address because at least two bonds must be cleaved for primary structure information to be forthcoming from the polypeptide chain within the loop defined by the disulfide bond. In this work, we employ somatostatin-14 as the model analyte species to compare the utility of various strategies for deriving sequence information from a peptide that is primarily present as a loop. Somatostatin-14, also known as “growth hormone inhibiting hormone”, regulates the endocrine system and affects neurotransmission and the release of secondary hormones38. It is a cyclic peptide with fourteen amino acids in the chain with a Cys-3 to Cys-14 disulfide linkage (H-AGCKNFFWKTFTSC-OH) and two lysine residues within the disulfide bridge. In this study, cations and anions of different charge states of the cyclic peptide somatostatin-14 were subjected to low energy beam type and ion trap collision-induced dissociation. Additionally, somatostatin cations were involved in gas phase cation switching ion/ion reactions with gold (I) dichloride anions followed by supplemental CID of the peptide-gold complex. Lastly, protonated somatostatin ions were involved in electron transfer dissociation (ETD) reactions with azobenzene radical anions.
Experimental Section
Materials
Acetonitrile, ammonium hydroxide, glacial acetic acid, hydrochloric acid, and methanol were purchased from Mallinckrodt Baker, Inc. (Phillipsburg, NJ). Azobenzene was obtained from Sigma-Aldrich (St. Louis, MO) and somatostatin-14 was purchased from Bachem (King of Prussia, PA). The somatostatin-14 solution was prepared at a concentration of approximately 50 μM in 49.5/49.5/1 (v/v/v) solvent mixture of water/methanol/acetic acid for positive ion formation and in 48/48/4 (v/v/v) solvent mixture of water/methanol/ammonium hydroxide for negative ion formation. Gold (III) chloride trihydrate (chloroauric (III) acid trihydrate) (Mallinckrodt Baker, Inc., Phillipsburg, NJ) was dissolved in acetonitrile with hydrochloric acid (1% by volume) at a concentration of approximately 20 mM for negative ESI. All chemicals were used without further purification.
Mass Spectrometry
All experiments were performed using either a prototype version of a QTRAP mass spectrometer39 (Applied Biosystems/MDS Sciex, Concord, ON, Canada) equipped with a home-built dual nano-electrospray ionization source40 or a prototype version of a QSTAR Pulsar XL mass spectrometer (Applied Biosystems/MDS Sciex, Concord, ON, Canada) that has also been modified for ion/ion reaction studies.41 Most experiments were conducted using the hybrid triple quadrupole/linear ion trap instrument (i.e., the QTRAP instrument) whereas a few CID experiments were conducted using the quadrupole/time-of-flight instrument (i.e., the QSTAR instrument). The ion path of each instrument includes at least three in-line quadrupole arrays, designated as Q0–Q2. A fourth quadrupole array, Q3, serves as the mass analyzer in the QTRAP, whereas an orthogonal acceleration time-of-flight mass spectrometer serves as the mass analyzer in the QSTAR. In both instruments it is possible to effect collisional activation in the Q2 quadrupole either via ion trap collisional activation or by injecting precursor ions into Q2 at relatively high kinetic energies. Ion trap collisional activation in the QTRAP instrument can also be effected in the Q3 array. The experiments were controlled using the research software Daetalyst 3.14 provided by MDS Sciex.
Cation Switching Ion/Ion Reactions
All ion/ion reaction experiments were conducted using the QTRAP instrument. In this work, Q0 was used as a transmission device to transport ions from the atmosphere/vacuum interface to Q1, which was operated in mass resolving mode to isolate the positive analyte ions of interest prior to injection and storage in Q2. The trapped ions were cooled for 50 ms in Q2 at a pressure of 6–8 mTorr of nitrogen, during which time, the high voltage on the positive emitter was turned off. After the cooling step, the power supply connected to the nano-ESI source that generated the negatively charged ions was triggered on and the negative reagent ions isolated in Q1 were passed through the Q2 collision cell where the positive peptide ions were stored. The ions in two polarities were allowed to react in transmission mode42 (i.e., the cations were stored while the anions were transmitted through Q2). The somatostatin-gold chloride product ion complex formed from the cation switching reaction in Q2 was transferred to Q3, which was held at a pressure of 3.5 × 10−5 Torr. In this transfer step, the products of the ion/ion reaction were accelerated between Q2 and Q3 to collisionally dissociate residual adduct ions via loss of one or more molecules of HCl. The dc offset difference, (RO2–RO3), where RO2 and RO3 are the dc potentials of the rods for Q2 and Q3, respectively, was between 15–25 V in this study and the kinetic energy of the accelerated ions was [(RO2–RO3) × Z] where, Z is the charge of the ions. The [M+Au+H]2+ product ions were isolated in Q3 prior to ion trap collisional activation in the rf/dc mode whereas only the lower mass ions were ejected, via a scan of the rf amplitude applied to Q3, in the case of the [M+Au]+ ions. Following an ion trap collisional activation period, the product ion spectrum was collected using mass–selective axial ejection (MSAE).39 The processes for formation of the gold-cationized somatostatin-14 species are summarized in reactions (1) and (2) for the [M+Au+H]2+ and [M+Au]+ ions, respectively.
| (1) |
| (2) |
ETD
A home built dual ion source including a nano-ESI emitter for generating multiply charged peptide cations and an atmospheric pressure chemical ionization (APCI) needle for producing azobenzene radical anions was coupled directly with the interface of the QTRAP instrument. The experimental procedure for electron transfer ion/ion reactions performed with this instrument has been reported in detail elsewhere.39 In brief, both polarity ions were isolated and stored in Q3 where they were allowed to react for various mutual storage times. After the reaction, the product ions were analyzed in Q3 using MSAE.
