Abstract
Surface recognition and penetration are among the most critical plant infection processes in foliar pathogens. In Magnaporthe oryzae, the Pmk1 MAP kinase regulates appressorium formation and penetration. Its orthologs also are known to be required for various plant infection processes in other phytopathogenic fungi. Although a number of upstream components of this important pathway have been characterized, the upstream sensors for surface signals have not been well characterized. Pmk1 is orthologous to Kss1 in yeast that functions downstream from Msb2 and Sho1 for filamentous growth. Because of the conserved nature of the Pmk1 and Kss1 pathways and reduced expression of MoMSB2 in the pmk1 mutant, in this study we functionally characterized the MoMSB2 and MoSHO1 genes. Whereas the Momsb2 mutant was significantly reduced in appressorium formation and virulence, the Mosho1 mutant was only slightly reduced. The Mosho1 Momsb2 double mutant rarely formed appressoria on artificial hydrophobic surfaces, had a reduced Pmk1 phosphorylation level, and was nonresponsive to cutin monomers. However, it still formed appressoria and caused rare, restricted lesions on rice leaves. On artificial hydrophilic surfaces, leaf surface waxes and primary alcohols-but not paraffin waxes and alkanes- stimulated appressorium formation in the Mosho1 Momsb2 mutant, but more efficiently in the Momsb2 mutant. Furthermore, expression of a dominant active MST7 allele partially suppressed the defects of the Momsb2 mutant. These results indicate that, besides surface hydrophobicity and cutin monomers, primary alcohols, a major component of epicuticular leaf waxes in grasses, are recognized by M. oryzae as signals for appressorium formation. Our data also suggest that MoMsb2 and MoSho1 may have overlapping functions in recognizing various surface signals for Pmk1 activation and appressorium formation. While MoMsb2 is critical for sensing surface hydrophobicity and cutin monomers, MoSho1 may play a more important role in recognizing rice leaf waxes.
Author Summary
The rice blast fungus is a major pathogen of rice and a model for studying fungal-plant interactions. Like many other fungal pathogens, it can recognize physical and chemical signals present on the rice leaf surface and form a highly specialized infection structure known as appressorium. A well conserved signal transduction pathway involving the protein kinase gene PMK1 is known to regulate appressorium formation and plant penetration in this pathogen. However, it is not clear about the sensor genes that are involved in recognizing various plant surface signals. In this study we functionally characterize two putative sensor genes called MoMSB2 and MoSHO1. Genetic and biochemical analyses indicated that these two genes have overlapping functions in recognizing different physical and chemical signals present on the rice leaf surface for the activation of the Pmk1 pathway and appressorium formation. We found that primary alcohols, a major component of leaf waxes in grasses, can be recognized by the rice blast fungus as chemical cues. While MoMSB2 is critical for sensing hydrophobicity and precursors of cutin molecules of rice leaves, MoSHO1 appears to be more important than MoMSB2 for recognizing wax components.
Introduction
The heterothallic ascomycete Magnaporthe oryzae is an important pathogen of rice throughout the world. In the past two decades, the rice-M. oryzae pathosystem has been developed as a model to study fungal-plant interactions [1], [2], [3]. M. oryzae initiates infection of rice leaves by the germination of conidia and differentiation of appressoria at the tip of germ tubes. The fungus then uses turgor pressure that develops within appressoria to penetrate the plant cuticle and cell wall. After penetration, the narrow penetration peg differentiates into invasive hyphae, which are enveloped by the host cytoplasmic membrane during the biotrophic phase [4]. As a hemibiotrophic pathogen, M. oryzae does not kill plant cells initially. At late infection stages, plant cells are killed due to extensive growth of infectious hyphae and blast lesions are normally visible within 7 days post-infection.
The surface of rice leaves is comprised of epicuticular waxes. Germ tubes of M. oryzae recognize the hydrophobicity of rice leaves. The fungus also forms appressoria on artificial hydrophobic surfaces. On hydrophilic surfaces, conidia produce long germ tubes without tip differentiation. Exogenous cAMP induces appressorium formation on hydrophilic surfaces. Molecular studies have confirmed the role of cAMP signaling in surface recognition and initiation of appressorium formation [5], [6]. Besides surface hydrophobicity, other factors including surface hardness, cutin monomers, and leaf waxes also affect appressorium formation in M. oryzae [7], [8], [9], [10], [11]. Various physical and chemical signals also have been shown to affect appressorium formation in other plant pathogenic fungi, including Ustilago maydis and Colletotrichum species.
While cAMP signaling controls surface recognition and tip deformation, the Pmk1 MAP kinase pathway regulates late stages of appressorium formation, penetration, and infectious growth in M. oryzae [12]. Pmk1 is orthologous to Kss1, which is a key MAP kinase involved in the filamentous growth pathway in Saccharomyces cerevisiae [13]. A number of genes functioning upstream from Pmk1, including the MEK (Mst7) and MEK kinase (Mst11), an adaptor protein Mst50, and Ras2 have been identified [14], [15]. One of the downstream transcription factors regulated by Pmk1 is Mst12, which is required for appressorial penetration and invasive growth [16]. The Pmk1 pathway is conserved in phytopathogenic fungi for regulating various infection processes [15]. Although key components of the cAMP signaling and Pmk1 pathways have been identified, the mechanisms for recognizing physical and chemical signals of plant surfaces have not been well studied in M. oryzae and other fungal pathogens. One putative receptor gene known to be involved in surface sensing is PTH11 [12], [17], [18]. The pth11 mutant is reduced in virulence and appressorium formation on hydrophobic surfaces, but it forms abundant appressoria in the presence of exogenous cAMP [19]. The M. oryzae genome contains about 60 putative GPCR genes, including several PTH11-like genes with the CEFM domain.
The filamentation MAPK pathway is well studied in yeast [20]. The downstream target of Kss1, Ste12, forms a heterodimer with Tec1 to regulate the expression of genes related to filamentous growth. Various genes, including RAS1, CDC42, SHO1, and MSB2, function upstream from the Kss1-dependent filamentation pathway [21], [22]. MSB2 encodes a surface mucin protein that interacts with Cdc42 for filamentous growth. Msb2 also interacts with Hkr1 and functions upstream from Sho1 for responses to hyperosmotic stresses [22]. SHO1 encodes a membrane sensor protein that is involved in the activation of the Ste11-Ste7-Kss1 pathway and the Hog1 osmoregulation pathway in yeast [21], [23]. During the preparation of this manuscript, msb2 and sho1 were shown to be essential for regulating appressorium formation and tumor development in the corn smut fungus Ustilago maydis [24]. The msb2 sho1 mutant failed to form appressoria on artificial and plant surfaces and was non-pathogenic.
Because of the conserved nature of the Pmk1 and Kss1 MAPK cascades and the importance of MSB2 and SHO1 in yeast filamentous growth, we examined the expression levels of their orthologs in M. oryzae (named MoMSB2 and MoSHO1 in this study). Both of them were reduced over 4-fold in the pmk1 mutant. Deletion of MoSHO1 had only minor effects on appressorium formation, but the Momsb2 deletion mutants rarely formed appressoria on artificial hydrophobic surfaces and failed to respond to cutin monomers. However, they were still pathogenic and recognized leaf surface waxes for appressorium formation. Further analyses indicated that primary alcohols in rice leaf epicuticular waxes induce appressorium formation. Expression of a dominant active allele of MST7 [14] also stimulated appressorium development in the Momsb2 mutant, which had a reduced level of Pmk1 phosphorylation. Overall, these results show that primary alcohols, a major component of epicuticular leaf waxes in grasses, are recognized by M. oryzae as signals for appressorium formation. Other fungal pathogens may also recognize primary alcohols or other wax components. Our data also indicate that MoMsb2 and MoSho1 may function as upstream sensors for the activation of the Pmk1 pathway and appressorium formation. While MoMsb2 is critical for sensing surface hydrophobicity and cutin monomers, MoSho1 may plays a more important role than MoMSB2 in recognizing rice leaf waxes.
Results
The MSB2 and SHO1 orthologs in Magnaporthe oryzae
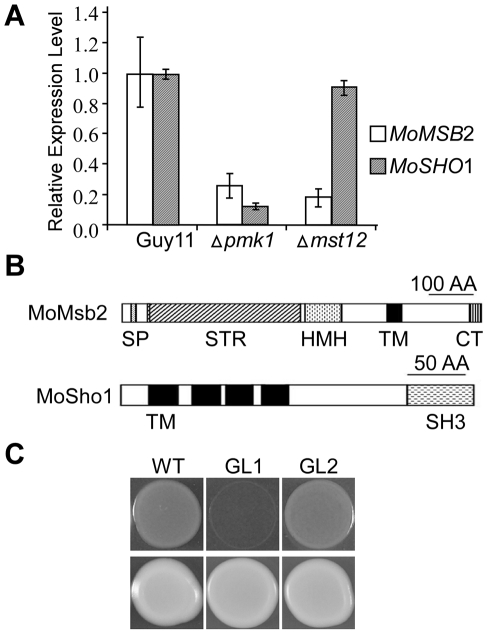
Like other filamentous fungi, M. oryzae has one distinct ortholog each for the yeast MSB2 and SHO1 genes (Fig. S1), which were named MoMSB2 and MoSHO1, respectively. When assayed by qRT-PCR with three independent biological replicates, both MoMSB2 and MoSHO1 transcripts had reduced expression levels (over 4-fold) in the pmk1 mutant but only MoMSB2 expression was significantly reduced in the mst12 mutant (Fig. 1A). The MoMsb2 protein has a signal peptide and one C-terminal transmembrane (TM) domain (Fig. 1B). Therefore, only the C-terminal portion behind the TM domain is inside the cytoplasm membrane. The bulk of the mature MoMsb2 protein is predicted to be extracellular and contains numerous putative glycosylation sites. In yeast, the promoter of MSB2 has two pheromone response elements (PRE) recognized by Ste12 and one TEA/ATTS consensus sequence (TCS) recognized by Tec1 [25]. The promoter region of MoMSB2 has two PRE-like sequences and two putative TCS elements (Fig. S2).
