Abstract
In contrast to large GWA studies based on thousands of individuals and large meta-analyses combining GWAS results, we analyzed a small case/control sample for uric acid nephrolithiasis. Our cohort of closely related individuals is derived from a small, genetically isolated village in Sardinia, with well-characterized genealogical data linking the extant population up to the 16th century. It is expected that the number of risk alleles involved in complex disorders is smaller in isolated founder populations than in more diverse populations, and the power to detect association with complex traits may be increased when related, homogeneous affected individuals are selected, as they are more likely to be enriched with and share specific risk variants than are unrelated, affected individuals from the general population. When related individuals are included in an association study, correlations among relatives must be accurately taken into account to ensure validity of the results. A recently proposed association method uses an empirical genotypic covariance matrix estimated from genome-screen data to allow for additional population structure and cryptic relatedness that may not be captured by the genealogical data. We apply the method to our data, and we also investigate the properties of the method, as well as other association methods, in our highly inbred population, as previous applications were to outbred samples. The more promising regions identified in our initial study in the genetic isolate were then further investigated in an independent sample collected from the Italian population. Among the loci that showed association in this study, we observed evidence of a possible involvement of the region encompassing the gene LRRC16A, already associated to serum uric acid levels in a large meta-analysis of 14 GWAS, suggesting that this locus might lead a pathway for uric acid metabolism that may be involved in gout as well as in nephrolithiasis.
Author Summary
There are a number of factors that contribute to renal stone formation, including diet and obesity, specific drugs, other diseases, climate changes, metabolic disorders, and genetic predisposition. In this article, we focus on identifying genomic regions that may be involved with nephrolithiasis associated with a uric acid component. We analyze data from a genetic isolate in Sardinia to take advantage of the potential improvement in power to detect association with complex traits when related, homogeneous affected individuals are selected. To take into account the correlations among our related sample of cases and controls, we applied a recently proposed method that corrects for both known and unknown population and pedigree structure using genome-wide data. In simulation studies for outbred populations with related individuals and population structure, the method has been demonstrated to provide a substantial improvement over a number of existing methods in terms of power and type 1 error. We investigate the properties of this new method, as well as other association methods, in our inbred sample. To our knowledge, this is the first application of this recently proposed method to a founder population. This study is also the first genome-wide association study carried out for uric acid nephrolithiasis.
Introduction
Nephrolithiasis is a multifactorial disorder of complex etiology characterized by the presence of stones in the urinary tract. Kidney stones are a common disorder, with a prevalence of urinary calculi between 4% and 10% in the adult population, with an increasing incidence in Western societies [1]. For instance, in the US the prevalence has risen from 3.2% to 5.2% in just over two decades from the mid-1970s to the mid-1990s [2]. Wide geographical variations exist in renal stone incidence and composition, and specific geographic belts have been identified [3], where increased incidence has been attributed to genetic and environmental factors, such as hot climate (fluid loss) and sun exposure that increases the rate of vitamin D.
Kidney stones are composed of inorganic and organic crystals amalgamated with proteins. Crystallisation and subsequent lithogenesis can happen with many solutes in the urine. Calcareous stones are still by far the most common type of nephrolithiasis, accounting for more than 80% of stones [4]. Uric acid nephrolithiasis (UAN) represent about 5–10% of the remaining stones, trailed by cystine, struvite, and ammonium acid urate stones.
Genetic contribution to renal stones formation has been extensively recognized, and a number of studies have established a link between several genes and predominantly oxalate kidney stones, including vitamin-D receptor gene (VDR) and calcitonin receptor (CTR) gene, heparan sulfate (HSPG2) gene, and fibronectin gene (FN1) [5], [6]. There are a number of factors that can contribute to the formation of renal stones, including diet and obesity, specific drugs, other diseases, climate changes, metabolic disorders, and genetic predisposition [7], [8]. The complexity of this disease has led researchers to consider nephrolithiasis as one feature of a broader systemic disease, rather than a disease specific to a single organic system. This is especially interesting in relation to gout and metabolic syndrome, which are both systemic disorders in close relation with nephrolithiasis [9], [10]. UAN primarily results from low urinary pH, which increases the concentration of the insoluble undissociated uric acid, causing formation of both uric acid and mixed uric acid/calcium oxalate stones. A persistently low urinary pH (<5.5, the pKa for uric acid is 5.35) is a distinctive feature of idiopathic UAN previously named gouty diathesis [11].