Ion Trap and Beam Type CID
In the ion trap CID experiments in the QTRAP, positive or negative ions of somatostatin were isolated in Q3 in the rf/dc mode and subjected to resonance excitation at sufficiently low amplitudes to minimize ejection while giving rise to CID. The products were analyzed in the Q3 mass analyzer using MSAE. The dc offset difference for the beam type CID, (Q0-RO3), where Q0 and RO3 are the dc potentials of the rods for Q0 and Q3 respectively, was between 13–35 V in this study and the kinetic energy of the accelerated ions was [(Q0-RO3) × Z], where Z is the charge of the ion. Note that this CID experiment in the QTRAP involves the accumulation and storage of both CID products and undissociated parent ions in Q3 until such time that they are analyzed by MSAE. In the case of the QSTAR, all CID products and undissociated precursor ions are stored in Q2 until they are released to the TOF. Hence, these experiments integrate products that form over the period of time between the activating collisions and eventual mass analysis, which takes place at least tens of milliseconds later. Therefore, the beam-type CID experiments described here integrate processes that occur over periods as long as tens of milliseconds. The main difference between the beam-type CID experiments and the ion trap CID experiments in this study is that the former integrate products formed from higher dissociation rate processes than the latter but both include dissociations that take place on the time-frame of milliseconds.
Results and Discussion
Ion trap and beam type CID of somatostatin anions and cations, CID of the cation-switching reaction products, and ETD of somatostatin cations with azobenzene were the tandem mass spectrometry techniques examined in this study to compare and contrast the structural information available for somatostatin-14 from these various approaches.
Ion Trap and Beam Type CID of Protonated Forms of Somatostatin
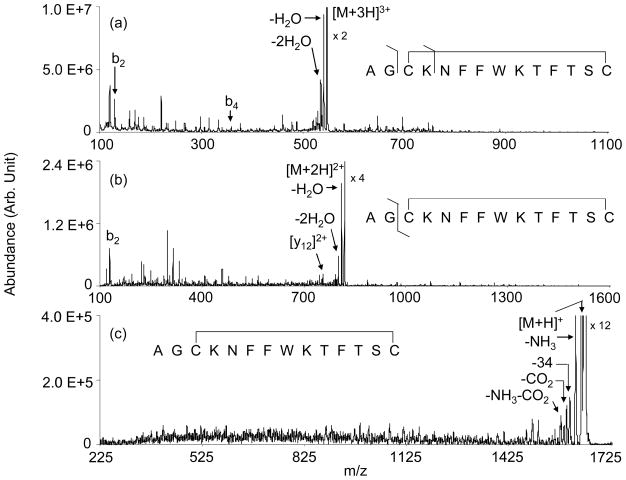
Figure 1 shows product ion spectra derived from ion trap collisional activation of the somatostatin [M+3H]3+ (Figure 1(a)), [M+2H]2+ (Figure 1(b)), and [M+H]+ ions (Figure 1(c)). Beam-type collisional activation (data not shown) yielded very similar product ion distributions. Aside from the abundant losses of small neutral molecules, most of the products in the spectra correspond to plausible internal fragments that must arise from the cleavage of at least two bonds. Few correspond to single backbone bond cleavages. The major exception is the b2 ion, which arises from cleavage outside the loop defined by the disulfide linkage. Products that require cleavage of peptide backbone bonds make the largest relative contribution in the data for the [M+3H]3+ ions. (A listing of the masses associated with singly protonated products, including both internal fragments associated with multiple backbone cleavages, as well as the conventional a-, b, c-, x-, y-, z-ions, is provided as supplementary material.) There is no clear evidence that cleavage of the disulfide linkage between Cys-3 and Cys-14 contributes in a significant way to product ion formation for any of the precursor ion charge states. However, the baselines for all three charge states show extensive “chemical noise” that appears only when the ion activation voltage is applied. This observation is consistent with small contributions from many combinations of multiple cleavages that give rise to many small signals that are difficult to interpret. It is clear from these data that CID under conditions that are accessible to many tandem mass spectrometers performs very poorly as a structural analysis tool for somatostatin cations.
Figure 1.
Ion trap CID spectra of the somatostatin (a) [M+3H]3+, (b) [M+2H]2+, and (c) [M+H]+ ions.