Figure 1. The MSB2 and SHO1 orthologs in M. oryzae.
A. qRT-PCR assay of MoMSB2 and MoSHO1 expression in the wild-type, pmk1, and mst12 mutant strains. The relative expression level of MoMSB2 and MoSHO1 in the mutants was compared to that of the wild-type strain (arbitrarily set to 1). Mean and standard error were calculated with data from three biological replicates. B. Schematic drawing of domains identified in MoMsb2 and MoSho1. SP, signal peptide; STR, serine/threonine rich region; HMH, Hkr1-Msb2 homology domain; TM, transmembrane domain; CT, cytoplasmic tail; SH3, and Src homology 3 domain. C. Colonies of XK1-25 (WT) and sho1 mutant transformed with pYES2 (GL1) or pMoSHO1 (GL2). Photos were taken after incubation for two days on YPGal plates with or without 1.5 M sorbitol.
Like MoMSB2, MoSHO1 is well conserved in filamentous fungi. It encodes a protein with four TM and one SH3 domains (Fig. 1B). Although MoSho1 and Sho1 share only 34% identity in amino acid sequences, expression of MoSHO1 in yeast functionally complemented the defects of the sho1 mutant in growth on medium with 1.5 M sorbitol (Fig. 1C). When MoMSB2 was expressed in yeast, similar to the msb2 gene from U. maydis [24], it failed to complement filamentation defects of the msb2 mutant.
MoMSB2 and MoSHO1 are involved in recognizing artificial hydrophobic surfaces
To determine their functions in M. oryzae, we used the gene replacement approach to delete the MoMSB2 and MoSHO1 genes. One Mosho1 and two Momsb2 mutants (Table 1) were identified and confirmed by Southern blot analysis (Fig. S3). The two Momsb2 mutants had the same phenotype although only data for mutant M6 were presented here. The Mosho1 mutant had no obvious defects in vegetative growth, but the growth rate of the Momsb2 mutant was slightly reduced (Table 2) in comparison with that of Ku80 [26], which was used to generate the mutants.
Table 1. Wild-type and mutant strains of Magnaporthe oryzae used in this study.
| Strain | Genotype description | Reference |
| Guy11 | Wild-type (MAT1-2, avr-Pita) | Chao and Ellingboe, 1991 |
| 70-15 | Wild-type (MAT1-1, AVR-Pita) | Chao and Ellingboe, 1991 |
| Ku80 | MgKu80 deletion mutant of Guy11 | Villalba et al., 2008 |
| nn78 | pmk1 mutant | Xu and Hamer, 1996 |
| mk23 | mst12 mutant | Park et al., 2004 |
| M2 | Momsb2 deletion mutant | This study |
| M6 | Momsb2 deletion mutant | This study |
| S72 | Mosho1 deletion mutant | This study |
| MS88 | Mosho1 MoMsb2 double mutant | This study |
| MS93 | Mosho1 MoMsb2 double mutant | This study |
| CM6 | Complemented strain of M6 (Momsb2/MoMSB2-eGFP) | This study |
| Cs12 | Transformant of 70-15 expressing MoSHO1-eGFP | This study |
| CS15 | Complemented strain of S72 (Mosho1/MoSHO1) | This study |
| CMS74 | Mosho1 MoMsb2/MoSHO1 MoMSB2 complemented strain | This study |
| Ect7 | Ectopic transformant for MoSHO1 deletion | This study |
| Ect12 | Ectopic transformant for MoSHO1 deletion | This study |
| Ect16 | Ectopic transformant for MoMSB2 deletion | This study |
| Ect20 | Ectopic transformant for Mosho1 MoMsb2 deletion | This study |
| DSSM | Transformant of M6 expressing MoMSB2 ΔSP-eGFP | This study |
| WDA2 | Transformant of 70-15 expressing the MST7 DA allele | This study |
| WDA12 | Transformant of M6 expressing the MST7 DA allele | This study |
Table 2. Phenotype characterization of the mutants generated in this study.
| Strain | Growth rate (mm/day)a | Conidiation (×106 spores/plate) | Appressorium formation (%)b | Lesion formationc (lesions/5cm leaf tip) |
| Ku80 | 5.6±0.1 A* | 23.3±5.4 A | 99.4±0.5 A | 24.8±14.7 A |
| M6 | 5.0±0.1 B | 22.9±5.5 A | 1.5±0.5 D | 2.1±1.1 C |
| S72 | 5.2±0.1 B | 4.1±0.8 C | 72.8±5.2 B | 9.2±5.3 B |
| MS88 | 5.0±0.1 B | 2.8±0.5 C | 0.9±0.4 D | 0.9±0.8 C |
| Ect20 | 5.7±0.3 A | 4.0±0.4 C | 98.5±0.4 A | N/Ad |
| CM6 | 5.5±0.2 A | 21.5±2.0 A | 96.0±2.6 A | N/A |
| CS15 | 5.6±0.2 A | 11.3±2.5 B | 96.7±2.1 A | N/A |
| CMS74 | 5.6±0.2 A | 11.0±3.0 B | 95.3±1.5 A | N/A |
Growth rate (extension in colony diameter) and conidiation were assayed with OA cultures.
Percentage of germ tubes formed appressoria on the hydrophobic side of Gelbond membranes by 24 h.
Lesion formation was examined on infected rice leaves 7 days after inoculation. Means and SD values were calculated from at least three independent experiments.
Not assayed.
*Data from three replicates were analyzed with the protected Fisher's Least Significant Difference (LSD) test. The same letter indicated that there was no significant difference. Different letters were used to mark statistically significant difference (P = 0.05).
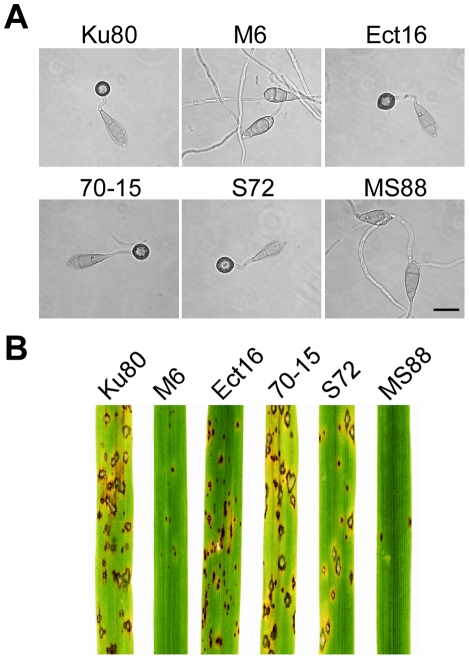
On hydrophobic surfaces, both the Momsb2 and Mosho1 mutants had no defects in conidium germination. However, the Momsb2 mutant was significantly reduced in appressorium formation. Less than 2% of its germ tubes formed melanized appressoria by 24 h (Fig. 2A). Under the same conditions, over 90% and 70% of the germ tubes formed appressoria in Ku80 and the Mosho1 mutant, respectively (Table 2), indicating that the Mosho1 mutant was only slightly reduced in appressorium formation.
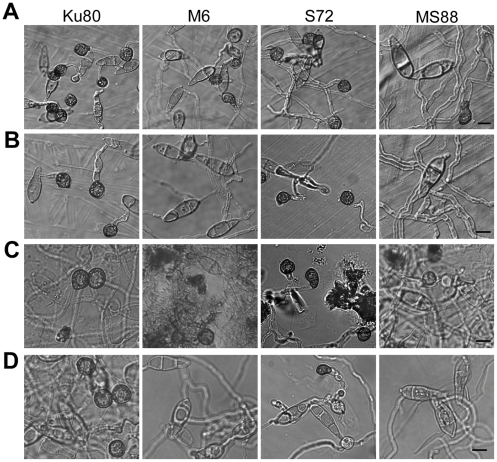
Figure 2. Appressorium formation and plant infection assays.
A. Conidia from wild-type strains 70-15 and Ku80, Momsb2 mutant M6, Mosho1 mutant S72, Mosho1 Momsb1 mutant MS88, and an ectopic transformant Ect16 were incubated on hydrophobic surfaces for 24 h. Bar = 20 µm. B. Rice leaves sprayed with conidia from the same set of strains. Typical leaves were photographed 7 dpi.
Because of the functions of Sho1 and Msb2 in yeast filamentous growth [22] and reduced appressorium formation in both Mosho1 and Momsb2 mutants, we deleted the MoMSB2 gene in the Mosho1 mutant. Transformants MS88 and MS93 (Table 1) were two Mosho1 Momsb2 mutants confirmed by Southern analysis (Fig. S3). On the hydrophobic surfaces, the double mutant produced long, curved germ tubes (Fig. 2A) that rarely (<1%) differentiated into appressoria (Table 2). These results indicate that MoMSB2 plays a critical role but MoSHO1 also plays a minor role in the recognition of surface hydrophobicity.