In this study we focused on a Sardinian isolated population, the village of Talana, located in a mountain area of the island. The Talana population has been extensively studied, and has been characterized by a limited number of original founders, a long-term, slow population growth rate and isolation [12], [13]. Studying founder, isolated populations like the Talana, allows to reduce genetic complexity underlying disease etiology and to increase environmental homogeneity, as inhabitants share a common and uniform lifestyle. In the extant population of Talana the frequency of nephrolithiasis is approximately 20%, with a strong prevalence of UAN stones (half of all renal stone formers). In our previous study, we performed a genome-wide linkage search in 14 closely-related affected individuals using 382 microsatellites. Suggestive regions were investigated in 37 individuals who were more distantly-related affecteds [14], allowing us to fine-map a susceptibility locus on the chromosomal region 10q21–q22, and to identify a possible candidate gene [15].
The advent of high-throughput technologies for single nucleotide polymorphisms (SNPs) genotyping has allowed for a rapid and an economical way to do GWA analysis, and it might now be possible to achieve adequate power for identifying risk variants associated to complex diseases such as nephrolithiasis. In this new study we perform a GWA scan in a larger sample of well characterized cases and controls from Talana, utilizing a highly-dense SNPs map. Association analysis of our cohort of cases and controls, all related through multiple lines of descent and belonging to a single, large, and well-characterized genealogy, is particularly challenging, due to the complex relatedness in the sample. A number of methods have been proposed in the recent years for case-control association testing in samples that include related individuals from a single population provided that the pedigrees are completely specified [16]–[19]. It is well known that in association studies, spurious association can arise if ancestry differences among the cases and controls are not properly accounted for. An improved association method, named ROADTRIPS, for samples with related individuals and population structure, has recently been implemented in a software program [20]. ROADTRIPS uses an empirical genotypic covariance matrix calculated from genome-screen data to allow for population structure and cryptic relatedness in a sample that may not be captured by the available genealogical information. This method is appropriate for sampled individuals (both cases and controls) from a founder population, who are related through multiple lines of descent, with pedigrees only partially specified. In simulation studies with related individuals from outbred populations and population structure, including admixture, ROADTRIPS has been demonstrated to provide a substantial improvement over a number of existing methods in terms of power and type 1 error. Furthermore, in a recent review investigating the current progresses on methods that correct for stratification while accounting for additional complexities, ROADTRIPS has been shown to have appropriate characteristics [21].
We applied ROADTRIPS to a sample of related cases, affected by UAN, and a sample of unaffected controls selected from the same isolated population, all related through a complex genealogy. We also investigated the properties of ROADTRIPS, as well as other association methods, in our highly inbred population. To our knowledge this is the first application of this recent method to a case/control sample of closely related individuals from a founder population with extended genealogical data. We then followed up on the more promising regions and the top associated SNPs identified in our initial sample from the genetic isolate and performed an association analysis in an independent sample collected from the Italian population (including a general Sardinian sub-sample).
Methods
Subjects
The study subjects were 861 individuals from Talana, linked through a multi-generation 4446-member pedigree, with a mean (median) kinship coefficient of 0.0201 (0.0115) (SD = 0.0231). During physical examination of each individual, a blood sample was collected for DNA extraction, and different phenotypic traits, and pathologies, were recorded. For this study, we collected information on age at diagnosis, medications, hospital admissions, and family history. Individuals with a history of urinary tract infection or with any secondary condition that might predispose to kidney stones (e.g., inflammatory bowel disease or gout) were not included. The diagnostic procedures have been carried out essentially as described elsewhere [14]. In brief, all subjects affected by renal stones and their family members underwent renal ultrasound examination to confirm reported disease occurrence and to identify asymptomatic cases. Clinical renal ultrasonography is used to image calculi, such as UAS, that are non-opaque on X-rays [22].