Ion Trap and Beam Type CID of Deprotonated Forms of Somatostatin
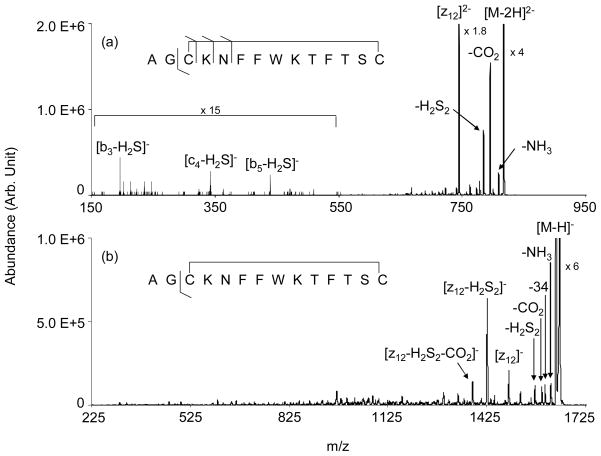
The beam-type CID of peptide anions, including those with disulfide linkages, has been reviewed.43,44 An earlier study has demonstrated the cleavage of the intermolecular disulfide linkage in digested somatostatin anions using beam type CID.16 The previous study also showed that cleavage of doubly deprotonated native somatostatin takes place almost exclusively at the disulfide linkage with subsequent cleavage to yield the z122− ion in an apparently slow process. Under beam-type CID conditions in a pure transmission mode experiment on a quadrupole/time-of-flight instrument, the process leading to the z122− ion was not observed. Rather, losses of CO2, CH3CHO, and H2S2 were observed.16 These losses are commonly observed in disulfide linked polypeptide anions.44 In this study, beam-type CID with storage of the products in Q3 showed the formation of the z122− ion to be a major process, which reflects the effect of allowing for a longer time for integration of product ions from relatively slow dissociation processes. Figure 2 shows the product ion spectra derived from ion trap collisional activation of the somatostatin [M−2H]2− (Figure 2(a)) and [M−H] − (Figure 2(b)) anions. The beam-type collisional activation data (not shown) exhibited the same products but the relative abundances of the z122− ion and its sequential dissociation products were lower. The ion trap CID spectrum of the dianion is dominated by loss of 44 Da, which can result from either CO2 loss, CH3CHO loss from the threonine side-chain, or both, loss of H2S2, and formation of the z122− anion. The latter ion is generated by cleavage of the CH2-S bond of Cys-3 followed by a rearrangement reaction to yield c- and z-fragments. (When two separate polypeptide chains are linked by a disulfide bond, the z-fragment arising from this process is missing the sulfur atom of the cysteine at the site of cleavage.) However, in the case of a cyclic peptide in which both cysteines are present in the z-fragment, provided there are no other cleavages associated with the disulfide linkage, both sulfur atoms are associated with the other cysteine residue such that the z-ion does not show a loss of sulfur. Aside from the z122− ion, little product ion signal associated with structurally informative fragments are noted. There are a few structurally informative fragments present at low relative abundance, such as the [b3-H2S] −, [c4-H2S] −, and [b5-H2S] − ions, that arise from sequential cleavages, one of which is cleavage of the Cys-3 CH2-S bond. (Note that the standard peptide nomenclature used here is not meant to imply the mechanisms for the processes that lead to these ions. The b-type ions and c-type ion presumably arise from the processes analogous to those referred to by Bowie et al. as β and γ, respectively.43,44)
Figure 2.
Product ion spectra derived from the ion trap CID of the somatostatin-14 (a) [M−2H]2− and (b) [M−H] − anions.
The product ion spectrum for the [M−H] − ion also shows losses common to cysteine linked polypeptide anions, such as losses of water, H2S, CO2 and/or CH3CHO, and H2S2. The process leading to the z12− ion also contributes. In fact, the most abundant product corresponds to the z12− ion that has also lost H2S2. Further losses of 44 Da from this product ion are also observed. There are many small fragments at lower mass-to-charge ratios and the spectrum appears to contain a relatively high degree of chemical noise. This likely reflects the contributions from many sequential fragmentations that can lead to a wide array of products. While there are product ion signals that may well correspond to structurally diagnostic ions, the complex nature of the spectrum makes it difficult to assign most of the small signals, even with the advantage of knowing the sequence of the polypeptide. Hence, while it is clear that disulfide bond cleavage is a primary process with the deprotonated forms of somatostatin, the sequence information that can be derived via CID is limited.