The Momsb2 and Momsb2 Mosho1 mutants are reduced in virulence
In infection assays with rice seedlings, the Mosho1 mutant was only slightly reduced in virulence (Fig. 2B, Table 2). In contrast, the Momsb2 and Mosho1 Momsb2 mutants were significantly reduced in virulence. Only rare lesions were observed on leaves inoculated with the double mutant (Table 2). The Momsb2 mutant caused a few more lesions than the double mutant MS88 but still much less than Ku80 and the Mosho1 mutant (Table 2). Lesions caused by the Momsb2 and Mosho1 Momsb2 mutants on rice leaves tended to be smaller than those caused by Ku80 (Fig. 2B) and have limited necrotic borders (Fig. S4).
Similar results were obtained in barley infection assays. Ku80 and the Mosho1 mutant caused numerous lesions on inoculated leaves. Under the same conditions, only a few lesions were formed on barley leaves infected with the Momsb2 and Momsb2 Mosho1 mutants, indicating a significant reduction in virulence. Results from these infection assays indicate that MoMSB2 plays a more critical role than MoSHO1 in pathogenesis. However, MoSHO1 also is required for full virulence because the Mosho1 mutant had reduced virulence (Table 2) and the Momsb2 mutant appeared to be more virulent than the Momsb2 Mosho1 double mutant.
For complementation assays, we re-introduced the wild-type MoSHO1 and MoMSB2 alleles to the mutants. Transformants CM6 (Momsb2/MoMSB2), CS15 (Mosho1/MoSHO1), and CMS74 (Mosho1 Momsb2/MoSHO1 MoMSB2) were normal in virulence (Fig. S5), vegetative growth, and appressorium formation (Table 2), indicating that reintroduction of the wild-type MoSHO1 and MoMSB2 genes complemented the defects of corresponding mutants.
Deletion of MoMSB2 does not affect appressorium formation on plant leaves
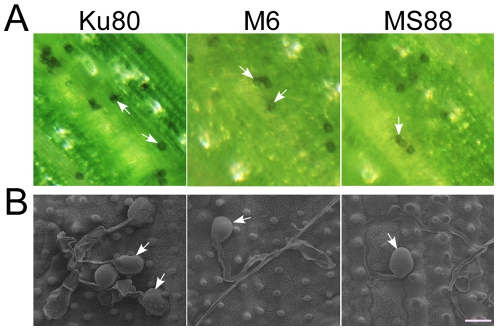
Because the double mutant rarely formed appressoria on artificial surfaces but still caused blast lesions, we assayed its ability to form appressoria on rice leaves. At 24 h, the Momsb2 and Momsb2 Mosho1 mutants produced abundant melanized appressoria (Fig. 3A). When examined by scanning electron microscopy (SEM), 68.7±7.3% and 57.7±8.4% of the Momsb2 and Mosho1 Momsb2 germ tubes, respectively, formed appressoria (Fig. 3B). Similar results were obtained in appressorium formation assays with barley leaves (Fig. S6), indicating that the Momsb2 and Mosho1 Momsb2 mutants still respond to chemical cues present on rice and barley leaf surfaces for appressorium formation.
Figure 3. Appressorium formation assays with intact rice leaves.
A. Rice leaves inoculated with conidia from strains Ku80, M6 (Momsb2), and MS88 (Mosho1 Momsb2). Melanized appressoria were observed 24 hpi. B. Appressoria formed by the same set of strains on rice leaf surface examined under SEM. The mutants produced appressoria at the tip of long germ tubes. Bar = 10 µm. Arrows marked appressoria.
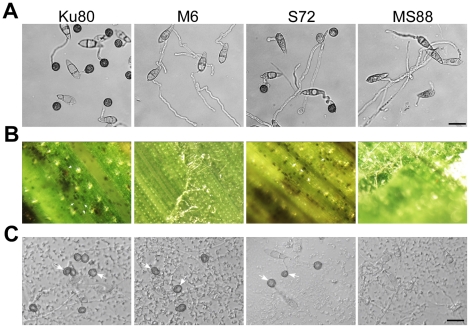
Cutin monomers fail to induce appressorium formation in the Momsb2 and Momsb2 Mosho1 mutants
Because cutin monomers, one type of plant surface molecules, are known to trigger appressorium formation in M. oryzae [27], we assayed the effects of two cutin monomers on mutants M6, S72, and MS88. In the presence of 10 µM 1, 16-hexadecanediol, over 95% of the wild-type and Mosho1 mutant germ tubes formed appressoria (Fig. 4A). Under the same conditions, appressorium formation was not induced in the Momsb2 and Mosho1 Momsb2 mutants (Fig. 4A). Similar results were obtained with 10 µM cis-9-octadecen-1-ol. Therefore, MoMSB2 is required for the recognition of these two cutin monomers in M. oryzae. Appressorium formation by the Momsb2 and Momsb2 Mosho1 mutants on rice or barley leaves is likely induced by plant surface molecules other than cutin monomers that were recognized by a MoMsb2-independent mechanism.
Figure 4. Assays for the effects of different treatments on appressorium formation.
Conidia from strains Ku80, M6 (Momsb2), S72 (Mosho1), and MS88 (Mosho1 Momsb2) mutants were incubated on: A) the hydrophilic surface in the presence of 10 µM 1,16-hexadecanediol; B) de-waxed leaves, and C) glass surface coated with the rice leaf wax extract. Representative germlings were photographed after 24 h incubation. Arrows marked appressoria. Bar = 20 µm.
Leaf surface waxes as chemical cues for appressorium formation
On rice leaves, germ tubes are in close contact with the epicuticular wax layer, which may play a role in surface recognition. To test this hypothesis, epicuticular waxes were removed by dipping rice leaves in hexane for 5 seconds. On de-waxed leaves, Ku80 and the Mosho1 mutant still efficiently formed melanized appressoria (Fig. 4B). However, the efficiency of appressorium formation by the Momsb2 and Mosho1 Momsb2 mutants was significantly reduced (Table S2). The vast majority of mutant germ tubes failed to form appressoria on de-waxed rice leaves (Fig. 4B). Similar results were obtained in appressorium formation assays with intact and de-waxed barley leaves (Fig. S6).
To further prove the role of surface waxes in stimulating appressorium formation in Momsb2 mutants, crude wax extracts were prepared from rice leaves and used to coat microscope glass slides. In Ku80 and the Mosho1 or Momsb2 mutant, about 90% of the germ tubes formed appressoria by 24 h in the presence of the rice leaf wax extract (Fig. 4C). Under the same conditions, less than 32% of the germ tubes formed appressoria in the Mosho1 Momsb2 double mutant (Fig. 4C), indicating that both MoSHO1 and MoMSB2 are important for appressorium formation on hydrophilic surfaces coated with leaf waxes.
Primary alcohols induce appressorium formation in M. oryzae
To test whether any rice leaf-specific wax compound is responsible for inducing appressorium formation, we also used bee and paraffin waxes to coat the glass surface. Similar to the rice leaf wax extract, bee wax induced appressorium formation more efficiently in Ku80 and the Mosho1 or Momsb2 mutant than in the Mosho1 Momsb2 double mutants (Fig. 5A). However, paraffin wax could only induce appressorium formation in Ku80 and the Mosho1 mutant (Table 3; Fig. 5B). In repeated experiments, only bee waxes but not paraffin waxes had stimulatory effects on appressorium formation in the MoMsb2 and Mosho1 MoMsb2 mutants. Because coating with paraffin wax changed the surface hydrophobicity (Fig. S7), these results were consistent with the defects of the Momsb2 and Mosho1 Momsb2 mutants in recognizing artificial hydrophobic surface. Paraffin wax must lack the components of rice leaf or bee waxes that are recognized by M. oryzae as chemical signals for appressorium formation.
Figure 5. Appressorium formation induced by different waxes or wax components.
Conidia from strains Ku80 (WT), M6 (Momsb2), Mosho1 (S72), and MS88 (Momsb2 Mosho1) were place over microscope glass slides coated with bee waxes (A), paraffin waxes (B), C28 primary alcohol 1-Octacosanol (C), and C31 alkane hentricacontane (D). Bar = 10 µm.
Table 3. Appressorium formation on glass slides coated with different waxes.
| Strain | Leaf waxesa | Parafin waxes | Bee waxes | Primary alcoholc | Alkane |
| Ku80 | 92.3±2.1 | 91.1±2.3 | 97.5±0.9 | 87.1±1.8 | 85.3±1.2 |
| M6 | 88.5±1.2 | NAb | 74.2±1.9 | 70.3±0.7 | NA |
| S72 | 90.7±0.9 | 87.4±1.9 | 96.3±1.3 | 81.4±1.1 | 50.3±0.9 |
| MS88 | 31.1±1.2 | NA | 49.2±0.7 | 41.1±2.1 | NA |
Percentage of germ tubes formed appressoria. Mean and standard error were calculated from three independent replicates.
No appressoria were observed.
The primary alcohol and alkane used in this study were 1-triacontanol (C30) and hentricacontane (C31), respectively.
Plant surface waxes contain primary and secondary alcohols, aldehydes, ketones, alkanes, esters, and long-chain fatty acids [28]. In contrast, paraffin wax mainly consists of long chain alkanes. Because primary alcohols are the major components of surface waxes in grasses, we tested the effects of primary alcohols and alkanes on appressorium formation in M. oryzae. On hydrophilic surfaces coated with 1-octacosanol (C28) and 1-triacontanol (C30), the Mosho1 Momsb2 mutant formed appressoria but less efficiently than Ku80 and the Mosho1 or Momsb2 mutant (Fig. 5C). In contrast, the C29 and C31 alkanes (nonacosane and hentricacontane alkanes) induced appressorium formation in the wild-type strain but not in the mutants (Table 3; Fig. 5D). These results indicate that primary alcohols of epicuticular waxes are among the chemical cues recognized by M. oryzae.
