Abstract
Background
Lactic acid bacteria (LAB) are important in the food industry for the production of fermented food products and in human health as commensals in the gut. However, the phylogenetic relationships among LAB species remain under intensive debate owing to disagreements among different data sets.
Results
We performed a phylogenetic analysis of LAB species based on 232 genes from 28 LAB genome sequences. Regardless of the tree-building methods used, combined analyses yielded an identical, well-resolved tree topology with strong supports for all nodes. The LAB species examined were divided into two groups. Group 1 included families Enterococcaceae and Streptococcaceae. Group 2 included families Lactobacillaceae and Leuconostocaceae. Within Group 2, the LAB species were divided into two clades. One clade comprised of the acidophilus complex of genus Lactobacillus and two other species, Lb. sakei and Lb. casei. In the acidophilus complex, Lb. delbrueckii separated first, while Lb. acidophilus/Lb. helveticus and Lb. gasseri/Lb. johnsonii were clustered into a sister group. The other clade within Group 2 consisted of the salivarius subgroup, including five species, Lb. salivarius, Lb. plantarum, Lb. brevis, Lb. reuteri, Lb. fermentum, and the genera Pediococcus, Oenococcus, and Leuconostoc. In this clade, Lb. salivarius was positioned most basally, followed by two clusters, one corresponding to Lb. plantarum/Lb. brevis pair and Pediococcus, and the other including Oenococcus/Leuconostoc pair and Lb. reuteri/Lb. fermentum pair. In addition, phylogenetic utility of the 232 genes was analyzed to identify those that may be more useful than others. The genes identified as useful were related to translation and ribosomal structure and biogenesis (TRSB), and a three-gene set comprising genes encoding ultra-violet resistance protein B (uvrB), DNA polymerase III (polC) and penicillin binding protein 2B (pbpB).
Conclusions
Our phylogenomic analyses provide important insights into the evolution and diversification of LAB species, and also revealed the phylogenetic utility of several genes. We infer that the occurrence of multiple, independent adaptation events in LAB species, have resulted in their occupation of various habitats. Further analyses of more genes from additional, representative LAB species are needed to reveal the molecular mechanisms underlying adaptation of LAB species to various environmental niches.
Background
Lactic acid bacteria (LAB) are Gram-positive bacteria that have been widely used as starter or nonstarter cultures in the plant, meat, and dairy fermentation, and also as probiotic bacteria in human gastrointestinal tract contributing to pathogen inhibition and immunomodulation. At present, nearly 400 LAB species have been recognized [1]. They are generally classified into four families and seven genera, as follows: family Lactobacillaceae (genera Lactobacillus and Pediococcus), family Leuconostocaceae (genera Oenococcus and Leuconostoc), family Enterococcaceae (genus Enterococcus) and family Streptococcaceae (genera Lactococcus and Streptococcus) [2-4]. Phylogenetic relationships among the LAB species have been hotly disputed. One of the foremost debates in LAB phylogeny concerns the species in the genera Lactobacillus, Pediococcus, Oenococcus, and Leuconostoc, which belong to family Lactobacillaceae and Leuconostocaceae, due to the severe disagreements arising from analyses of different data sets [2-11]. In the genus Lactobacillus, there are uncertainties about the interspecies affinities within the acidophilus complex [8] that consists of five species Lb. gasseri, Lb. johnsonii, Lb. acidophilus, Lb. helveticus and Lb. delbrueckii. In particular, the divergence between Lb. gasseri/Lb. johnsonii, Lb. acidophilus/Lb. helveticus and Lb. delbrueckii remains unresolved. Based on the analyses of a 16 S rRNA gene and a few nuclear genes [3,5,7,10,12] and that of 32 ribosomal proteins [9], Lb. delbrueckii was found to be more closely associated with Lb. acidophilus/Lb. helveticus than with Lb. gasseri/Lb. johnsonii. However, a recent study using 141 core proteins from 17 LAB species suggested that Lb. delbrueckii diverged earliest within the acidophilus complex, while Lb. acidophilus/Lb. helveticus and Lb. gasseri/Lb. johnsonii clustered into a sister group [8].
Although the paraphyly of Lactobacillus species is well-established, a general consensus for the placement of the Lactobacillus species, e.g., Lb. salivarius, Lb. plantarum, Lb. brevis, Lb. reuteri, Lb. sakei, and Lb. casei, and their relationship to the genera Pediococcus, Oenococcus, and Leuconostoc has not yet emerged in the 'salivarius' subgroup. For example, in the analysis of four subunits of RNA polymerase, the clade uniting Lb. sakei and Lb. casei is placed at the most basal position, followed by Lb. salivarius. Pediococcus is sister to the clade containing Lb. plantarum and Lb. brevis, while Oenococcus/Leuconostoc clusters with Lb. reuteri [7]. In contrast, an analysis of 141 core proteins suggested that the Lb. sakei/Lb. casei clade is more related to acidophilus complex, while the other Lactobacillus species and Pediococcus, Oenococcus, as well as Leuconostoc group together, in which Oenococcus/Leuconostoc diverged earliest, followed by Lb. salivarius, Pediococcus, Lb. reuteri, and lastly the species most recently diverged, Lb. plantarum and Lb. brevis [8].