From an initial set of 173 renal stone formers, we selected 80 severe cases that showed uric acid as the principal component. Disease severity was established on the basis of the presence of stones during ultrasonography and past history of kidney stones, with more than one episode of abdominal colic. Subjects with mild to moderate disease symptoms (e.g., having only a single episode or spots but no episodes) were not classified as affected in the present study. We identified 94 control subjects, who were examined by ultrasonography to exclude individuals with asymptomatic disease. The mean age at observation of unaffected controls was sufficiently high (∼55 years) to have given an elevated probability of developing stones.
All subjects gave written informed consent, and all samples were taken in accordance with the Helsinki declaration.
Genotyping
Genotyping for the initial GWA study was carried out using the Affymetrix 500K chips using standard protocols, and the 50K chips with SNPs distributed around known genes. SNPs genotyping was performed on the Affymetrix Gene-Chip platform. We used the GeneChip Human Mapping to genotype the 500K Array Set that comprises two arrays (the Nsp and Sty arrays) capable of genotyping ∼262,000 and ∼238,000 SNPs, respectively. We followed the recommended protocol described in the Affymetrix manual. In total, 861 individuals were genotyped for the 500K set and 528 individuals for the 50K set.
Genotypic quality control
Details on QC analysis in Talana are provided in the Text S1. Briefly, we first checked for gender mismatches to make sure that individuals in our database align with individual DNA samples in the genetic data, dropping problematic samples. Individuals with a missing rate >90% were removed, and SNPs showing a missing rate >95% and a MAF <0.05 were dropped in both the 50K and 500K sample sets. HWE was tested and two different thresholds (due to the different number of SNPs) were used to exclude SNPs that showed extreme deviation from HWE (threshold of p<1E-6 for the 500K, and of p<1E-4 for the 50K). Furthermore, we estimated the proportion of IBD sharing derived from the genome between each pair of genotyped individuals and compared it with the proportion expected based on the genealogical information. Relatedness between examinees was estimated from an LD-pruned dataset of SNPs derived from the whole genome data using PLINK [23]. From this analysis we identified and excluded individuals that showed recurrent inconsistencies between the two IBD sharing proportions.
Statistical analysis
For the statistical analysis of our sample of related cases and controls derived from a single large genealogy, we used the recently proposed method implemented in the ROADTRIPS software [20]. This program allows for single SNP (currently just for autosomes) case-control association testing in samples from isolated founder populations with partially or completely unknown genealogies. A significant improvement over the previously proposed tests for founder populations, implemented in the CC-QLS and the MQLS software packages [16], [18], is that ROADTRIPS uses an empirical covariance matrix, denoted by Ψ, calculated from genome-wide SNP data to correct for unknown population and pedigree structure, while maintaining high power by taking advantage of known pedigree information when it is available. The structure matrix estimated from genome-wide data is used in the variance calculation to account for structure that may not be captured by the kinship coefficient matrix, denoted by Φ, derived from the known genealogy. Additional advantages of this approach are that it allows for two different types of controls, unaffected controls and controls of unknown phenotype (e.g., general population controls), to be included in the same analysis, and it can incorporate phenotype information on relatives with missing genotype data at the SNP being tested.
We now give a brief overview of the different test statistics used in the analysis. The ROADTRIPS extension of the statistics implemented in the MQLS software, namely, MQLS, WQLS, and the corrected Pearson's χ2 test, are RM, RW, and Rχ, respectively. The MQLS, WQLS, and the corrected Pearson's χ2 tests were developed for related samples from a single population with known pedigrees, and ROADTRIPS extends these statistics to allow for population structure and pedigrees that are partially or completely unknown. For two-allele disease models, optimal properties of the MQLS test (and the RM test when the individuals are from a single population) is that it is most powerful in a general class of linear statistics for general two-allele disease models in outbreds and for additive disease models in inbreds, as effect size tends to 0. The MQLS and RM tests improve power by taking advantage of the enrichment of predisposing alleles in affected individuals with affected relatives. The WQLS (and the RW test when the individuals are from a single population) is optimal when the true genetic trait model is a rare, fully penetrant dominant allele. The corrected Pearson's χ2 test and Rχ are extensions of the Pearson's χ2 test for independence of trait value and marker genotype. The Rχ statistic has a correction factor that is similar to the correction factor used in genomic control [24]. When the aforementioned test statistics have been applied to various association studies in the context of complex trait mapping, where the traits of interest are influenced by numerous genes as well as environmental factors, the tests have given complimentary as well different results, with the MQLS (and RM) test often having slightly higher power to detect association than the corrected Pearson's χ2 test (and Rχ) and with WQLS (and RW) having the lowest power [16], [18], [20]. A summary of the characteristics of the statistics that were used is shown in Table 1.