MoMSB2 is important for appressorium penetration
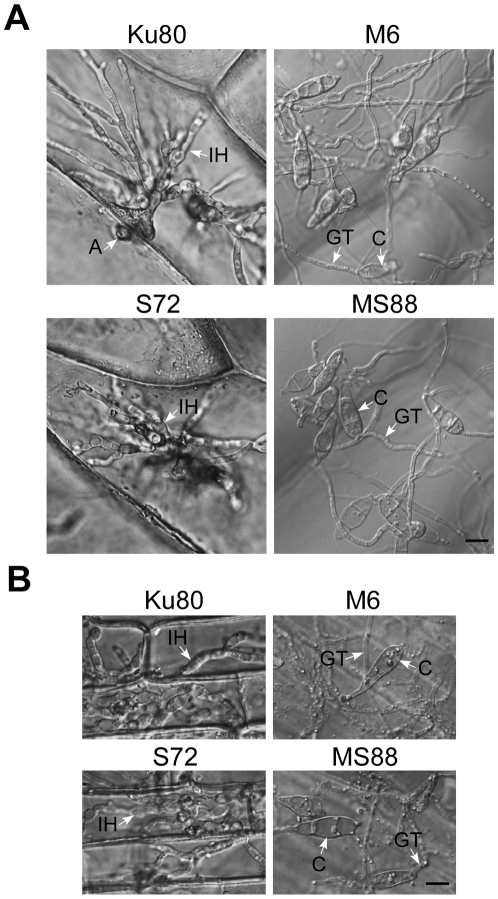
In penetration assays with onion epidermises (Fig. 6A), appressoria formed by Ku80 and the Mosho1 mutant produced invasive hyphae inside plant cells by 48 hpi. Under the same conditions, most (over 99%) conidia from the Momsb2 and Mosho1 Momsb2 mutants produced long germ tubes without tip differentiation. Rare appressoria formed by these two mutants failed to penetrate onion epidermal cells. Similar results were obtained in penetration assays with rice leaf sheaths (Fig. 6B). Because the inner surface of rice leaf sheaths lacks epicuticular waxes [29], efficient formation of appressoria by the Momsb2 and Mosho1 Momsb2 mutants on rice leaves but not on leaf sheaths further shows that epicuticular waxes are recognized by M. oryzae as one of the surface signals. While appressorium formation was not observed in the double mutant, rare appressoria formed by the Momsb2 mutant failed to penetrate rice leaf sheath cells, indicating that MoMSB2 is important for appressorium penetration, which may explain why the Momsb2 mutants formed abundant melanized appressoria but rarely caused lesions on rice leaves.
Figure 6. Appressorium penetration assays with onion and rice epidermal cells.
A. Onion epidermal cells inoculated with Ku80, M6 (Momsb2), S72 (Mosho1), and MS88 (Momsb2 Mosho1) were examined 48 hpi. B. Epidermal cells of rice leaf sheaths were inoculated with the same set of strains and examined 48 hpi. A, appressorium; C, conidium; GT, germ tube; IH, infectious hypha. Bar = 10 µm.
MoMsb2 functions upstream from the Pmk1 pathway
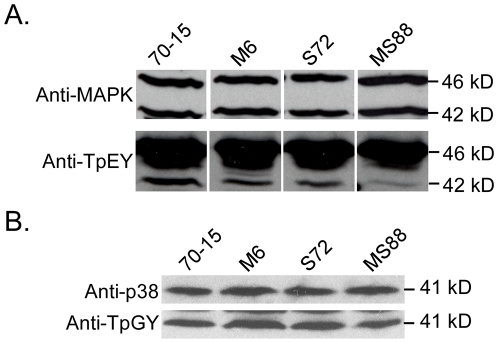
Although Pmk1 expression was not affected, the activation of Pmk1 as detected with an anti-TpEY antibody was reduced in the Momsb2 and Mosho1 Momsb2 mutants (Fig. 7A). In the same western blot analysis, the expression and phosphorylation of Mps1 [30] was normal in these three mutants (Fig. 7A). Osmoregulation is mediated by Osm1, the third MAPK in M. oryzae [30]. When detected with an anti-TpGY antibody, the wild type and mutants had similar levels of Osm1 phosphorylation (Fig. 7B), suggesting that MoMSB2 and MoSHO1 play only a minor role, if any, in the osmoregulation pathway. Overall, Pmk1 was the only MAPK with a reduced phosphorylation level in Momsb2 deletion mutants, indicating that MoMSB2 may function upstream from the Pmk1 MAP kinase.
Figure 7. Assays for MAP kinase phosphorylation.
Western blots were conducted with proteins isolated from vegetative hyphae of the wild-type (70-15) and the Momsb2 (M6), Mosho1 (S72), and Mosho1 Momsb2 (MS88) mutant strains. A. The anti-MAPK and anti-TpEY antibodies detected the expression and phosphorylation levels of Pmk1 (42-kD) and Mps1 (46-kD), respectively. B. The 41-kD Osm1 band was detected with both an anti-P38 MAPK antibody and the anti-TpGY antibody.
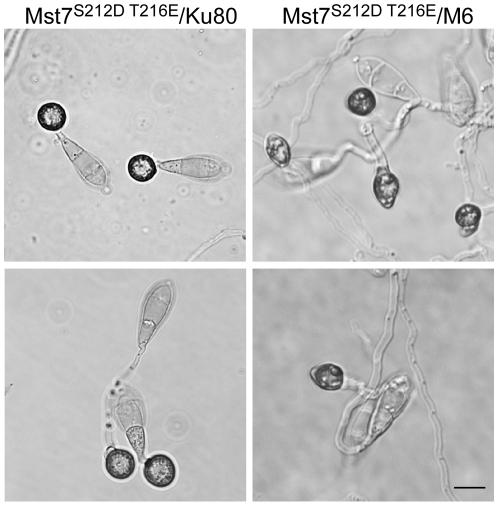
The MST7 gene encodes a MEK that activates Pmk1. Expressing a dominant active allele of MST7 induces appressorium formation on hydrophilic surfaces [14]. We introduced this MST7 DA allele into the Momsb2 mutant. In the resulting transformants WDA2 and WDA12 (Table 1), appressorium formation was observed on hydrophilic and hydrophobic surfaces (Fig. 8), indicating that expression of the MST7 DA allele induced appressorium formation in the Momsb2 mutant. Therefore, MoMsb2 may function upstream from the Pmk1 MAPK cascade for appressorium formation.
Figure 8. Effects of expressing the dominant active allele of MST7 on appressorium formation.
Conidia from transformants of the wild-type strain 70-15 (WDA2) and the Momsb2 mutant M6 (WDA12) expressing the MST7 DA allele were incubated on hydrophobic (upper) or hydrophilic (lower) surface of Gelbond membranes for 24 h. Bar = 10 µm.
Expression and localization of MoMsb2
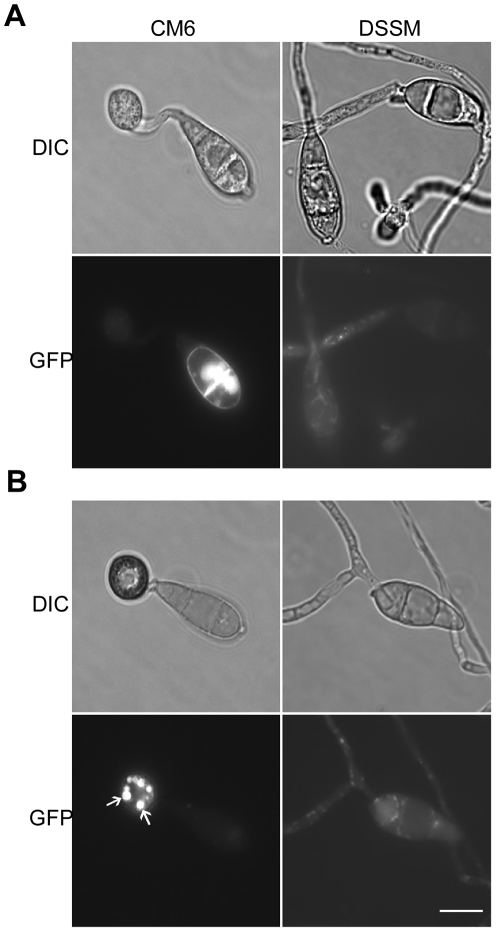
In transformant CM6 expressing the MoMSB2-eGFP fusion construct, GFP signals were detected mainly in vacuole-like structures but also on the cytoplasmic membrane in vegetative hyphae and conidia (Fig. 9; Fig. S8). During conidium germination and appressorium formation, fluorescent signals were detected on the cytoplasmic membrane and in vacuoles that were visible under DIC microscopy (Fig. 9A). Interestingly, germ tubes and young appressoria had no or very weak fluorescent signals. In mature appressoria (24 h), GFP signals mainly localized to vacuole-like structures (Fig. 9B).
Figure 9. Expression and subcellular localization of MoMsb2-eGFP.
A. Conidia, germ tubes, and young appressoria of the MoMSB2-eGFP (CM6) and MoMSB2 ΔSP-eGFP (DSSM) transformants were examined by DIC or epifluoresence microscopy. B. In mature appressoria (24 h) of transformant CM6, GFP signals localized to small vacuole-like structures (marked with arrows). In transformant DSSM, appressorium formation was not observed and GFP signals localized in the cytoplasm. Bar = 10 µm. The same fields were examined under DIC (left) and epifluoresence microscopy (right).