These findings highlight the need to gather and analyze larger sequence data sets in order to unravel the phylogenetic relationships among LAB species and clarify specifically those within genera Lactobacillus, Pediococcus, Oenococcus, and Leuconostoc. The increasing availability of LAB genome sequence data provides a good opportunity to understand the evolutionary history of LAB species. In the present study, we studied LAB phylogeny by gathering and analyzing 232 orthologous genes from 28 LAB genome sequences representing all genera from four families. Our objectives were to provide new insights into the relationships of LAB species and to examine the utility of such an analysis in the context of LAB phylogeny, and develop new potential genetic markers for study of LAB systematics. This study not only contributes to clarifying the currently obscure LAB species relationship, but also lays a foundation for further studies on adaptive evolution of LAB species in different environmental niches.
Results and Discussions
Identification of orthologous genes
The use of accurate and reliable methods for the identification of orthologous genes is essential for phylogenetic reconstruction based on analyses of large data sets, especially for those using whole genome sequences [13]. In the present study, the strategy of developing potential orthologous gene sets for LAB phylogenomic studies was different from those used in previous LAB analyses. First, in previous studies of LAB phylogeny, less stringent clusters of orthologous groups (COGs) [6] and reciprocal best hits [8] methods were applied to identify putative orthologs. Here, we applied both Inparanoid [14] and MultiParanoid [15] programs to serve this purpose. Inparanoid [14] exploits a BLAST-based strategy to identify orthologs as reciprocal best hits between two species, and applies additional rules to accommodate in-paralogs that arise from recent duplication events after speciation. Compared with other methods, including COGs [16] and OrthoMCL [17], Inparanoid's superiority lies in the ability to distinguish orthologs from in-paralogs and out-paralogs (those that arose via ancient duplication event before speciation) [17-21]. MultiParanoid software [15] performs clustering of orthologs and in-paralogs that are shared by more than two species. By using the conservative searching algorithms, we obtained a total of 310 one-to-one protein coding orthologs (Additional file 1 Table S1).
To make our dataset more conservative, we further excluded potentially problematic orthologs such as those with short sequence lengths and those involved in horizontal gene transfer (HGT). These criteria have not yet been used in previous LAB studies. In the end, a total of 232 orthologous genes, including 225 genes that have clear functional definition and 7 genes that have been annotated as hypothetical proteins (Additional file 2 Table S2), were used to reconstruct LAB phylogeny in this study. This dataset of 232 genes included those encoding 135 out of the 141 core proteins of the Claesson's study [8] that were identified by phylogenomic analyses of 17 LAB species genomes. Noticeably, 6 core proteins included in Claesson's study [8] were discovered as in-paralogs here and hence excluded from further analyses. This suggests that our dataset is more conservative and reliable than those from previous studies aimed at inferring LAB phylogeny.
Reconstruction of LAB phylogenomic tree
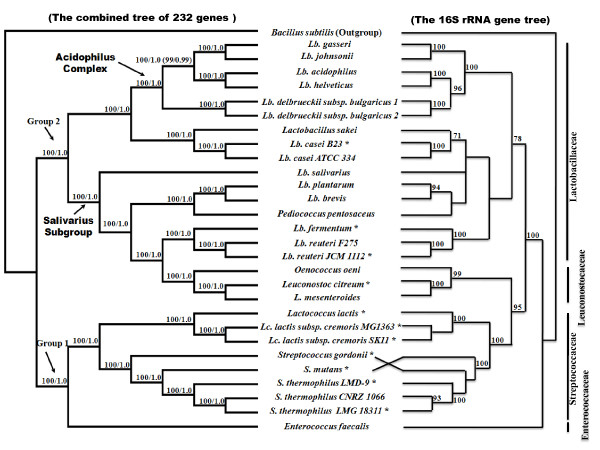
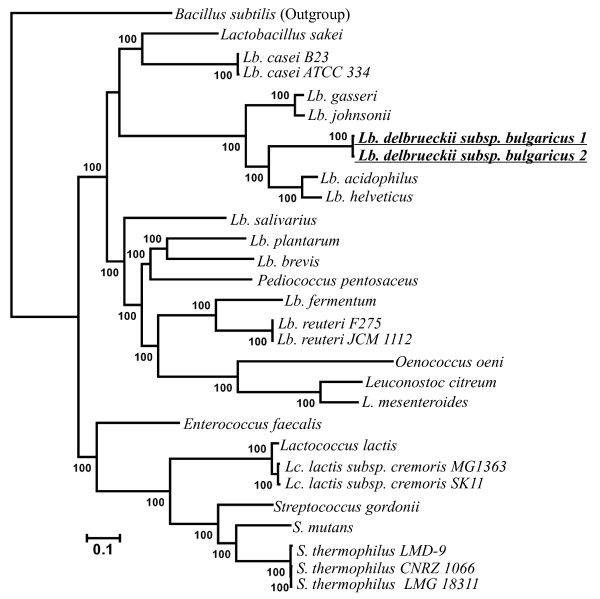
Based on the concatenated amino acid alignment of 232 genes, phylogenetic analyses using two gap selection criteria (see Methods) and two tree-building methods, partitioned maximum likelihood (ML) and Bayesian analyses, yielded an identical, well-resolved tree topology with strong supports for all nodes (BS > 99% and PP > 0.99) (Figure 1), suggesting that the accuracy of our phylogenetic inference is independent of tree-building methods. As revealed in Figure 1 the monophyly for families Leuconostocaceae, Enterococcaceae and Streptococcaceae were strongly supported. For Lactobacillaceae, some species were more closely related to Leuconostocaceae than the other Lactobacillaceae species, supporting the paraphyly for family Lactobacillaceae, providing a possibility that Leuconostocaceae and Lactobacillaceae can be combined into a family.