CID of Gold-cationized Somatostatin Ions Formed via Cation Switching Reactions
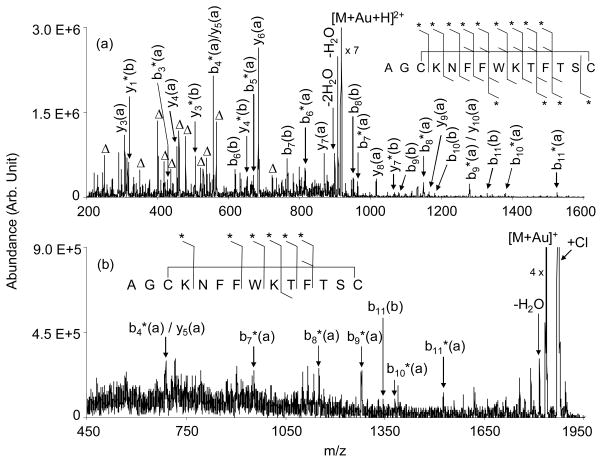
An earlier report demonstrated that the incorporation of a gold (I) cation into trypsin digestion products of somatostatin-14 resulted in the cleavage of the disulfide bond when the protonated form of the molecule showed essentially no evidence for disulfide bond cleavage.37 The gold (I) cation was incorporated into the digest products via ion/ion cation switching using AuCl2− as the reagent. In this study, the effect of the incorporation of gold (I), an ion of d10 configuration that forms linear or tetrahedral complexes, into intact cyclic somatostatin-14 was examined. The somatostatin-gold chloride complexes initially generated from the ion/ion reaction were first subjected to beam type CID to eliminate two molecules of HCl,37 and the resulting peptide-gold complex was subjected to low energy ion-trap CID conditions to obtain sequence information. The CID spectrum of the [M+Au+H]2+ ion generated by the reaction of [M+3H]3+ with AuCl2− is shown in Figure 3(a). The Au(I)-containing form of somatostatin showed no propensity for loss of H2S2, which is similar to the behavior noted for species cationized with Ni(II), Zn(II), and Cu(II),34 but stands in contrast with sodium and alkaline earth metal cationized disulfide-linked peptides.35,36 Likewise, no H2S2 loss was noted in the dissociation of the Au(I)-cationized trypsin digestion products either.37
Figure 3.
Product ion spectra derived from the ion trap CID of the somatostatin-14 (a) [M+Au+H]2+ and (b) [M+Au]+ cations. The Δ symbol represents internal fragments from within the loop defined by the disulfide bridge that result from two consecutive amide bond cleavages. The * symbol indicates the presence of Au in the fragment. The designations (a) and (b) associated with fragments relates to the nominal structures of Scheme 1. E.g., b*5(a) represents the Au-containing b5-ion from structure (a) of Scheme 1.
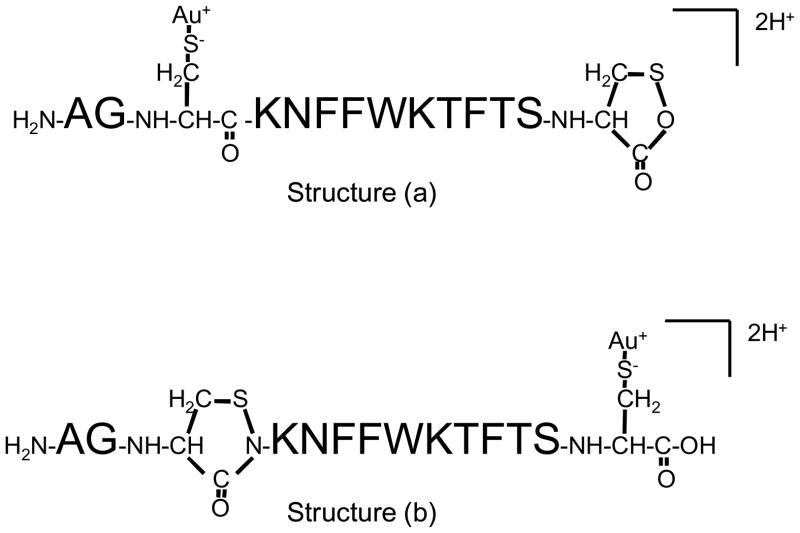
CID of the [M+Au+H]2+ species results in a rich product ion spectrum that reflects contributions from a wide array of sequential cleavages of bonds initially present in the loop defined by the disulfide linkage. These include combinations of two or more cleavages of the polypeptide chain, which give rise to “internal fragments” because they contain neither the N-terminus nor the C-terminus, as well as cleavage of the S-S bond and an amide bond cleavage. None of the most abundant internal fragments appears to contain the gold cation, which is consistent with the gold being associated with one or both of the cysteine residues. Cleavage of the CH2-S bond does not appear to compete significantly with S-S bond dissociation. Most of the major products greater than 500 in mass-to-charge could be assigned as arising from S-S bond cleavage in conjunction with amide bond cleavage to yield a b- or y-type fragment, in some cases coupled with one or more losses of a small molecule (i.e., water or ammonia). All of the major sequence informative product ions are consistent with the Au(I) ion being associated either with Cys-3 (Structure (a) of Scheme 1) or Cys-14 (Structure (b) of Scheme 1). Interestingly, most of the product ion signal suggests that the gold ion is preferentially associated with Cys-3. The structures of Scheme 1 are consistent with the hypothesis offered by Lioe et al. that Au+ attaches to the disulfide bond making it susceptible to nucleophilic attack by the C-terminal amide nitrogen, in the case of Cys-3, or by the hydroxyl oxygen of the C-terminus, in the case of Cys-14.32 The apparent dominance of Structure (a) is reflected by the series of b-type ions that contain gold, i.e, b4(a)*–b11(a)*, and the complementary set of y-type ions, y3(a)–y10(a). Collectively, these ions comprise a much larger fraction of total product ion abundance than those accounted for by Structure (b) (e.g., the b6(b)–b11(b) series and the y(b) ions). The results suggest that the amide nitrogen of Cys-3 is a less effective nucleophile than the oxygen of the C-terminus. Methyl esterification of the C-terminus yielded data consistent with this conclusion in that the product ion spectra showed extensive sequential cleavages leading to uninformative ions (data not shown), much like the CID spectra of protonated somatostatin-14. Far fewer structurally diagnostic ions were noted, which suggests that methyl esterification led to less S-S bond cleavage.