The bulk of the mature MoMsb2 protein is predicted to be extracellular. To test the importance of the signal peptide, we generated the MoMSB2 ΔSP-eGFP allele and introduced it into mutant M6. Transformant DSSM (Table 1) expressing this mutant allele, similar to the original Momsb2 mutant, was defective in appressorium formation (Fig. 9) and plant infection. Weak GFP signals were detected in vegetative hyphae, conidia, and germ tubes. However, the subcellular localization pattern of GFP signals in transformant DSSM differed from that of transformant CM6. In transformant DSSM, GFP signals mainly localize to the cytoplasm (Fig. 9B). Therefore, the signal peptide is essential for the localization and function of MoMsb2.
Functional characterization of different domains of MoMsb2
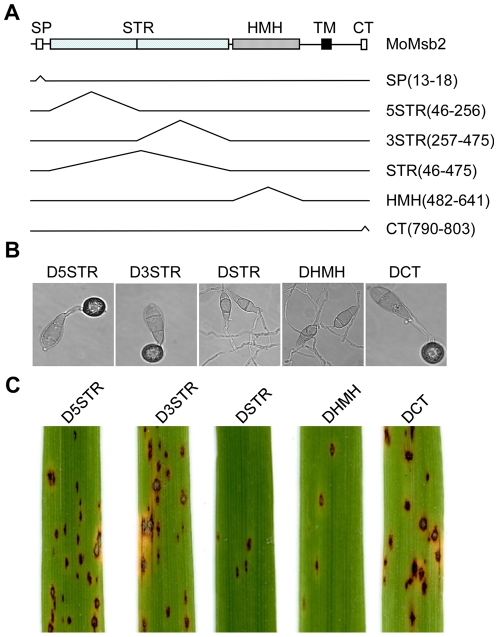
In addition to the signal peptide, MoMsb2 has a STR region, a HMH domain, and a CT domain. We generated mutant alleles of MoMSB2-GFP deleted of different regions (Fig. 10A) and transformed them into the Momsb2 mutant M6. The resulting transformants (Table 1) were confirmed by PCR analysis. Transformants expressing the MoMSB2 ΔHMH- and MoMSB2 ΔSTR-eGFP alleles were, similar to the original Momsb2 mutant, defective in appressorium formation (Fig. 10B) and plant infection (Fig. 10C), indicating that the STR and HMH domain are essential for MoMsb2 function. In contrast, the CT domain was dispensable for appressorium formation (Fig. 10B) and virulence (Fig. 10C). Transformants expressing the MoMSB2 Δ5STR-, and MoMSB2 Δ3STR-eGFP alleles were only slightly reduced in appressorium formation and plant infection (Fig. 10B and 10C). Therefore, the N-terminal and C-terminal regions of the Ser- and Thr-rich mucin domain have redundant functions in MoMsb2.
Figure 10. Functional characterization of different domains of the MoMSB2 gene.
A. Schematic drawing of different mutant alleles of MoMSB2. The numbers in the bracket indicate the amino acid residues deleted in each allele. B. Appressorium formation assays with transformants D5STR, D3STR, DSTR, DHMH, and DCT that expressed the MoMSB2 Δ5STR-, MoMSB2 Δ3STR-, MoMSB2 ΔSTR-, MoMSB2 ΔHMH-, and MoMSB2 ΔCT-eGFP alleles. Bar = 10 µm. C. Rice leaves inoculated with conidia from the same set of strains. Like the original Momsb2 mutant, transformants DSTR and DHMH were significantly reduced in appressorium formation on hydrophobic surfaces and virulence.
Discussion
Mucin proteins are characterized by the Ser- and Thr-rich mucin domain and divided into secreted and cell surface (signaling) mucins [31]. Muc1 is the most extensively studied cell surface mucin that affects various cellular functions in mammalian cells, including MAPK signaling. In yeast, MSB2 was first isolated as a multiple copy suppressor gene of a temperature sensitive allele of CDC24. Although it is dispensable for yeast growth under normal conditions, the Msb2 surface mucin interacts with Cdc42 and Sho1 for regulating filamentous growth [22], [32]. One other mucin gene in S. cerevisiae is HKR1 that is not related to the filamentation pathway [33]. In M. oryzae, MoMsb2 has structural components conserved in cell surface mucins (Fig. 1). The other mucin-like protein in M. oryzae is the chitin-binding protein Cbp1 [34], which shares limited homology (15% identity) with Hkr1 but lacks the mucin and TM domains. Therefore, MoMSB2 likely is the only cell surface mucin gene in M. oryzae.
In M. oryzae, MoMSB2 appears to play a minor role in regulating vegetative growth. The Momsb2 mutant was slightly reduced in the growth rate but it was normal in conidiation (Table 2). In contract, the Mosho1 and Mosho1 Momsb2 mutants were reduced over 5-fold in conidiation (Table 2), suggesting that MoSHO1 is involved in the regulation of conidium production in M. oryzae. However, transformants of Mosho1 mutants expressing the wild-type allele of MoSHO1 were increased in conidiation about 3-fold but they still produced fewer conidia than Ku80 (Table 2). These results indicate that ectopic integration of MoSHO1 did not fully complement the defect of Mosho1 mutant in conidiation. Nevertheless, the Mosho1/MoSHO1 complemented strain was complemented in the growth rate and appressorium formation (Table 2). Therefore, it is likely that the Mosho1 mutant was reduced in conidiation due to deletion of MoSHO1 together with an un-related event during transformation.
Surface hydrophobicity and cutin monomers are two well known surface signals recognized by M. oryzae [30]. The Momsb2 mutant was significantly reduced in appressorium formation on hydrophobic surfaces (Table 2). It also failed to respond to two cutin monomers. These results indicate that MoMsb2 is involved in sensing surface hydrophobicity and cutin monomers. Interestingly, the msb2 and sho1 genes were recently shown to be essential for appressorium formation on artificial hydrophobic and plant surfaces in U. maydis [24]. However, appressoria formed by U. maydis lack the distinct morphological features of M. oryzae appressoria. It may be difficult to observe rare appressoria formed by the sho1 msb2 mutant. The Momsb2 mutant still formed appressoria efficiently on intact rice or barley leaves, suggesting that MoMSB2 is not essential for recognizing leaf surface waxes. Therefore, surface hydrophobicity, cutin monomers, and waxes are sensed by different mechanisms in M. oryzae. In comparison with the single mutants, the Mosho1 Momsb2 double mutant had more severe defects in appressorium formation and virulence. The MoMSB2 and MoSHO1 genes must have overlapping functions in appressorium development and plant infection. In yeast, the sho1 msb2 mutant displays more severe defects in filamentous growth than the msb2 mutant [22]. In M. oryzae, MoSho1 may play a role in the recognition of rice leaf waxes.
On glass surfaces coated with bee or rice leaf waxes, 90% of Momsb2 germ tubes differentiated appressoria, but less than 32% of the germ tubes formed appressoria in the Mosho1 Momsb2 mutant. We also observed that primary alcohols were more efficient in inducing appressorium formation in the Momsb2 mutant than in the Mosho1 Momsb2 double mutant. These results further prove that MoMSB2 is not important for appressorium formation induced by leaf waxes. The difference between the Momsb2 and Mosho1 Momsb2 mutants in the efficiency of wax-induced appressorium formation suggests that MoSHO1 plays a more important role than MoMSB2 in recognizing surface waxes as chemical signals for appressorium formation. Coating with waxes changed the surface hydrophobicity, which may be recognized by MoMsb2 and resulted in induced appressorium formation in the Mosho1 mutant. However, the Mosho1 Momsb2 mutant still formed a few appressoria on plant leaves or wax-coated glass slides. Additional sensor genes must exist in M. oryzae for recognizing wax components. Different genes may be responsible for responding to specific physical or chemical signals in the rice blast fungus.
Bee and rice leaf waxes, but not paraffin wax, induced appressorium formation in the Momsb2 deletion mutants. Because coating with paraffin wax changed the surface hydrophobicity, these results indicate that physical signals related to hydrophobicity are not sufficient to trigger appressorium development in mutants deleted of MoMSB2. Some components of bee and rice leaf waxes must be recognized as chemical signals by the Momsb2 deletion mutants. Unlike rice leaf waxes comprised of alcohols, aldehydes, ketones, alkanes, and esters [28], paraffin wax mainly consists of long chain alkanes. In further experiments, two C29 and C31 alkanes, nonacosane and hentricacontane, failed to induce appressorium formation in mutant M6 or MS88. In contrast, these mutants formed melanized appressoria on hydrophilic surfaces coated with two primary alcohols 1-octacosanol (C28) and 1-triacontanol (C30). Therefore, primary alcohols but not alkanes in leaf waxes may be responsible for inducing appressorium formation in the Momsb2 and Mosho1 Momsb2 mutants. One major component of leaf epicuticular waxes in grass species is primary alcohols [35]. Other plant pathogenic fungi may also recognize primary alcohols for regulating infection-related morphogenesis. Because grapes (one of the native environments for the budding yeast) also are covered with waxes, it is possible that waxes play a role in inducing filamentous growth in S. cerevisiae.
Deletion of MoMSB2 resulted in defects in appressorium formation on artificial surfaces and plant penetration, two processes regulated by Pmk1 [36]. Although its expression was not affected, the phosphorylation of Pmk1 was reduced in the MoMsb2 and Mosho1 Momsb2 mutants. To further prove that MoMsb2 and MoSho1 function upstream from the Pmk1 cascade, the dominant active allele of MST7 [14] was transformed into the Momsb2 mutant. The resulting transformants formed appressoria on hydrophilic surfaces (Fig. 8). Overall, these results further confirm that many components of the yeast filamentation MAPK pathway are involved in the regulation of infection-related morphogenesis in M. oryzae. In U. maydis, Msb2 and Sho1 also function upstream a MAPK pathway and are important for plant infection [24]. In nature, filamentation may be important for the budding yeast to colonize the substrates, such as grapes.