Figure 1.
Partitioned Bayesian/ML tree topology inferred from the selected 232 genes and the 16 S rRNA gene tree of 29 species. For the concatenated tree of 232 genes, ML bootstrap supports and Bayesian posterior probabilities are shown above the branches. The stars imply newly added species in this study compared with that of Claesson et al. [8]. Lb. delbrueckii subsp. bulgaricus 1 refers to Lb. delbrueckii subsp. bulgaricus ATCC BAA-365; Lb. delbrueckii subsp. bulgaricus 2 refers to Lb. delbrueckii subsp. bulgaricus ATCC 11842; NJ analysis under 1000 bootstrap runs of 16 S rRNA genes from the study by Ventura et al [12] and Kawamura et al 's study [22]. ML bootstrap supports higher than 50 are shown above the branches.
The LAB species were divided into two groups. Group 1 included Enterococcaceae and Streptococcaceae. Group 2 included Lactobacillaceae and Leuconostocaceae. Within Group 1, the monophyly of the genera Enterococcus, Lactococcus and Streptococcus were strongly supported. In Streptococcus, S. mutans and S. thermophilus were grouped together, and S. gordonii was their sister taxon. The relationships within Group 1 observed here were congruent with two other studies [5,10], but disagreed with the 16 S rRNA gene tree [22] (Figure 1). Within Group 2, LAB species were divided into two clades. One clade composed of acidophilus complex of genus Lactobacillus and two other Lactobacillus species, Lb. sakei and Lb. casei. This result is in contradiction with the RNA polymerase-based study of Liu [7] that suggested that Lb. sakei and Lb. casei are more closely related to other Lactobacillus species and the genera Pediococcus, Oenococcus as well as Leuconostoc. However, our results are in agreement with the RNA polymerase trees [5,10], ribosomal-protein tree [9] and the 141-core proteins tree [8]. Of the five recognized Lactobacillus species in the acidophilus complex, our results strongly support the notion that Lb. delbrueckii separated first, while Lb. acidophilus/Lb. helveticus and Lb. gasseri/Lb. johnsonii clustered into a sister group. This finding is in accordance with the result derived from the 141-core proteins analyses [8], but disagrees with those derived the single 16 S rRNA gene [3,8,12] and the nuclear gene analyses [5,7,10,23] as well as that of 32 ribosomal proteins [9], in which Lb. delbrueckii was seen to be more closely associated with Lb. acidophilus/Lb. helveticus than Lb. gasseri/Lb. johnsonii. Five Lactobacillus species, including Lb. salivarius, Lb. plantarum, Lb. brevis, Lb. reuteri, Lb. fermentum, and the genera Pediococcus, Oenococcus, and Leuconostoc constitute the other clade, the 'salivarius' subgroup within Group 2. In this clade, Lb. salivarius was positioned most basally, followed by two distinct clusters, one corresponding to Lb. plantarum/Lb. brevis group and Pediococcus, and the other including Oenococcus/Leuconostoc group and Lb. reuteri/Lb. fermentum group. The basal position of Lb. salivarius in this clade is consistent with the RNA polymerase tree inferred by Makarova and Koonin [5] as well as by Liu [7], but not with the 16 S rRNA gene tree [12] and studies by Claesson [8] and Cai [10] that indicated that Oenococcus/Leuconostoc group diverged first. In addition, the grouping of Lb. plantarum/Lb. brevis and Pediococcus observed here is supported in most current studies, but is in contradiction with the recent proposal of the connecting of Lb. plantarum/Lb. brevis and Lb. reuteri. In the present study, the close relatedness of Oenococcus/Leuconostoc group and Lb. reuteri/Lb. fermentum is in agreement with RNA polymerase tree inferred by Liu et al. [7]. The possible placement of Oenococcus/Leuconostoc group as the first diverging taxa [8,10] or as the diverging taxa subsequent to Lb. salivarius [5] was not supported here.
Taken together, our study provides new insights into the evolutionary relationships of these LAB species, and helps to resolve the current controversial issues in LAB phylogeny. Depending on the gene segments or genomes and the tree-building methods used, different phylogenetic hypotheses can be obtained. Interestingly, our study demonstrated that different evolutionary rates among sites may also affect LAB phylogenetic reconstruction. When we repeated the phylogenetic analyses by setting a fixed alpha value of gamma distribution in the optimal amino acid substitution model, the species relationships within acidophilus complex, i.e., that among Lb. gasseri/Lb. johnsonii, Lb. acidophilus/Lb. helveticus and Lb. delbrueckii, became unstable and were poorly supported in partitioned ML and Bayesian analyses (data not shown). Therefore, our study revealed that different evolutionary rate among sites is also an important factor in tracing the evolutionary history of LAB species.
Besides the contribution of phylogenetic resolution, our results revealed the presence of independent adaptation to four types of habitat niches in LAB species (Figure 1), involving human gastrointestinal tract, human oral flora, dairy fermentation and other fermentations of beer, wine, plants, or meat (Table 1). For example, within acidophilus complex, Lb. acidophilus that is isolated from human gastrointestinal tract and Lb. helveticus that is widely applied to dairy fermentation are more closely related to each other than to the other three Lactobacillus species, suggesting an independent adaptation to their respective niches. The independent adaptation events of Lb. plantarum to human gastrointestinal tract were also evidenced by transcriptome analyses [24], although Lb. plantarum strains isolated from the gastrointestinal tract or feces may be derived from human diet and may in fact reflect earlier adaption to other environmental niches such as fermentations of meat, plant, cheese or wine [25]. Otherwise, Lb. brevis is most suitable for meat fermentation in our phylogenetic tree. Given that strains of many LAB species occur in a multitude of ecological niches, further analyses of more genes and functional assays of additional LAB species are needed to reveal the molecular mechanisms underlying the adaptation of LAB species to various environmental survival niches.