Table 1. Summary of the test statistics used in the analysis.
| Test statistic | Population controls | Corrected for Ψ | Corrected for Φ | Program |
| RM | ✓ | ✓ | ✓ | ROADTRIPS |
| Rχ | ✗ | ✓ | ✗ | ROADTRIPS |
| RW | ✗ | ✓ | ✓ | ROADTRIPS |
| MQLS | ✓ | ✗ | ✓ | MQLS |
| Corrected χ2 | ✗ | ✗ | ✓ | MQLS |
| WQLS | ✗ | ✗ | ✓ | CC-QLS |
The p-values for the test statistics in the ROADTRIPS software are based on a χ2 asymptotic null distribution with 1 degree of freedom. To assess whether or not the p-value is “exact”, the ROADTRIPS software uses a similar criterion to what is commonly used for Pearson's χ2 test for independence between trait and marker genotype, where the expected counts in each cell for a 2×2 table should be at least 5 in order for the χ2 distribution assumption to hold. The asymptotic null distribution assumption will hold for SNPs with rare alleles provided that there are enough minor allele counts observed for the SNPs in the sample. The ROADTRIPS software provides a warning message “The p-value might not be exact because of the small number of type 1 alleles in …” referring to cases, controls, or both, when the asymptotic null distribution assumption for the statistics may not be satisfied, which can occur for SNPs with low minor allele counts.
The Rχ test is calculated using naïve allele frequency estimates, i.e., allele frequency estimates based on giving equal weights to the sample individuals, while both the RM, and RW tests use BLUE estimates [25]. The latter allele frequency estimator is the best linear unbiased estimator and is calculated conditioned on the genealogy of the sample individuals. The BLUE takes into account relatedness in the sample and the estimator allows for inbreeding and for sample individuals to be related through multiple lines of descent.
Replication study
We collected an independent sample from the Italian general population, and in particular 69 cases from the Department of Nephrology and Dialysis of Bergamo, and 98 controls deriving from randomly ascertained blood donors in the same area. We also collected 56 affected individuals, and 59 controls from randomly ascertained blood donors in Sardinia. The Sardinian affected individuals were collected from the Clinics of Urology of Cagliari and Lanusei. All cases were selected to have pure uric acid stones or uric acid as the principal component. In total we analyzed 282 individuals (125 cases and 157 controls) in the replication study, but we considered the two population samples (continental Italy and Sardinia) as two different clusters, in order to exclude potential bias in the analysis derived from the geographical origin of the samples.
We genotyped 96 SNPs in the independent replication sample as well as in the 73 cases and 93 controls from Talana analyzed in the initial study. A total of 28 SNPs were selected either from the top results in the initial study (10 SNPs), or in the candidate regions on chromosomes 2, 6 and 10, based on a Rχ p-value <0.05 and RM p-value<0.01 (18 SNPs). For 11 out of 28 of these SNPs, only 48 cases and 67 controls were genotyped in the initial set (i.e. these SNPs belonged to the 50K set). We also genotyped 4 cSNPs (missense) in the candidate genes SLC17A1, ADAMTS14, and UNC5B, selected from Hapmap with a MAF in CEU >0.01. The remaining SNPs (64) were selected using Tagger [26] to cover the candidate regions on chromosomes 2, 6 and 10. We selected tSNPs with the criteria of “pick only the N best tags”, where N was based on the specific size and recombination pattern of each region. We used the “pairwise tagging only” mode, providing the Illumina design score for preferential picking of the tSNPs, to capture only SNPs with MAF>0.05. The tagged regions and the resulting coverage based on r2 are shown in Table S1. The initial set of cases and controls from Talana was also genotyped for the SNPs typed in the replication cohort.