Domain deletion analysis with MoMSB2 indicates that the signal peptide is essential for its function. Interestingly, the cbp1 and Momsb2 mutants had similar defects in appressorium formation although the chitin-binding protein Cbp1 protein lacks the transmembrane domain. Both MoMsb2 and Cbp1 are predicted to be heavily glycosylated according to analyses with the (http://cbs.dtu.dk/services/NetNGlyc/) and (http://ogpet.utep.edu/OGPET/). It will be important to generate and characterize the Momsb2 cbp1 double mutant and determine the relationship between these two genes. MoMsb2 and Cbp1 may be functionally related by forming a surface complex for recognizing different extracellular signals to activate the Pmk1 pathway.
Based on GFP signals observed in transformant CM6, MoMSB2 is constitutively expressed. The MoMsb2-eGFP protein may localize to the cytoplasmic membrane in its inactive form. Localization to the vacuoles or vacuole-like structures may be related to the internalization of the fusion proteins. In yeast, the cleavage of Msb2 at the cleavage domain (located upstream from the TM domain) is essential for its activity [37]. MoMsb2 has the sequence element adjacent to the TM domain that is similar to the Msb2 cleavage domain. The activation of MoMsb2 may involve protein cleavage at this site and result in its dissociation from the cytoplasmic membrane and diffusion into cell wall and extracellular space, which may explain the absence of GFP signals in germ tubes and young appressoria of transformant CM6. It is possible that only the cleaved MoMsb2 (the active form) interacts with other extracellular proteins (such as Cbp1) to sense various physical and chemical signals for appressorium formation. Therefore, characterizing the role of protein cleavage in the activation and localization of MoMsb2 may provide critical information about surface recognition mechanisms in M. oryzae.
PTH11 encodes a putative GPCR that is involved in recognifzing surface hydrophobicity in M. oryzae [19]. In the pth11 mutant, about 10–15% of the germ tubes still form appressoria on hydrophobic surfaces [19], which is approximately five times higher than that of the Momsb2 mutant. In addition, the pth11 mutant, unlike the Momsb2 mutant, still responds to cutin monomers for appressorium formation. PTH11 likely functions upstream from cAMP signaling [19] but its role in the activation of the PMK1 MAPK pathway has not been studied. PTH11 and MoMSB2 may be functionally related in recognizing different chemical and physical signals present on the rice leaf surface. It will be important to determine their relationship in the activation of the cAMP-PKA and PMK1 MAPK pathways for regulating appressorium formation and penetration.
Materials and Methods
Strains and culture conditions
All the wild-type and mutant strains of M. oryzae (Table 1) were cultured on oatmeal agar plates (OA) at 25°C. Culture preservation, genetic crosses, transformation, and measurements of conidiation and growth rate were performed as described [38], [39], [40]. For nucleic acid and protein isolation, vegetative hyphae were harvested from two-day-old liquid CM cultures [41]. Lesions formed on 5-cm rice leaf tip segments were counted as described [42], [43].
Deletion of MoSHO1 and MoMSB2
Approximately 0.8-kb upstream and downstream flanking sequences of MoMSB2 were amplified with primers 11F/12R and 13F/14R (Table S1) and ligated with the hph cassette. The MSB2 gene replacement construct was amplified with primers 11F and 14R and transformed into Ku80 [26]. To delete the MoSHO1 gene, a DNA fragment containing the entire MoSHO1 and its 710-bp upstream and 329-bp downstream flanking sequences were amplified with primers 1F and 2R, and cloned into pGEM-T easy vector (Promega, Madison, WI). The MoSHO1 gene replacement construct pLL9 (Fig. S2) was generated by replacing a 355-bp BamHI/BsiWI fragment of MoSHO1 with the hph cassette amplified from pCB1003 with primers HPH5F and HPH4R.
To generate the Mosho1 MoMsb2 double mutant, the MoMSB2 replacement construct was generated with a bleomycin-resistant cassette and transformed into the Mosho1 deletion mutant S72. Transformants resistant to both hygromycin and bleomycin were screened by PCR. For complementation assays, the MoMSB2 gene was cloned into pYP1 that was generated by replacing the hph gene in pDL1 [44] with the bleomycin-resistance gene. The resulting construct pXY130 was transformed into the Momsb2 mutant M6. The MoSHO1 complementation vector pKNSHO1 was transformed into mutant S72 or co-transformed with pXY130 into the double mutant MS88.
Complementation of yeast sho1 and msb2 mutants
The MoSHO1 ORF was amplified with primer Sho1F/Sho1R and cloned into pYES2 as pMoSHO1. The same procedure was used to generate plasmid pMoMSB2. The resulting constructs were transformed into the sho1 mutant obtained from Open Biosystems (Huntsville, AL) and the msb2 mutant [45] with the alkali-cation yeast transformation kit (MP Biomedicals, Solon, OH). Ura3+ transformants were assayed for invasive growth and sensitivity on YPD and YPGal plates with 1.5 M sorbitol as described [32], [46].
Appressorium formation and penetration assays
Conidia were harvested from 10-day-old OA cultures, resuspended to 5×104 conidia/ml in sterile water, and used for appressorium formation assays with glass cover slips (Fisher Scientific, Pittsburgh, PA) or Gelbond membranes (FMC, Philadelphia, PA) as described [40], [47]. Penetration assays were conducted with onion epidermal cells and rice leaf sheaths [4], [48].
Infection assays with detached barley leaves
Conidia were harvested from 10-day-old OA cultures and resuspended to 5×105 conidia/ml in 0.25% gelatin. Two-week-old seedlings of rice cultivars Nipponbare and CO-39 were used for spray or injection infection assays as described [49]. Eight-day-old seedlings of barley cultivar Golden Promise were used for spray or drop inoculation assays as described [50]. Lesion formation was examined 5–7 days post-inoculation (dpi). For assaying changes in virulence, lesions formed on 5-cm leaf segments were counted as described [38], [51].
Wax extraction and treatments
Rice leaves of two-week-old seedlings were dipped in hexane for 20 s. Epicuticular waxes dissolved in hexane were recovered by evaporation under a nitrogen stream [52]. For coating glass surface, 10 mg of the rice leaf surface wax was dissolved in 3 ml chloroform. Aliquot of 50 µL were dropped onto microscope glass slides (Gold Seal, Portsmouth, NH). Bee wax (Stakich Inc., Bloomfield Hills, MI) and paraffin wax (Oak Sales, San Diego, CA) were directly applied on to glass surface. Drops of 25 µl conidium suspensions were placed on wax-coated areas and assayed for appressorium formation. The C28 (1-octacosanol, C28H58O) and C30 (1-triacontanol, C30H62O) primary alcohols and C29 (nonacosane, C29H60) and C31 (hentricacontane, C31H64) alkanes (Sigma) were dissolved to 4 mg/ml in chloroform.
Scanning electron microscopy (SEM) examination
Barley and rice leaves inoculated with the wild-type and mutant conidia were sampled at 24 hpi and immediately frozen in liquid nitrogen. After being sputter-coated with gold in the presence of argon in a Hexland CT-1000 cryo-system (Gatan, Pleasanton, CA), leaf samples were examined at −140°C in a JEOL JSM-840 scanning electron microscope for germ tube growth and appressorium formation [49].
Construction of MoMSB2-eGFP and MoSHO1-eGFP
A 4.2-kb fragment of the MoMSB2 gene was amplified with primers MGFPF and MGFPR (Table S1) and co-transformed with XhoI-digested pYP1 into XK1-25 [42]. For MoSHO1, a 2.8-kb fragment was amplified with primers SGFPF and SGFPR and co-transformed into XK1-25 with XhoI-digested pDL2 [44]. Plasmids pXY130 and pXY122 containing the MoMSB2-eGFP and MoSHO1-eGFP fusion constructs, respectively, were recovered from Trp+ yeast transformants and transformed into protoplasts of 70-15. For complementation assays, the MoMSB2-eGFP and MoSHO1-eGFP constructs were transformed into the Momsb2 mutant M6 and the Mosho1 mutant S72, respectively. Zeocin-resistant transformants were examined for GFP signals in conidia, appressoria, and infectious hyphae as described [42], [53].
Domain deletion analyses of MoMSB2
To delete the signal peptide, PCR products amplified with primers MGFPF/ΔSSR1 and ΔSSF2/MGFPR were cotransformed with XhoI-digested pYP1 into S. cerevisiae strain XK1-25. Plasmid pXY148 was recovered from yeast Trp+ transformants and confirmed by sequencing analysis to contain the MoMSB2 ΔSP construct. The same yeast gap repair approach [44] was used to generate the MoMSB2 ΔHMH, MoMSB2 ΔCT, MoMSB2 Δ5STR, MoMSB2 Δ3STR, and MoMSB2 ΔSTR alleles. PCR primers for generating these mutant alleles were listed in supplemental table 1. All domain deletion constructs were transformed into the Momsb2 mutant M6.
qRT-PCR analysis
RNA samples were isolated from mycelia from two-day-old liquid CM cultures and 24 h appressoria with the Trizol Reagent (Invitrogen, CA). First-strand cDNA was synthesized with the AccuScript 1st strand cDNA synthesis kit (Stratagene, La Jolla, CA). RT-PCR was performed with the Stratagene Gene MX 3000 PM using the RT2 Real-TimeTM SYBR Green/ROX PCR master mix (SABiosciences, MD). Primer pairs Msb2QF/Msb2QR and AQF/AQR were used to amplify the MoMSB2 and actin (MGG_03982) genes, respectively. The relative quantification of each transcript was calculated by the 2-ΔΔCT method [54] with the actin gene as the internal control.