Scheme 1.
Structures that show Au(I) associated with Cys-3 (Structure (a)) and Cys-14 (Structure (b)).
The product ion spectrum of the [M+Au]+ ion (Figure 3(b)) shows several of the sequence informative sequential cleavages that are also apparent in the [M+Au+H]2+ data. Hence, sequential cleavages that include S-S bond dissociation take place with the singly charged ion. However, the spectrum is even more complex than that of the doubly charged gold-containing ion, as reflected by what appears to be relatively high levels of chemical noise. This complex array of signals reflects a diverse set of sequential decomposition products, many of which do not include S-S bond cleavage. In order for sequence informative cleavages to dominate the spectrum, S-S bond dissociation must accompany amide bond cleavage, as occurs with the many sequence informative products noted for the doubly charged ion. Clearly, the product ion spectrum of the [M+Au]+ ion is less readily interpreted than that of the [M+Au+H]2+ due to the greater relative contributions from cleavages that lead to internal fragments.
Electron Transfer Dissociation
Digested versions of somatostatin that resulted in two polypeptide chains linked with a disulfide bond have previously been subjected to electron transfer dissociation using SO2−• ions22 or azobenzene anions24. Both disulfide bond cleavage and N-Cα bond cleavage were noted. The previous studies showed that the two types of cleavages compete and that relative contributions of each type depend upon the cation charge state. Disulfide bond cleavage was the dominant process for all charge states with the relative contribution from N-Cα bond cleavage increasing with increasing peptide charge. For native somatostatin, the generation of sequence informative fragments requires that both types of cleavages take place following a single electron transfer. It has been demonstrated in the ECD of cyclic peptides45 that multiple bond cleavages can take place following a single electron capture event via radical migration mechanisms.46 ECD of the +5 charge state of insulin showed evidence for the cleavage of two disulfide bonds upon capture of a single electron, as reflected in the appearance of separated A-chain and B-chain ions.22 No evidence for the cleavage of more than one bond upon electron transfer to the +6-+3 charge states of insulin from azobenzene anions was noted.23 Collisional activation of the charge reduced products (i.e., ions that underwent charge state reduction without undergoing dissociation), however, did show formation of separate A- and B-chain ions. This observation might be accounted for by ETD of one disulfide linkage followed by CID of the other or cleavage of both disulfide bonds as a result of electron transfer with collisional activation being required to separate the non-covalently bonded fragments.
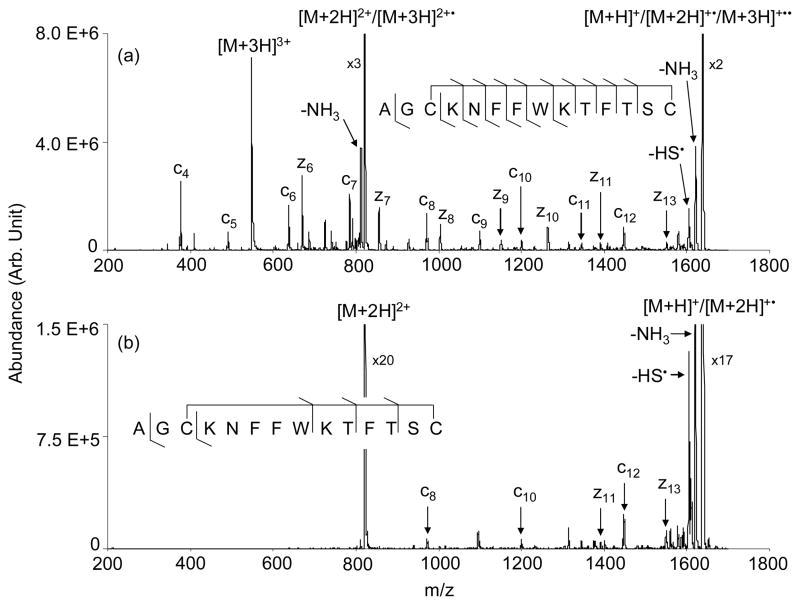
Somatostatin system is smaller than insulin and provides a less complex system for identifying multiple cleavages upon the transfer of a single electron. Figure 4 presents the ETD product spectra of triply (Figure 4(a)) and doubly (Figure 4(b)) protonated somatostatin reacting with azobenzene radical anions in Q3 of the QTRAP instrument. No means were used to collisionally activate the first generation product ions in the data for Figure 4. Doubly protonated somatostatin-14 largely underwent charge state reduction both via electron transfer and proton transfer. A few c- and z-type ions were observed but only at abundances of a few percent or less of the charge reduced products (see Figure 4(b)). Nevertheless, the results for the doubly protonated species indicate that cleavage of both the disulfide bond and an N-Cα can occur following a single electron transfer reaction. Triply protonated somatostatin-14, on the other hand, showed relatively abundant c4–c12, z6–z11 and z13 fragment ions, thereby yielding the most primary sequence information of all of the approaches examined in this study. A further advantage for the ETD results is that they are much less complicated by competing sequential backbone cleavages than the CID-based approaches and are therefore more readily interpretable.