SNPs were typed by using the VeraCode GoldenGate Genotyping Assay from Illumina according to the manufacturer's protocol (Illumina, San Diego, CA). Briefly, the technology is based on allele-specific primer extension. Genomic DNA (250 ng) was activated chemically with biotin and then hybridized to a pool of locus-specific oligos (OPA, OligoPool All; Illumina). After removal of nonspecific unbound oligos, a PCR reaction was performed by using fluorescent-labeled primers (Cy3 and Cy5). PCR products were cleaned and denatured, and single-stranded fluorescent-labeled DNAs were hybridized to VeraCode beads, which were scanned on a BeadXpress reader by using Illumina VeraScan V1.1 software. Raw data, consisting of intensities of fluorescence, were then imported into the analysis software GenomeStudio and the automatic allele calling was done using GeneCall threshold of 0.25. The final SNP call rate (the number of SNP successfully genotyped for each sample) was >0.97. Standard QC was performed and only 1 SNP was excluded due to extreme deviation from HWE, where this SNP had only the two homozygous genotypes.
For the replication set, the sample of unrelated cases and controls was analyzed with PLINK using standard methods, based on allele frequencies differences. The Cochran-Mantel-Haenszel (CMH) tests for stratified tables, which allow for tests of association conditional on cluster of samples was used for merged sets (we clustered individuals based on the geographic origins, namely continental Italy and Sardinia). The Breslow-Day (BD) test was computed to test the homogeneity of odds ratios within clusters.
We also performed a global association test by including to the replication set a Talana sub-sample that consisted of distantly related cases/controls, as an additional cluster using the CMH and BD tests. The Talana sub-sample was extracted from the whole sample of cases and controls using a pairwise sampling approach [27] basing on a kinship<0.125 between each pair (resulting in 41 cases and 38 controls).
The 73 cases and 93 controls from Talana, used in the initial study, were all typed for the 96 SNPs and analyzed with ROADTRIPS. For 11 SNPs identified in the initial study (28 SNPs), only 48 cases and 67 controls were genotyped in the initial set (i.e. these SNPs belonged to the 50K set), and the remaining 66 tSNPs were not typed in the initial GWAS. It should also be noted that for these SNPs we could not use the option in ROADTRIPS to include all unknown population controls in the analysis as was done in the initial GWAS, since only cases and unaffected controls were typed for these SNPs, while the remaining 668 sample individuals from Talana were not. This smaller sample size can lead to a reduction in power for the replication analysis, since samples sizes strongly influence the power of the test.
Results
Evaluation of the test statistics' properties
For SNPs that have a low minor allele count in either the cases or the controls (unaffected and unknown phenotype) such that the asymptotic χ2 null distribution assumption with 1 degree of freedom for the statistics may not be valid, ROADTRIPS provides a warning message. In our GWAS we observed 22,502 warning messages for RM (∼7% of the tests), and 26,772 for Rχ (∼8% of the tests). We investigated the occurrences of warning messages for the RM and Rχ statistics in relation to MAF. Since in our GWAS we used all available information, thus also including in the analysis 668 unknown population controls, the RM statistics did not show any warning message referring to controls, but only to cases. In Figure S1 we show box-plots of MAF (naïve estimates) for the RM and Rχ statistics tagged by a warning message for each specific group (for RM in controls only; for Rχ in cases, controls, or both), stratified by the 500K and 50K sets as a different number of individuals were genotyped in the two sets (829 and 514 subjects, respectively).
Figure S2 shows the Q-Q plots for the RM and Rχ statistics stratified by the presence of a warning message in the ROADTRIPS output. This figure illustrates that the asymptotic null distribution assumption does not hold for SNPs with low minor allele frequency and counts in our sample (due to the small sample size used in this analysis), particularly for the RM statistic. We also investigated whether the lower minor allele count SNPs are contributing to the excess of smaller p-values of the RM and Rχ statistics than what is expected under the null. From the figure it is evident that for RM these SNPs do not contribute to any inflation of the type 1 error, as none of the SNPs with warning messages for the RM test are anywhere near the significance level threshold, and for the Rχ test the vast majority of the SNPs with warning messages are also not close to being significant.