Protein isolation and western blot analysis
Vegetative hyphae were harvested from 2-day-old CM cultures and used for protein extraction as described [42], [55]. Total proteins (approximately 20 mg) were separated on a 12.5% SDS-PAGE gel and transferred to nitrocellulose membranes for western blot analysis [56]. TEY- and TGY-specific phosphorylations of MAP kinases were detected with the PhophoPlus p44/42 and p38 MAP kinase antibody kits (Cell Signaling Technology, Danvers, MA) following the manufacturer's instructions.
GenBank accession numbers
Sequence data for genes described in this article can be found in the GenBank under the following accession numbers: MSB2 (NP_011528), SHO1 (NP_011043), MoMSB2 (MG06033), MoSHO1 (MG09125), Aspergillus nidulans AnMSB2 (ANID_07041), A. nidulans AnSHO1 (ANID_07698), Neurospora crassa NcMSB2 (NCU04373), N. crassa NcSHO1 (NCU08067), Candida albicans CaMSB2 (XP_722538), C. albicans CaSHO1 (CAC81238).
Supporting Information
Alignment of MoMsb2 (A) and MoSho1 (B) with corresponding orthologs from Neurospora crassa (Nc), Aspergillus nidulans (An), Candida albicans (Ca), and Saccharomyces cerevisiae. Identical and similar residues were shaded in black and gray, respectively. STR, serine/threonine rich region; HMH, Hkr1-Msb2 homology domain; TM, transmembrane domain; CT, cytoplasmic tail; SH3, and Src homology 3 domain. The STR region was not well conserved in Msb2 orthologs.
(0.46 MB RTF)
Nucleotide sequence of the MoMSB2 promoter region. Two PRE-like sequences were underlined. Two putative TCS elements were shaded in gray.
(0.02 MB DOC)
The Momsb2 and Mosho1 deletion mutants. A. The MoSHO1 gene replacement event and Southern blot analysis. Genomic DNA samples of 70-15 (WT), S72 (Mosho1), Ect7, and Ect12 (ectopic) were digested with BclI. The blot on the left was hybridized with probe 1 amplified with 1F and 6R. On the right was the same blot stripped and re-hybridized with probe 2 amplified with 5F and 7R. B. The MoMSB2 gene replacement events and Southern blot analysis of the Momsb2 (M6) and Mosho1 Momsb2 (MS88) mutants. When hybridized with a fragment of the MoMSB2 gene (probe 3), the wild-type 5.7-kb NcoI band was absent in mutants M6 and MS88. When hybridized with a downstream fragment of MoMSB2 (probe 4), mutants M6 and MS88 lacked the wild-type 5.7-kb band but had the expected 3.8-kb and 4.3-kb bands, respectively.
(0.70 MB TIF)
Lesions formed by the Momsb2 and Mosho1 Momsb2 mutants on rice leaves. A representative leaf tips sprayed with 5×104 conidia/ml conidia from the Momsb2 and Mosho1 Momsb2 mutants. Typical leaves were photographed 7 dpi. B. Close view of lesions caused by the Momsb2 and Mosho1 Momsb2 mutants.
(1.18 MB TIF)
Appressorium formation and plant infection assays with the complemented transformants. A. Melanized appressoria formed by transformants CM6 (Momsb2/MoMSB2) and CMS74 (Mosho1 Momsb2/MoSHO1 MoMSB2) on hydrophobic surfaces. Bar = 10 µm. B. Blast lesions formed on rice leaves inoculated with CM6 and CMS74.
(0.57 MB TIF)
Barley leaves were inoculated with strains Ku80, M6 (Momsb2), and MS88 (Momsb2 Mosho1) and examined under SEM. The mutants formed appressoria on intact barley leaves (upper panels) but not on de-waxed leaves. Bar = 10 µm.
(0.50 MB TIF)
Surface hydrophobicity of waxed microscope glass slides. Drops of 20 µl of 0.02% bromophenol blue (BPB) in distilled water were placed onto the surface of glass slides that were untreated or coated with bee waxes.
(0.85 MB TIF)
Time course assays for the expression and localization of MoMsb2-eGFP during appressorium formation. Representative images were presented for each time point (labeled on top). Bar = 10 µm. The same fields were examined under DIC (left) and epifluoresence microscopy (right).
(0.28 MB TIF)
PCR primers used in this study.
(0.05 MB DOC)
Appressorium formation on intact and de-waxed rice leaves.
(0.03 MB DOC)
Acknowledgments
We thank Drs. Larry Dunkle and Janna Beckerman for critical reading of this manuscript and Dr. Paul Cullen at SUNY Buffalo for providing the msb2 mutant. We also thank Drs. Matthew Jenks and Shiyou Lu for assistance with rice leaf wax experiments and Dr. Yang Wang for assistance in phenotype characterization.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by a grant from the National Research Initiative of the USDA CSREES (#2007-35319-102681) and the 111 Project from the Ministry of Education of China (B07049). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Valent B, Chumley FG. Molecular genetic analysis of the rice blast fungus Magnaporthe grisea. Annu Rev Phytopathol. 1991;29:443–467. doi: 10.1146/annurev.py.29.090191.002303. [DOI] [PubMed] [Google Scholar]
- 2.Xu JR, Peng YL, Dickman MB, Sharon A. The dawn of fungal pathogen genomics. Annu Rev Phytopathol. 2006;44:337–366. doi: 10.1146/annurev.phyto.44.070505.143412. [DOI] [PubMed] [Google Scholar]
- 3.Wilson RA, Talbot NJ. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nature Rev Micriobiol. 2009;7:185–195. doi: 10.1038/nrmicro2032. [DOI] [PubMed] [Google Scholar]
- 4.Kankanala P, Czymmek K, Valent B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell. 2007;19:706–724. doi: 10.1105/tpc.106.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi WB, Dean RA. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell. 1997;9:1973–1983. doi: 10.1105/tpc.9.11.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell TK, Dean RA. The cAMP-dependent protein kinase catalytic subunit is required for appressorium formation and pathogenesis by the rice blast pathogen Magnaporthe grisea. Plant Cell. 1995;7:1869–1878. doi: 10.1105/tpc.7.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uchiyama T, Okuyama K. Participation of Oryza-Sativa leaf wax in appressorium formation by Pyricularia oryzae. Phytochemistry. 1990;29:91–92. [Google Scholar]
- 8.Xiao JZ, Watanabe T, Sekido S, Choi WB, Kamakura T, et al. An anti-hydrotactic response and solid surface recognition of germ tubes of the rice blast fungus, Magnaporthe grisea. Biosci Biotechnol Biochem. 1997;61:1225–1229. [Google Scholar]
- 9.Ohtake M, Yamamoto H, Uchiyama T. Influences of metabolic inhibitors and hydrolytic enzymes on the adhesion of appressoria of Pyricularia oryzae to wax-coated cover-glasses. Biosci Biotechnol Biochem. 1999;63:978–982. doi: 10.1271/bbb.63.978. [DOI] [PubMed] [Google Scholar]
- 10.Jelitto TC, Page HA, Read ND. Role of external signals in regulating the pre-penetration phase of infection by the rice blast fungus Magnaporthe grisea. Planta. 1994;194:471–477. [Google Scholar]
- 11.Liu H, Suresh A, Willard FS, Siderovski DP, Lu S, et al. Rgs1 regulates multiple G alpha subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. Embo Journal. 2007;26:690–700. doi: 10.1038/sj.emboj.7601536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao X, Mehrabi R, Xu J-R. Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot Cell. 2007;6:1701–1714. doi: 10.1128/EC.00216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Bioch Biophys Acta. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao XH, Kim Y, Park G, Xu JR. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell. 2005;17:1317–1329. doi: 10.1105/tpc.104.029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park G, Xue C, Zhao X, Kim Y, Orbach M, et al. Multiple upstream signals converge on an adaptor protein Mst50 to activate the PMK1 pathway in Magnaporthe grisea. Plant Cell. 2006;18:2822–2835. doi: 10.1105/tpc.105.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park G, Xue GY, Zheng L, Lam S, Xu JR. MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol Plant-Microbe Interact. 2002;15:183–192. doi: 10.1094/MPMI.2002.15.3.183. [DOI] [PubMed] [Google Scholar]
- 17.Kramer B, Thines E, Foster AJ. MAP kinase signalling pathway components and targets conserved between the distantly related plant pathogenic fungi Mycosphaerella graminicola and Magnaporthe grisea. Fungal Genet Biol. 2009;46:667–681. doi: 10.1016/j.fgb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Rispail N, Soanes DM, Ant C, Czajkowski R, Grünler A, et al. Comparative genomics of MAP kinase and calcium–calcineurin signalling components in plant and human pathogenic fungi. Fung Genet Biol. 2009;46:287–298. doi: 10.1016/j.fgb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 19.DeZwaan TM, Carroll AM, Valent B, Sweigard JA. Magnaporthe grisea Pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell. 1999;11:2013–2030. doi: 10.1105/tpc.11.10.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bardwell L. Mechanisms of MAPK signalling specificity. Biochem Soc Transact. 2006;34:837–841. doi: 10.1042/BST0340837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cullen PJ, Schultz J, Horecka J, Stevenson BJ, Jigami Y, et al. Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics. 2000;155:1005–1018. doi: 10.1093/genetics/155.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitoniak A, Birkaya B, Dionne HM, Vadaie N, Cullen PJ. The signaling mucins Msb2 and Hkr1 differentially regulate the filamentation mitogen-activated protein kinase pathway and contribute to a multimodal response. Mol Biol Cell. 2009;20:3101–3114. doi: 10.1091/mbc.E08-07-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raitt DC, Posas F, Saito H. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 2000;19:4623–4631. doi: 10.1093/emboj/19.17.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanver D, Mendoza-Mendoza A, Brachmann A, Kahmann R. Sho1 and Msb2-related proteins regulate appressorium development in the smut fungus Ustilago maydis. Plant Cell. 2010;22:2085–2101. doi: 10.1105/tpc.109.073734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 26.Villalba F, Collemare J, Landraud P, Lambou K, Brozek V, et al. Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genet Biol. 2008;45:68–75. doi: 10.1016/j.fgb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Gilbert RD, Johnson AM, Dean RA. Chemical signals responsible for appressorium formation in the rice blast fungus Magnaporthe grisea. Physiol Mol Plant Pathol. 1996;48:335–346. [Google Scholar]