Figure 4.
ETD of (a) [Somatostatin + 3H]3+ and (b) [Somatostatin + 2H]2+ reacting with azobenzene radical anions.
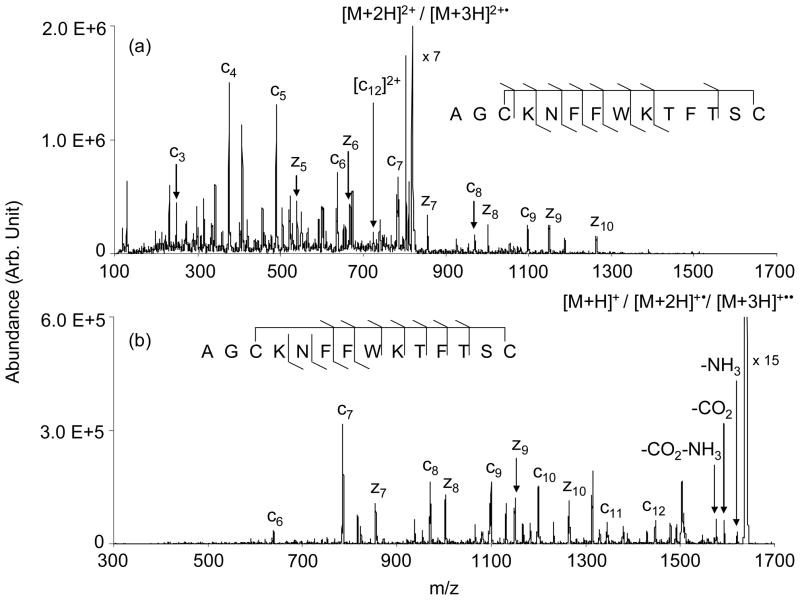
The data of Figure 4(b) suggest that sequential cleavages can result from a single electron transfer, although the yield of such products was only a few percent. A much greater yield of structurally informative products was generated from the [M+3H]3+ ion (Figure 4(a)). However, at least some of the products might have been generated from sequential electron transfer reactions of the [M+3H]3+ ion. The appearance of relatively abundant intact singly charged ions, which are comprised of species generated from sequential proton transfer to yield the [M+H]+ ion, one electron transfer and one proton transfer to yield the [M+2H]+• species, and two electron transfers to yield the [M+3H]+•• ion, make this a possibility. However, experiments using shorter reaction times that yielded much lower intact singly charged ion signals generated the same dissociation products observed in Figure 4(a) (data not shown), which suggests that contributions from fragmentations that result from sequential ion/ion reactions are minimal. Ion trap collisional activation of the intact ion/ion reaction products, the results for which are shown in Figure 5, yielded predominantly c- and z-ions. As is commonly the case for CID of surviving [M+2H]+• ions in ETD, relatively high m/z products tend to be favored upon CID of the singly-charged somatostatin ions (see Figure 5(b)), presumably because they are better able to compete for the excess charge than the complementary product of lower mass. In the case of surviving [M+3H]2+• ions, competition for charge is less of an issue such that relatively low m/z products can be observed, provided they are higher in m/z than the low m/z cut-off of the ion trap. Indeed, low m/z products tend to dominate in the CID of the somatostatin doubly charged survivors (see Figure 5(a)), which likely reflects higher detection efficiencies for the lower m/z products in this instrument. The fact that some relatively low m/z products are prominent in the direct ETD data for the [M+3H]3+ ion (e.g., the c4 ion in Figure 4(a)) is further evidence that two bond cleavages result from a single electron transfer. If all of the c- and z-type ions were to require two consecutive electron transfer steps, the resulting singly charged ion would not likely yield abundant low m/z fragment ions for the same reason that CID of the surviving singly charged ions does not.
Figure 5.
CID product ion spectra of the surviving ion/ion reaction products (a) [M+2H]2+/[M+3H]2+• and (b) [M+H]+/[M+2H]2+•/[M+3H]+••.