Table 1.
Summary of 28 LAB taxa and one outgroup (Bacillus subtilis)
| Species-Organisms | Association | NCBI RefSeq |
|---|---|---|
| Bacillus subtilis subsp. subtilis str. 168 | Outgroup | NC_000964 |
| Enterococcus faecalis V583 | gastrointestinal tract bacteria | NC_004668 |
| Lactobacillus acidophilus NCFM | gastrointestinal tract bacteria | NC_006814 |
| Lactobacillus brevis ATCC 367 | other fermentation such as beer, wine, plants, or meat | NC_008497 |
| Lactobacillus casei ATCC 334 | dairy fermentation | NC_008526 |
| Lactobacillus casei BL23 | dairy fermentation | NC_010999 |
| Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842 | dairy fermentation | NC_008054 |
| Lactobacillus delbrueckii subsp. bulgaricus ATCC BAA-365 | dairy fermentation | NC_008529 |
| Lactobacillus fermentum IFO 3956 | other fermentation such as beer, wine, plants, or meat | NC_010610 |
| Lactobacillus gasseri ATCC 33323 | gastrointestinal tract bacteria | NC_008530 |
| Lactobacillus helveticus DPC 4571 | dairy fermentation (Swiss cheese isolate) | NC_010080 |
| Lactobacillus johnsonii NCC 533 | gastrointestinal tract bacteria | NC_005362 |
| Lactobacillus plantarum WCFS1 | Human saliva (first), gut, dairy, wine, plants, or meat | NC_004567 |
| Lactobacillus reuteri F275 | gastrointestinal tract bacteria | NC_009513 |
| Lactobacillus reuteri JCM 1112 | gastrointestinal tract bacteria | NC_010609 |
| Lactobacillus sakei subsp. sakei 23K | other fermentation such as beer, wine, plants, or meat | NC_007576 |
| Lactobacillus salivarius UCC118 | gastrointestinal tract bacteria | NC_007929 |
| Lactococcus lactis subsp. cremoris MG1363 | dairy fermentation | NC_009004 |
| Lactococcus lactis subsp. cremoris SK11 | dairy fermentation | NC_008527 |
| Lactococcus lactis subsp. lactis Il1403 | dairy fermentation | NC_002662 |
| Leuconostoc citreum KM20 | other fermentation such as beer, wine, plants, or meat | NC_010471 |
| Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 | other fermentation such as beer, wine, plants, or meat | NC_008531 |
| Oenococcus oeni PSU-1 | other fermentation such as beer, wine, plants, or meat | NC_008528 |
| Pediococcus pentosaceus ATCC 25745 | dairy fermentation | NC_008525 |
| Streptococcus gordonii str. Challis substr. CH1 | human oral flora (dental plaque) | NC_009785 |
| Streptococcus mutans UA159 | oral streptococci (leading cause of dental caries) | NC_004350 |
| Streptococcus thermophilus CNRZ1066 | dairy fermentation | NC_006449 |
| Streptococcus thermophilus LMD-9 | dairy fermentation | NC_008532 |
Utilities of different genes in LAB phylogeny
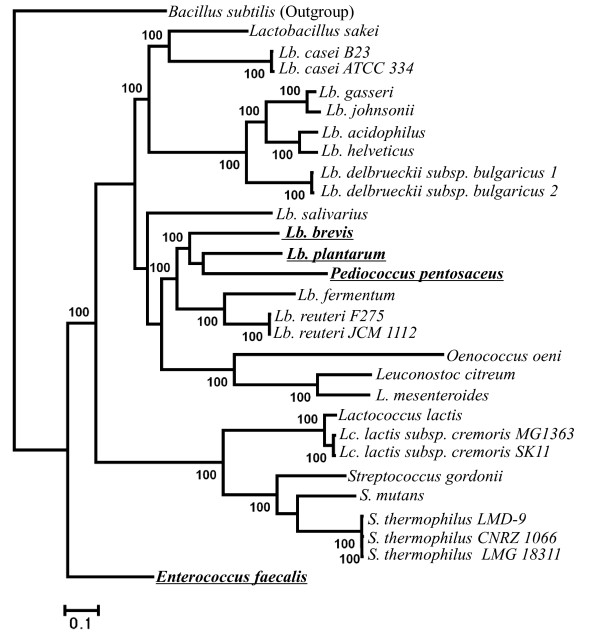
We also evaluated the phylogenetic utility of different genes used here. According to COG annotation [16], we classified 232 genes into four functional categories (Additional file 2 Table S2) relating to: information storage and processing (ISP; 135 genes), cellular processes and signaling (CPS; 49 genes), metabolism (41 genes), and hypothetical proteins (HP; 7 genes). Among them, the genes with ISP function were further divided into translation, ribosomal structure and biogenesis (TRSB; 69 genes), replication/repair/recombination (RRR; 51 genes), and transcription (15 genes). The phylogenetic analyses of LAB were repeated using each of the above six categories of genes individually. Our results suggested that the analyses of RRR (Figure 2), transcription (Figure 3), CPS (Figure 4), metabolism (Figure 5) and HP (Figure 6) genes produced different tree topologies from that of all concatenated genes (Figure 1), while the analyses of TRSB genes yielded identical tree topologies to those shown in Figure 1 suggesting that the TRSB genes are better indicators of LAB phylogeny than are other subsets of genes. The Robinson-Foulds distances analysis (Additional file 3 Table S3) also showed that there are no differences between the tree of TRSB genes and that of all concatenated genes. The differences among tree topologies based on these functional categories can be caused by various factors, including different selective constraints imposed by the functional categories that were involved in various metabolic networks [26-31].