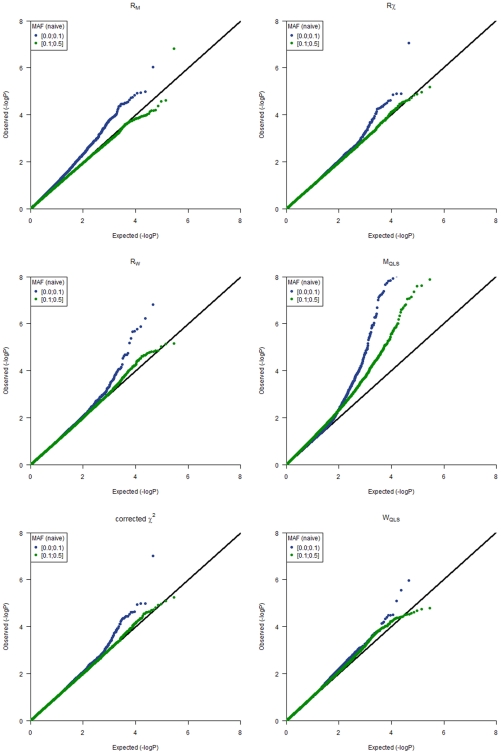
Q-Q plots for the different statistics (RM, Rχ, RW, MQLS, corrected χ2 and WQLS) obtained from the GWAs are shown in Figure 1, where we stratified by naïve allele frequencies of the SNPs in the whole sample. From Figure 1, it is clear that for the SNPs with lower minor allele frequency, the type 1 error distribution is in general inflated for all statistics based on the BLUE estimation. In particular, the RM test may be quite sensitive to allele frequencies, and therefore hard to calibrate. Figure 1 also illustrates that for SNPs with a minor allele frequency of at least 0.1 the asymptotic null distribution assumption for the RM test appears to be adequate for this sample, and the test may actually be conservative for this particular sample in the right tail of the distribution based on the −log(p-values), which may result in a slight loss of power for the RM test for the analysis of this sample.
Figure 1. Q-Q plots for the different test statistics used in the analyses.
Q-Q plots are stratified by the naïve allele frequencies observed in the whole dataset. Namely, MAF> = 0.1 (green points) or MAF<0.1 (blue points).
It is evident that ROADTRIPS provides a significant improvement of the RM test over MQLS test in this dataset, in terms of type 1 error, since there appears to be cryptic relatedness in this study that is not being accounted for in the MQLS statistic, and for which inflated p-values are observed over the whole genome (independently from MAF). In contrast, not much difference is observed between Rχ (which corrects for both population and pedigree structure using genome-wide data) and the corrected χ2 implemented in MQLS (which corrects for relatedness using the genealogical data) in our data.
Finally, we were interested in gaining a better understanding for why some of the SNPs give large p-value differences for the RM and Rχ statistics in our data. We investigated SNPs with discordant RM and Rχ results for the analyses that included the unknown population controls. Specifically we investigated SNPs for which the RM test gives a p-value <1E-4 and the Rχ p-value is not close to significance level (>0.05), and vice versa. In the Text S2 we present a formal investigation of the different behavior of the test statistics for the specific SNPs. The large difference that is observed for RM and Rχ for these SNPs is due to the small number of founders and the high degree of relatedness among the sample individuals. Even though there are 842 individuals in the sample, when comparing the allele frequency variance of the BLUE for this sample to the number of independent (i.e., unrelated non-inbred) individuals that would give the same variance, we estimate the number of independent alleles in the sample [28] to be equivalent to having approximately 61 founders in the sample, i.e., 61 independent individuals (60.52 to be more precise). This estimate is based on the kinship and inbreeding coefficients for the 842 individuals that were calculated from the available genealogical data. The number of independent alleles in this sample may actually be less than our estimate since there is evidence of cryptic relatedness in this sample, as we previously mentioned.