Conclusions
Somatostatin-14 illustrates the range of peptide fragmentation phenomena that can be observed, depending upon ion type and dissociation method. It is an example of a polypeptide with an intramolecular disulfide linkage that renders cyclic part of the molecule. Such species can be difficult to characterize structurally via tandem mass spectrometry because two bonds, one of which should preferably be part of the disulfide linkage, must be cleaved for structurally diagnostic product ions to arise from within the cyclic portion of the molecule. Ion trap and low energy beam-type CID of protonated forms of the molecule were ineffective for structural characterization due to the limited degree of fragmentation of the disulfide linkage between Cys-3 with Cys-14. This result is significant due to the fact that CID of protonated and multiply protonated species is the most common tandem mass spectrometry approach for polypeptides. Several alternative approaches to structural characterization that are more likely to result in gas-phase cleavage of the disulfide linkage have been examined. The CID of deprotonated forms of somatostatin (i.e., [M−2H]2− and [M−H]−) showed cleavage of the disulfide linkage to be a dominant process. However, relatively limited structural information was forthcoming from regions within the original loop of somatostatin. The CID of gold (I) containing cations, however, proved capable of yielding primary structure information. Particularly in the case of the [M+Au+H]2+ ion, preferential cleavage of the S-S bond was noted such that disulfide bond cleavage in conjunction with amide bond cleavage yielded many sequence-related ions. Evidence for similar informative processes was also noted for the [M+Au]+ species but more extensive contributions from less informative internal cleavages was also apparent. ETD of the [M+3H]3+ ion yielded the most extensive and readily interpretable sequence information of all the approaches examined here. The data suggest that a single electron transfer event can give rise to consecutive disulfide bond and N-Cα bond cleavages. Further study is required to determine if there is a favored order. However, previous studies with separate polypeptide chains bound by a disulfide linkage have indicated that the two processes compete, thereby suggesting that both sequences likely contribute. Based on the observation noted here, electron transfer and gold (I) cationization are two distinct approaches that appear to hold merit for the selective cleavage of disulfide bonds in disulfide linked polypeptide cations.
Acknowledgments
This research was supported by the National Institute of General Medical Sciences under Grant GM 45372 and MDS Sciex, an industrial Associate of the Department of Chemistry.
References
- 1.Brinkworth CS, Dua S, McAnoy AM, Bowie JH. Rapid Commun Mass Spectrom. 2001;15:1965. doi: 10.1002/rcm.457. [DOI] [PubMed] [Google Scholar]
- 2.McLafferty FW, Turecek F. Interpretation of Mass Spectra. 4. University Science Books; Sausalito, CA: 1993. p. 371. [Google Scholar]
- 3.Loo JA, Ogorzalek-Loo RR, Udseth HR, Edmonds CG, Smith RD. Rapid Commun Mass Spectrom. 1991;5:101. doi: 10.1002/rcm.1290050303. [DOI] [PubMed] [Google Scholar]
- 4.Huang Y, Triscari JM, Pasa-Tolic L, Anderson GA, Lipton MS, Smith RD, Wysocki VH. J Am Chem Soc. 2004;126:3034. doi: 10.1021/ja038041t. [DOI] [PubMed] [Google Scholar]
- 5.Lin T, Payne AH, Glish GL. J Am Soc Mass Spectrom. 2001;12:497. doi: 10.1016/S1044-0305(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 6.Newton KA, Amunugama R, McLuckey SA. J Phys Chem A. 2005;109:3608. doi: 10.1021/jp04416i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton KA, He M, Amunugama R, McLuckey SA. Phys Chem Chem Phys. 2004;6:2710. doi: 10.1021/jp04416i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton KA, McLuckey SA. J Am Soc Mass Spectrom. 2004;15:607. doi: 10.1016/j.jasms.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 9.Newton KA, McLuckey SA. J Am Chem Soc. 2003;125:12404. doi: 10.1021/ja036924e. [DOI] [PubMed] [Google Scholar]
- 10.Pingitore F, Wesdemiotis C. Anal Chem. 2005;77:1796. doi: 10.1021/ac048469q. [DOI] [PubMed] [Google Scholar]
- 11.Wells JM, McLuckey SA. Methods Enzymol. 2005;402:148. doi: 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- 12.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc Natl Acad Sci USA. 2004;101:9528. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zubarev RA, Kelleher NL, McLafferty FW. J Am Chem Soc. 1998;120:3265. [Google Scholar]