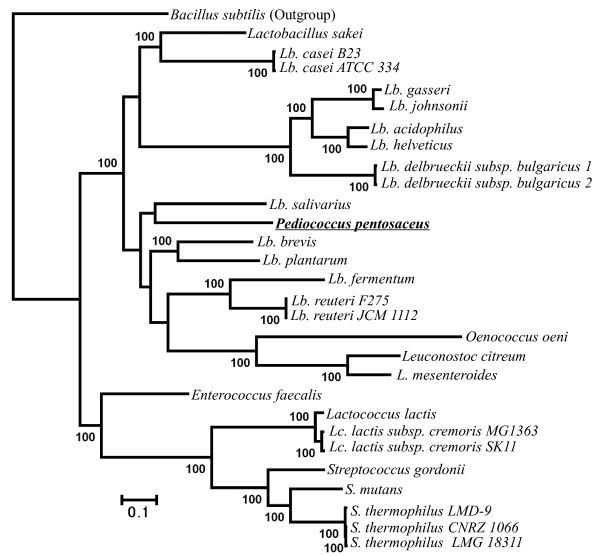
Figure 2.
Tree topologies inferred from 51 replication/repair/recombination genes using ML analyses with 1,000 bootstrap runs. Bootstrap supports higher than 50 are shown above the branches. Compared to the combined tree of 232 genes (Figure 1), taxa with topological differences are underlined. Lb. delbrueckii subsp. bulgaricus 1 = Lb. delbrueckii subsp. bulgaricus ATCC BAA-365; Lb. delbrueckii subsp. bulgaricus 2 = Lb. delbrueckii subsp. bulgaricus ATCC 11842.
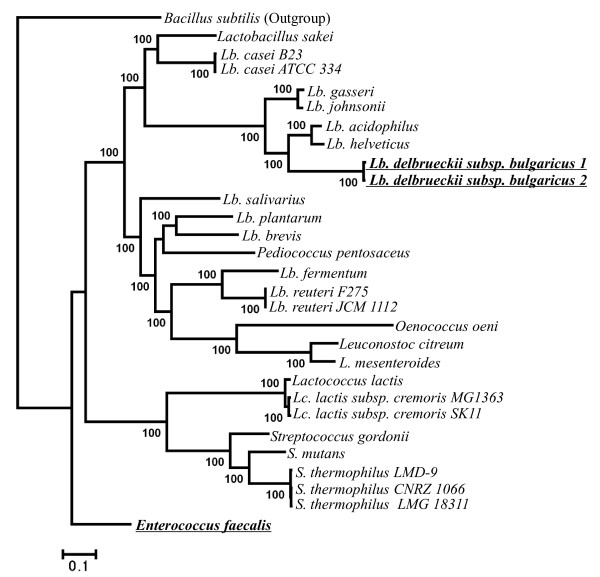
Figure 3.
Tree topologies inferred from 15 transcription genes using ML analyses with 1,000 bootstrap runs. Bootstrap supports higher than 50 are shown above the branches. Compared to the combined tree of 232 genes (Figure 1), taxa with topological differences are underlined. Lb. delbrueckii subsp. bulgaricus 1 = Lb. delbrueckii subsp. bulgaricus ATCC BAA-365; Lb. delbrueckii subsp. bulgaricus 2 = Lb. delbrueckii subsp. bulgaricus ATCC 11842.
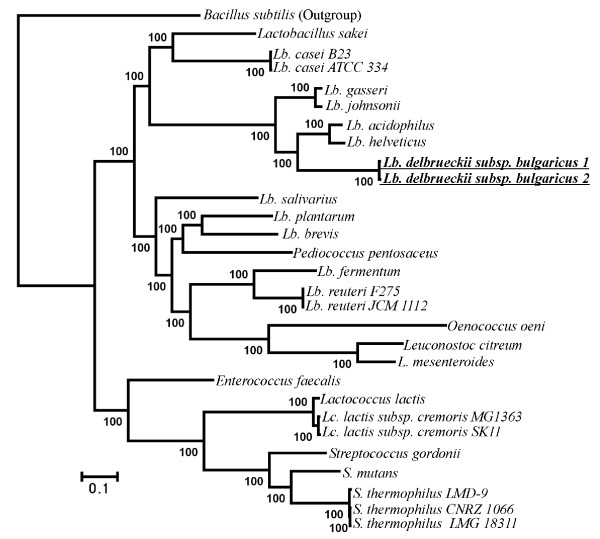
Figure 4.
Tree topologies inferred from 49 cellular processes and signaling genes using ML analyses with 1,000 bootstrap runs. Bootstrap supports higher than 50 are shown above the branches. Compared to the combined tree of 232 genes (Figure 1), taxa with topological differences are underlined. Lb. delbrueckii subsp. bulgaricus 1 = Lb. delbrueckii subsp. bulgaricus ATCC BAA-365; Lb. delbrueckii subsp. bulgaricus 2 = Lb. delbrueckii subsp. bulgaricus ATCC 11842.