- 28.Kunst L, Samuels AL. Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res. 2003;42:51–80. doi: 10.1016/s0163-7827(02)00045-0. [DOI] [PubMed] [Google Scholar]
- 29.Yu D, Ranathunge K, Huang H, Pei Z, Franke R, et al. Wax Crystal-Sparse Leaf1 encodes a beta-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta. 2008;228:675–685. doi: 10.1007/s00425-008-0770-9. [DOI] [PubMed] [Google Scholar]
- 30.Dixon KP, Xu JR, Smirnoff N, Talbot NJ. Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell. 1999;11:2045–2058. doi: 10.1105/tpc.11.10.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol. 2008;70:431–457. doi: 10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- 32.Cullen PJ, Sabbagh W, Jr, Graham E, Irick MM, van Olden EK, et al. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatebayashi K, Tanaka K, Yang HY, Yamamoto K, Matsushita Y, et al. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 2007;26:3521–3533. doi: 10.1038/sj.emboj.7601796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamakura T, Yamaguchi S, Saitoh K, Teraoka T, Yamaguchi I. A novel gene, CBP1, encoding a putative extracellular chitin- binding protein, may play an important role in the hydrophobic surface sensing of Magnaporthe grisea during appressorium differentiation. Mol Plant-Microbe Interact. 2002;15:437–444. doi: 10.1094/MPMI.2002.15.5.437. [DOI] [PubMed] [Google Scholar]
- 35.Zabka V, Stangl M, Bringmann G, Vogg G, Riederer M, et al. Host surface properties affect prepenetration processes in the barley powdery mildew fungus. New Phytol. 2008;177:251–263. doi: 10.1111/j.1469-8137.2007.02233.x. [DOI] [PubMed] [Google Scholar]
- 36.Xu JR, Hamer JE. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 37.Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, et al. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol. 2008;181:1073–1081. doi: 10.1083/jcb.200704079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweigard JA, Carroll AM, Farrall L, Chumley FG, Valent B. Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol Plant-Microbe Interact. 1998;11:404–412. doi: 10.1094/MPMI.1998.11.5.404. [DOI] [PubMed] [Google Scholar]
- 40.Xue CY, Park G, Choi WB, Zheng L, Dean RA, et al. Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell. 2002;14:2107–2119. doi: 10.1105/tpc.003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sweigard JA, Chumley FG, Valent B. Disruption of a Magnaporthe grisea cutinase gene. Mol Gen Genet. 1992;232:183–190. [PubMed] [Google Scholar]
- 42.Bruno KS, Tenjo F, Li L, Hamer JE, Xu JR. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot Cell. 2004;3:1525–1532. doi: 10.1128/EC.3.6.1525-1532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu WD, Xie SY, Zhao XH, Chen X, Zheng WH, et al. A homeobox gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae. Mol Plant-Microbe Interact. 2010;23:366–375. doi: 10.1094/MPMI-23-4-0366. [DOI] [PubMed] [Google Scholar]
- 44.Bourett TM, Sweigard JA, Czymmek KJ, Carroll A, Howard RJ. Reef coral fluorescent proteins for visualizing fungal pathogens. Fungal Genet Biol. 2002;37:211–220. doi: 10.1016/s1087-1845(02)00524-8. [DOI] [PubMed] [Google Scholar]
- 45.Cullen PJ, Sabbagh W, Graham E, Irick MM, van Olden EK, et al. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zarrinpar A, Bhattacharyya RP, Nittler MP, Lim WA. Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol Cell. 2004;14:825–832. doi: 10.1016/j.molcel.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 47.Tucker SL, Thornton CR, Tasker K, Jacob C, Giles G, et al. A fungal metallothionein is required for pathogenicity of Magnaporthe grisea. Plant Cell. 2004;16:1575–1588. doi: 10.1105/tpc.021279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koga H. Hypersensitive death, autofluorescence, and ultrastructural-changes in cells of leaf sheaths of susceptible and resistant near-isogenic lines of rice (PI-Z(T)) in relation to penetration and growth of Pycularia oryza. Can J Bot. 1994;72:1463–1477. [Google Scholar]
- 49.Park G, Bruno KS, Staiger CJ, Talbot NJ, Xu JR. Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol Microbiol. 2004;53:1695–1707. doi: 10.1111/j.1365-2958.2004.04220.x. [DOI] [PubMed] [Google Scholar]
- 50.Li L, Xue CY, Bruno K, Nishimura M, Xu JR. Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol Plant-Microbe Interact. 2004;17:547–556. doi: 10.1094/MPMI.2004.17.5.547. [DOI] [PubMed] [Google Scholar]
- 51.Talbot NJ, Kershaw MJ, Wakley GE, deVries OMH, Wessels JGH, et al. MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell. 1996;8:985–999. doi: 10.1105/tpc.8.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen XB, Goodwin SM, Boroff VL, Liu XL, Jenks MA. Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell. 2003;15:1170–1185. doi: 10.1105/tpc.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li L, Ding SL, Sharon A, Orbach M, Xu JR. Mir1 is highly upregulated and localized to nuclei during infectious hyphal growth in the rice blast fungus. Mol Plant-Microbe Interact. 2007;20:448–458. doi: 10.1094/MPMI-20-4-0448. [DOI] [PubMed] [Google Scholar]
- 54.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 55.Ding S, Liu W, LLiuk A, Ribot C, Vallet J, et al. The Tig1 HDAC complex regulates infectious growth in the rice blast fungus Magnaporthe oryzae. Plant Cell. 2010;22:2495–2508. doi: 10.1105/tpc.110.074302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Russell D. Molecular Cloning - A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of MoMsb2 (A) and MoSho1 (B) with corresponding orthologs from Neurospora crassa (Nc), Aspergillus nidulans (An), Candida albicans (Ca), and Saccharomyces cerevisiae. Identical and similar residues were shaded in black and gray, respectively. STR, serine/threonine rich region; HMH, Hkr1-Msb2 homology domain; TM, transmembrane domain; CT, cytoplasmic tail; SH3, and Src homology 3 domain. The STR region was not well conserved in Msb2 orthologs.
(0.46 MB RTF)
Nucleotide sequence of the MoMSB2 promoter region. Two PRE-like sequences were underlined. Two putative TCS elements were shaded in gray.
(0.02 MB DOC)
The Momsb2 and Mosho1 deletion mutants. A. The MoSHO1 gene replacement event and Southern blot analysis. Genomic DNA samples of 70-15 (WT), S72 (Mosho1), Ect7, and Ect12 (ectopic) were digested with BclI. The blot on the left was hybridized with probe 1 amplified with 1F and 6R. On the right was the same blot stripped and re-hybridized with probe 2 amplified with 5F and 7R. B. The MoMSB2 gene replacement events and Southern blot analysis of the Momsb2 (M6) and Mosho1 Momsb2 (MS88) mutants. When hybridized with a fragment of the MoMSB2 gene (probe 3), the wild-type 5.7-kb NcoI band was absent in mutants M6 and MS88. When hybridized with a downstream fragment of MoMSB2 (probe 4), mutants M6 and MS88 lacked the wild-type 5.7-kb band but had the expected 3.8-kb and 4.3-kb bands, respectively.
(0.70 MB TIF)
Lesions formed by the Momsb2 and Mosho1 Momsb2 mutants on rice leaves. A representative leaf tips sprayed with 5×104 conidia/ml conidia from the Momsb2 and Mosho1 Momsb2 mutants. Typical leaves were photographed 7 dpi. B. Close view of lesions caused by the Momsb2 and Mosho1 Momsb2 mutants.
(1.18 MB TIF)
Appressorium formation and plant infection assays with the complemented transformants. A. Melanized appressoria formed by transformants CM6 (Momsb2/MoMSB2) and CMS74 (Mosho1 Momsb2/MoSHO1 MoMSB2) on hydrophobic surfaces. Bar = 10 µm. B. Blast lesions formed on rice leaves inoculated with CM6 and CMS74.
(0.57 MB TIF)
Barley leaves were inoculated with strains Ku80, M6 (Momsb2), and MS88 (Momsb2 Mosho1) and examined under SEM. The mutants formed appressoria on intact barley leaves (upper panels) but not on de-waxed leaves. Bar = 10 µm.
(0.50 MB TIF)
Surface hydrophobicity of waxed microscope glass slides. Drops of 20 µl of 0.02% bromophenol blue (BPB) in distilled water were placed onto the surface of glass slides that were untreated or coated with bee waxes.
(0.85 MB TIF)
Time course assays for the expression and localization of MoMsb2-eGFP during appressorium formation. Representative images were presented for each time point (labeled on top). Bar = 10 µm. The same fields were examined under DIC (left) and epifluoresence microscopy (right).
(0.28 MB TIF)
PCR primers used in this study.
(0.05 MB DOC)
Appressorium formation on intact and de-waxed rice leaves.
(0.03 MB DOC)