- 14.Zubarev RA. Mass Spectrom Rev. 2003;22:57. doi: 10.1002/mas.10042. [DOI] [PubMed] [Google Scholar]
- 15.Wu J, Watson JT. Methods in Molecular Biology. In: Kannicht C, editor. Posttranslational Modifications of Proteins. Vol. 194. Humana Press; 2002. pp. 1–22. [Google Scholar]
- 16.Chrisman PA, McLuckey SA. J Proteome Res. 2002;1:549. doi: 10.1021/pr025561z. [DOI] [PubMed] [Google Scholar]
- 17.Zhang MX, Kaltashov IA. Anal Chem. 2006;78:4820. doi: 10.1021/ac060132w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lioe H, O’Hair RAJ. Anal Bioanal Chem. 2007;389:1429. doi: 10.1007/s00216-007-1535-1. [DOI] [PubMed] [Google Scholar]
- 19.Jones MD, Patterson SD, Lu HS. Anal Chem. 1998;70:136. doi: 10.1021/ac9707693. [DOI] [PubMed] [Google Scholar]
- 20.Kalli A, Hakansson K. Int J Mass Spectrom. 2007;263:71. [Google Scholar]
- 21.Kocher T, Engstrom A, Zubarev RA. Anal Chem. 2005;77:172. doi: 10.1021/ac0489115. [DOI] [PubMed] [Google Scholar]
- 22.Zubarev RA, Kruger NA, Fridriksson EK, Lewis MA, Horn DM, Carpenter BK, McLafferty FW. J Am Chem Soc. 1999;121:2857. [Google Scholar]
- 23.Chrisman PA, Pitteri SJ, Hogan JM, McLuckey SA. J Am Soc Mass Spectrom. 2005;16:1020. doi: 10.1016/j.jasms.2005.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Gundawardena HP, Huang T-Y, McLuckey SA. Int J Mass Spectrom. 2008;276:160. [Google Scholar]
- 25.Gunawardena HP, Gorenstein L, Erickson DE, Xia Y, McLuckey SA. Int J Mass Spectrom. 2007;265:130. [Google Scholar]
- 26.Fung YME, Kjeldsen F, Silivra OA, Chan TWD, Zubarev RA. Angew Chem Int Edn. 2005;44:57. doi: 10.1002/anie.200501533. [DOI] [PubMed] [Google Scholar]
- 27.Stephenson JL, Cargile BJ, McLuckey SA. Rapid Commun Mass Spectrom. 1999;13:2040. doi: 10.1002/(SICI)1097-0231(19991030)13:20<2040::AID-RCM754>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 28.Hogan JM, McLuckey SA. J Mass Spectrom. 2003;38:245. doi: 10.1002/jms.458. [DOI] [PubMed] [Google Scholar]
- 29.Wells JM, Stephenson JL, Jr, McLuckey SA. Int J Mass Spectrom. 2000;203:A1. [Google Scholar]
- 30.Sallans L, Lane KR, Freiser BS. J Am Chem Soc. 1989;111:865. [Google Scholar]
- 31.Payne AH, Glish GL. Int J Mass Spectrom. 2001;204:47. doi: 10.1016/S1044-0305(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 32.Kleinnijenhuis AJ, Mihalca R, Heeren RMA, Heck AJR. Int J Mass Spectrom. 2006;253:217. [Google Scholar]
- 33.Lioe H, Duan M, O’Hair RAJ. Rapid Commun Mass Spectrom. 2007;21:2727. doi: 10.1002/rcm.3122. [DOI] [PubMed] [Google Scholar]
- 34.Mihalca R, van der Burgt YEM, Heck AJR, Heeren RMA. J Mass Spectrom. 2007;42:450. doi: 10.1002/jms.1175. [DOI] [PubMed] [Google Scholar]
- 35.Kim HI, Beauchamp JL. J Am Chem Soc. 2008;130:1245. doi: 10.1021/ja075698w. [DOI] [PubMed] [Google Scholar]
- 36.Kim HI, Beauchamp JL. J Am Soc Mass Spectrom. 2009;20:157. doi: 10.1016/j.jasms.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 37.Gunawardena HP, O’Hair RAJ, McLuckey SA. J Proteome Res. 2006;5:2087. doi: 10.1021/pr0602794. [DOI] [PubMed] [Google Scholar]
- 38.Brazeau P, Vale W, Burgus R, Ling N, Butcher M, Rivier J, Guillemin R. Science. 1973;179:77. doi: 10.1126/science.179.4068.77. [DOI] [PubMed] [Google Scholar]
- 39.Hager JW. Rapid Commun Mass Spectrom. 2002;16:512. [Google Scholar]
- 40.Liang X, Xia Y, McLuckey SA. Anal Chem. 2006;78:3208. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia Y, Chrisman PA, Erickson DE, Liu J, Liang X, Londry FA, Yang MJ, McLuckey SA. Anal Chem. 2006;78:4146. doi: 10.1021/ac0606296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang X, McLuckey SA. J Am Soc Mass Spectrom. 2007;18:882. doi: 10.1016/j.jasms.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bowie JH, Brinkworth CS, Dua S. Mass Spectrom Rev. 2002;21:87. doi: 10.1002/mas.10022. [DOI] [PubMed] [Google Scholar]
- 44.Bilusich D, Bowie JH. Mass Spectrom Rev. 2009;28:20. doi: 10.1002/mas.20206. [DOI] [PubMed] [Google Scholar]
- 45.Leymarie N, Costello CE, O’Connor PB. J Am Chem Soc. 2003;125:8949. doi: 10.1021/ja028831n. [DOI] [PubMed] [Google Scholar]
- 46.O’Connor PB, Lin C, Cournoyer JJ, Pittman JL, Belyayev M, Budnik BA. J Am Soc Mass Spectrom. 2006;17:576. doi: 10.1016/j.jasms.2005.12.015. [DOI] [PubMed] [Google Scholar]