Figure 5.
Tree topologies inferred from 41 metabolism genes using ML analyses with 1,000 bootstrap runs. Bootstrap supports higher than 50 are shown above the branches. Compared to the combined tree of 232 genes (Figure 1), taxa with topological differences are underlined. Lb. delbrueckii subsp. bulgaricus 1 = Lb. delbrueckii subsp. bulgaricus ATCC BAA-365; Lb. delbrueckii subsp. bulgaricus 2 = Lb. delbrueckii subsp. bulgaricus ATCC 11842.
Figure 6.
Tree topologies inferred from 7 hypothetical genes using ML analyses with 1,000 bootstrap runs. Bootstrap supports higher than 50 are shown above the branches. Compared to the combined tree of 232 genes (Figure 1), taxa with topological differences are underlined. Lb. delbrueckii subsp. bulgaricus 1 = Lb. delbrueckii subsp. bulgaricus ATCC BAA-365; Lb. delbrueckii subsp. bulgaricus 2 = Lb. delbrueckii subsp. bulgaricus ATCC 11842.
Ranking single genes in six function categories by their respective phylogenetic resolution to LAB species reveals that 3 of 232 genes, including the ultra-violet resistance protein B gene (uvrB) and the DNA polymerase III gene (polC) from RRR category, and the penicillin binding protein 2B gene (pbpB) from CPS category (Additional file 2 Table S2), produced ML tree topology (Additional file 4 Figure S1a-1c) that was largely consistent with that of the complete analyses (Figure 1), albeit with low supports for some branches (BS < 70%). When we conducted the phylogenetic analyses by combining the three genes, a completely identical tree topology to that shown in Figure 1 with high supports for most of nodes was obtained. Therefore, a combined analysis using uvrB, polC and pbpB together seems to be a better indicator for inferring LAB phylogeny than the other subset of genes including the ribosomal protein families or RNA polymerase subunits that have been widely used in previous LAB phylogenetic studies [5-7,9,10]. The Robinson-Foulds distances analysis (Additional file 3 Table S3) also showed that there are no differences between the tree of combined uvrB, polC and pbpB genes and that of all concatenated genes. In the present study, the assessment of phylogenetic utility and limits of the individual genes makes it possible to preselect subsets of genes for future molecular studies of LAB phylogeny when the complete genome sequences are unavailable.
Conclusions
In this study, phylogenetic relationships among LAB species are presented based on 232 genes from 28 LAB genome sequences. The concatenation of all these genes allowed the recovery of a strongly supported phylogeny, providing a maximum and decisive resolution of the relationships among the LAB species examined. Our phylogenomic analyses provide important insights into not only LAB phylogeny, but also the phylogenetic utility of different genes suggesting that the genes relating to translation, ribosomal structure and biogenesis (TRSB) function and a three-gene set consisting of uvrB, polC and pbpB, may be better indicators for LAB phylogenetic studies than the other subsets of genes. In addition, our study demonstrates the presence of multiple independent adaption events of LAB species to different survival habitats, indicating that further analyses of more genes from representatives of additional LAB species are needed in order to reveal the molecular mechanisms underlying the adaptation of LAB species to various environmental survival niches.
Methods
Sequence Data
A total of 28 available LAB genomes [6,9,32-44] representing seven genera of four families were used (Table 1). In addition, the genome sequence from Bacillus subtilis was used as an outgroup to root the tree.
Identification of one-to-one orthologs for LAB phylogenetic inference
Based on protein coding genes (pseudogenes are not included) downloaded from 28 LAB and one B. subtilis genome sequences, a search for orthologs was conducted with the program Inparanoid version 2.0 [14]. Several stringent criteria were employed: (1) using a BLAST score cut-off of 50 bits; (2) using an overlap cut-off of 50%; (3) using a confidence value of 95% when searching in-paralogs; (4) using BLOSUM45 amino acid substitution matrix [45]. Automatic clustering of orthologs and inparalogs identified by the program Inparanoid was then performed by program Multiparanoid [15].
Among the candidate orthologous genes selected as above, we excluded those that met the following criteria from subsequent analyses: (1) lesser than 100 amino acid sequence length; (2) involved in potential horizontal gene transfer (HGT) events, as predicted by Horizontal Gene Transfer Database (HGT-DB) http://genomes.urv.es/HGT-DB/ and http://www.tinet.org/~debb/HGT/welcomeOLD.html and by previous studies [6]. In the end, a total of 232 orthologous genes, including 225 that have clear functional definition and 7 that have been annotated to be hypothetical proteins, were used to reconstruct LAB phylogeny in this study (Additional file 2 Table S2).
Phylogenetic Reconstruction of LAB species
In total 232 orthologous genes were concatenated into two supermatrices according to two gap selection criteria in Gblocks [allowed gap positions = none (61,020 amino acids in length) and with half (only positions where 50% or more of the sequences have a gap are treated as a gap position in the final alignment) (63,910 amino acids in length)] [46]. Optimal substitution models were selected by using the program ProtTest version 2.4 [47] according to Akaike Information Criterion (AIC) [48]. The selected substitution models were used in partitioned Bayesian analysis implemented MrBayes v3.2.1 [49-51] and partitioned maximum likelihood (ML) analysis implemented in RAxML v7.0.4 [52]. The reliability of ML tree topology was evaluated by bootstrapping sampling (BP) of 1000 replicates. For Bayesian analyses, three independent runs of one-million generations each were used. The trees sampled prior to reaching convergence were discarded as burn-in and the remaining trees were used to construct the consensus tree and posterior probabilities (PP).