Based on the more stable characteristics of the Rχ statistics that we observed in our sample over the entire range of minor allele counts (low to high), we focused on results obtained with the Rχ statistics (p-value<1E-4), but we also required validation of the SNPs by the RM statistic with a p-value <1E-2. We therefore included in our follow up analysis potentially interesting SNPs with small p-values that did not necessarily reach the conservative Bonferroni genome-wide significance threshold.
GWAS in the initial set
The final SNPs dataset used in this study consisted of a 334,674 SNPs in the merged set (500K and 50K) on the autosomes. The final sample set, that passed the QC, consisted of 73 affected and 92 controls. The characteristics of the case/control sample used in this analysis are summarized in Table 2. The 73 affected subjects are all related through a large pedigree of 1666 individuals. In total 80 cases and 94 controls were analyzed with ROADTRIPS, which allows additional phenotyped relatives that are not genotyped (namely 7 cases and 2 controls) to contribute to the analysis.
Table 2. Inbreeding and kinship of the case/control sample used in this analysis.
| Inbreeding | Kinship | |||||||||||
| sample size | pedigree sizea | Median | Mean | SD | Min | Max | Median | Mean | SD | Min | Max | |
| affected subjects | 73 | 1666 | 0.0092 | 0.0095 | 0.0058 | 0.00004 | 0.0316 | 0.0187 | 0.0242 | 0.0285 | 0.00034 | 0.2783 |
| control subjects | 92 | 2341 | 0.0082 | 0.0091 | 0.0070 | 0.00054 | 0.0371 | 0.0163 | 0.0223 | 0.0293 | 0.00001 | 0.2916 |
aPedigree connecting all subjects (all affected and all unaffected, separately).
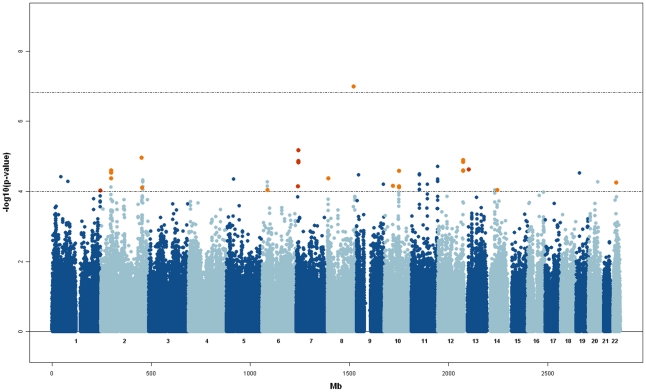
Results from the GWAS for Rχ, are shown in Figure 2, where consistent results obtained with the RM statistics are also highlighted (RM p-value<1E-2). In Table 3 we summarize the top results obtained for SNPs that have a Rχ p-value<1E-4 and RM p-value<1E-2.
Figure 2. Manhattan plot for the Rχ statistic.
Orange points above the −log10(p-value)>4 threshold indicate SNPs that have a p-value<1E-2 for the RM statistic. Significant threshold after Bonferroni correction is shown.
Table 3. Top results obtained for SNPs that showed a Rχ p-value<1E-4 and RM p-value<1E-2.