Tree topology comparison
The differences between tree topologies were compared using Robinson-Foulds distances that were calculated with program Treedist from the PHYLIP v3.69 package [53].
Authors' contributions
ZZ and PS designed the study. ZZ, ZY, LY and PS analyzed the data and wrote the manuscript. All the authors have read and approved the final manuscript.
Supplementary Material
Table S1. Summary of 310 one-to-one orthologs from 28 LAB species and one outgroup (Bacillus subtilis).
Table S2. Summary of 232 one-to-one orthologs used in LAB phylogenomic inference and their functional categories based on COG annotation.
Table S3. Robinson-Foulds distances between different tree topologies.
Figure S1. Single gene trees inferred from ML analyses with 1,000 replicates.
Contributor Information
Zhi-Gang Zhang, Email: zhzhg2007@gmail.com.
Zhi-Qiang Ye, Email: zhiqiang.yes@gmail.com.
Li Yu, Email: yuli1220@yahoo.com.cn.
Peng Shi, Email: ship@mail.kiz.ac.cn.
Acknowledgements
Special thanks to members of the Shi lab and two anonymous reviewers for valuable comments and Dong-Qiang Chen for technical assistance. This work was supported by grants from General Program of Natural Science Foundation of Yunnan Province of China (Grant No. 2009CD108) and by a start-up fund of "Hundreds Talent Program" from Chinese Academy of Sciences to P.S.
References
- Euzéby JP. List of bacterial names with standing in nomenclature: a folder available on the internet. Int J Syst Bacteriol. 1997;47:590–592. doi: 10.1099/00207713-47-2-590. http://www.bacterio.cict.fr/index.html [DOI] [PubMed] [Google Scholar]
- Salminen S, von Wright A, Ouwehand Ae. Lactic Acid Bacteria: Microbiological and Functional Aspects (3rd ed.) Revised and Expanded Edition. Marcel Dekker Inc, New York; 2004. [Google Scholar]
- Collins MD, Rodrigues U, Ash C, Aguirre M, Farrow JAE, Martinez-Murcia A, Phillips BA, Williams AM, Wallbanks S. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16 S rRNA. FEMS Microbiology Letters. 1991;77(1):5–12. doi: 10.1111/j.1574-6968.1991.tb04313.x. [DOI] [Google Scholar]
- Carr FJ, Chill D, Maida N. The lactic acid bacteria: a literature survey. Crit Rev Microbiol. 2002;28(4):281–370. doi: 10.1080/1040-840291046759. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV. Evolutionary genomics of lactic acid bacteria. J Bacteriol. 2007;189(4):1199–1208. doi: 10.1128/JB.01351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N. et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA. 2006;103(42):15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Nauta A, Francke C, Siezen RJ. Comparative genomics of enzymes in flavor-forming pathways from amino acids in lactic acid bacteria. Appl Environ Microbiol. 2008;74(15):4590–4600. doi: 10.1128/AEM.00150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, van Sinderen D, O'Toole PW. Lactobacillus phylogenomics - towards a reclassification of the genus. Int J Syst Evol Microbiol. 2008;58(12):2945–2954. doi: 10.1099/ijs.0.65848-0. [DOI] [PubMed] [Google Scholar]
- Callanan M, Kaleta P, O'Callaghan J, O'Sullivan O, Jordan K, McAuliffe O, Sangrador-Vegas A, Slattery L, Fitzgerald GF, Beresford T. et al. Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J Bacteriol. 2008;190(2):727–735. doi: 10.1128/JB.01295-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Thompson R, Budinich M, Broadbent JR, Steele JL. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol Evol. 2009. evp019. [DOI] [PMC free article] [PubMed]
- Berger B, Pridmore RD, Barretto C, Delmas-Julien F, Schreiber K, Arigoni F, Brussow H. Similarity and differences in the Lactobacillus acidophilus group identified by polyphasic analysis and comparative genomics. J Bacteriol. 2007;189(4):1311–1321. doi: 10.1128/JB.01393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, O'Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, van Sinderen D, O'Toole PW. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Micro. 2009;7(1):61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6(5):361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Remm M, Storm CEV, Sonnhammer ELL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. Journal of Molecular Biology. 2001;314(5):1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Alexeyenko A, Tamas I, Liu G, Sonnhammer ELL. Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics. 2006;22(14):e9–15. doi: 10.1093/bioinformatics/btl213. [DOI] [PubMed] [Google Scholar]
- Tatusov R, Fedorova N, Jackson J, Jacobs A, Kiryutin B, Koonin E, Krylov D, Mazumder R, Mekhedov S, Nikolskaya A. et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4(1):41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Research. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Marcotte EM. A probabilistic view of gene function. Nat Genet. 2004;36(6):559–564. doi: 10.1038/ng1370. [DOI] [PubMed] [Google Scholar]
- Kopelman NM, Lancet D, Yanai I. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat Genet. 2005;37(6):588–589. doi: 10.1038/ng1575. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Tomlins SA, Varambally S, Mahavisno V, Barrette T, Kalyana-Sundaram S, Ghosh D, Pandey A, Chinnaiyan AM. Probabilistic model of the human protein-protein interaction network. Nat Biotech. 2005;23(8):951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S. et al. A Human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122(6):957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Hou X-G, Sultana F, Miura H, Ezaki T. Determination of 16 S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45(2):406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- Naser SM, Dawyndt P, Hoste B, Gevers D, Vandemeulebroecke K, Cleenwerck I, Vancanneyt M, Swings J. Identification of lactobacilli by pheS and rpoA gene sequence analyses. International Journal of Systematic and Evolutionary Microbiology. 2007;57(12):2777–2789. doi: 10.1099/ijs.0.64711-0. [DOI] [PubMed] [Google Scholar]
- Marco ML, de Vries MC, Wels M, Molenaar D, Mangell P, Ahrne S, de Vos WM, Vaughan EE, Kleerebezem M. Convergence in probiotic Lactobacillus gut-adaptive responses in humans and mice. ISME J. 2010. [DOI] [PubMed]
- Siezen RJ, Tzeneva VA, Castioni A, Wels M, Phan HTK, Rademaker JLW, Starrenburg MJC, Kleerebezem M, Molenaar D, Van Hylckama Vlieg JET. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environmental Microbiology. 2010;12(3):758–773. doi: 10.1111/j.1462-2920.2009.02119.x. [DOI] [PubMed] [Google Scholar]
- Tourasse NJ, Li W-H. Selective constraints, amino acid composition, and the rate of protein evolution. Molecular Biology and Evolution. 2000;17(4):656–664. doi: 10.1093/oxfordjournals.molbev.a026344. [DOI] [PubMed] [Google Scholar]
- KIMURA M. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, England; 1983. [Google Scholar]
- LI W-H. Molecular evolution. Sinauer, Sunderland, Mass; 1997. [Google Scholar]
- NEI M. Molecular evolutionary genetics. Columbia University Press, New York; 1987. [Google Scholar]
- Greenberg AJ, Stockwell SR, Clark AG. Evolutionary constraint and adaptation in the metabolic network of drosophila. Molecular Biology and Evolution. 2008;25(12):2537–2546. doi: 10.1093/molbev/msn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Ramirez S, Gonzalez V. Factors affecting the concordance between orthologous gene trees and species tree in bacteria. BMC Evolutionary Biology. 2008;8(1):300. doi: 10.1186/1471-2148-8-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdić D, McShan WM, McLaughlin RE, Savić G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H. et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002;99(22):14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M. et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci USA. 2005;102(11):3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Renault P, Sorokin A, Ehrlich SD, Kulakauskas S, Lapidus A, Goltsman E, Mazur M, Pusch GD. et al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat Biotech. 2004;22(12):1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillou S, Champomier-Verges MC, Cornet M, Crutz-Le Coq AM, Dudez AM, Martin V, Beaufils S, Darbon-Rongere E, Bossy R, Loux V. et al. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23 K. Nat Biotechnol. 2005;23(12):1527–1533. doi: 10.1038/nbt1160. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Li Y, Leahy S, Canchaya C, van Pijkeren JP, Cerdeño-Tárraga AM, Parkhill J, Flynn S, O'Sullivan GC, Collins JK. et al. Multireplicon genome architecture of Lactobacillus salivarius. Proc Natl Acad Sci USA. 2006;103(17):6718–6723. doi: 10.1073/pnas.0511060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JF, Jeong H, Lee J-S, Choi S-H, Ha M, Hur C-G, Kim J-S, Lee S, Park H-S, Park Y-H. et al. Complete genome sequence of Leuconostoc citreum KM20. J Bacteriol. 2008;190(8):3093–3094. doi: 10.1128/JB.01862-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW. et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci USA. 2003;100(4):1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S. et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390(6657):249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Morita H, Toh H, Fukuda S, Horikawa H, Oshima K, Suzuki T, Murakami M, Hisamatsu S, Kato Y, Takizawa T. et al. Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res. 2008;15(3):151–161. doi: 10.1093/dnares/dsn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF. et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299(5615):2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- Pridmore RD, Berger B, Desiere F, Vilanova D, Barretto C, Pittet A-C, Zwahlen M-C, Rouvet M, Altermann E, Barrangou R. et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc Natl Acad Sci USA. 2004;101(8):2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Guchte M, Penaud S, Grimaldi C, Barbe V, Bryson K, Nicolas P, Robert C, Oztas S, Mangenot S, Couloux A. et al. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc Natl Acad Sci USA. 2006;103(24):9274–9279. doi: 10.1073/pnas.0603024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP. et al. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J Bacteriol. 2007;189(8):3256–3270. doi: 10.1128/JB.01768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Akaike H. Information theory and an extension of the maximum likelihood principle. Proceedings of 2nd International Symposium on Information Theory, Budapest, Hungary. 1973. pp. 267–281.
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Parallel Metropolis-coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20(3):407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294(5550):2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey J. Bayesian phylogenetic analysis of combined Data. Syst Biol. 2004;53(1):47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
- Ott M, Zola J, Stamatakis A, Aluru S. Proceedings of the 2007 ACM/IEEE conference on Supercomputing. Reno, Nevada: ACM; 2007. Large-scale maximum likelihood-based phylogenetic analysis on the IBM BlueGene/L; pp. 1–11. full_text. [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author Department of Genome Sciences, University of Washington, Seattle. 2005.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of 310 one-to-one orthologs from 28 LAB species and one outgroup (Bacillus subtilis).
Table S2. Summary of 232 one-to-one orthologs used in LAB phylogenomic inference and their functional categories based on COG annotation.
Table S3. Robinson-Foulds distances between different tree topologies.
Figure S1. Single gene trees inferred from ML analyses with 1,000 replicates.