| ROADTRIPS | MQLS | naive frequencies | BLUE frequencies | ||||||||||||||
| ch | SNP | bp | genes | alleles | RM | Rχ | RW | Corr. χ2 | MQLS | cases | cont's | all sample | cases | cont's | unkn cont's | all sample | Hapmap (CEU) |
| 1 | rs12404212 | 244348886 | SMYD3 | A/G | 0.004620 | 0.000094 | 0.548685 | 0.000080 | 0.000061 | 0.22 | 0.16 | 0.11 | 0.15 | 0.11 | 0.07 | 0.07 | 0.08 |
| 2 | rs1915174 | 50451419 | NRXN1 | G/A | 0.002606 | 0.000028 | 0.000116 | 0.000023 | 0.000819 | 0.30 | 0.47 | 0.48 | 0.34 | 0.51 | 0.51 | 0.53 | 0.57 |
| 2 | rs12465806 | 50451831 | NRXN1 | A/G | 0.002432 | 0.000029 | 0.000119 | 0.000025 | 0.000755 | 0.30 | 0.47 | 0.48 | 0.34 | 0.51 | 0.52 | 0.53 | 0.56 |
| 2 | rs11125301 | 50452354 | NRXN1 | T/A | 0.001982 | 0.000025 | 0.000080 | 0.000020 | 0.000582 | 0.30 | 0.47 | 0.48 | 0.34 | 0.53 | 0.52 | 0.53 | 0.57 |
| 2 | rs1829534 | 50464580 | NRXN1 | C/T | 0.002602 | 0.000028 | 0.000122 | 0.000023 | 0.000818 | 0.30 | 0.47 | 0.48 | 0.34 | 0.51 | 0.51 | 0.53 | 0.58 |
| 2 | rs1402129 | 50470343 | NRXN1 | T/C | 0.003125 | 0.000042 | 0.000313 | 0.000035 | 0.001035 | 0.30 | 0.47 | 0.48 | 0.34 | 0.51 | 0.51 | 0.52 | 0.57 |
| 2 | rs1864466 | 203564702 | ALS2CR8 | A/G | 0.000657 | 0.000011 | 0.000125 | 0.000008 | 0.000001 | 0.32 | 0.14 | 0.17 | 0.26 | 0.10 | 0.09 | 0.09 | 0.08 |
| 2 | rs2359681 | 207274185 | DYTN, ADAM23 | T/C | 0.002540 | 0.000078 | 0.018889 | 0.000072 | 0.000140 | 0.30 | 0.18 | 0.17 | 0.22 | 0.18 | 0.13 | 0.13 | 0.15 |
| 6 | rs10946741 | 25118978 | LRRC16A-SLC17A4 | A/G | 0.000256 | 0.000091 | 0.064420 | 0.000087 | 0.000169 | 0.21 | 0.35 | 0.41 | 0.27 | 0.43 | 0.51 | 0.48 | 0.44 |
| 7 | rs16872208 | 7957012 | GLCCI1 | A/T | 0.009254 | 0.000071 | 0.000014 | 0.000061 | 0.002070 | 0.29 | 0.19 | 0.17 | 0.26 | 0.17 | 0.13 | 0.14 | 0.08 |
| 7 | rs17528240 | 10716439 | NDUFA4, PHF14 | G/A | 0.003906 | 0.000013 | 0.000010 | 0.000011 | 0.001173 | 0.57 | 0.38 | 0.39 | 0.50 | 0.37 | 0.36 | 0.36 | 0.28 |
| 7 | rs12673360 | 10802614 | NDUFA4, PHF14 | A/G | 0.006443 | 0.000015 | 0.000032 | 0.000012 | 0.002306 | 0.57 | 0.38 | 0.39 | 0.51 | 0.37 | 0.38 | 0.37 | 0.30 |
| 7 | rs6460751 | 10811628 | NDUFA4, PHF14 | G/A | 0.005312 | 0.000007 | 0.000007 | 0.000006 | 0.001823 | 0.58 | 0.37 | 0.39 | 0.53 | 0.37 | 0.38 | 0.38 | 0.30 |
| 8 | rs1484190 | 2879144 | CSMD1 | T/A | 0.002964 | 0.000042 | 0.024529 | 0.000040 | 0.001511 | 0.60 | 0.39 | 0.40 | 0.62 | 0.35 | 0.35 | 0.36 | 0.42 |
| 8 | rs12707927a | 129274206 | PVT1 | A/G | 0.006739 | 8.95E-08 | 0.031231 | 6.49E-08 | 0.004214 | 0.16 | 0.05 | 0.05 | 0.08 | 0.04 | 0.07 | 0.06 | 0.06 |
| 10 | rs11239832 | 42739373 | BMS1 | T/G | 0.006675 | 0.000069 | 0.064082 | 0.000057 | 0.000746 | 0.23 | 0.09 | 0.12 | 0.22 | 0.09 | 0.08 | 0.10 | 0.12 |
| 10 | rs12784847 | 72181534 | ADAMTS14 | A/G | 0.004672 | 0.000072 | 0.331840 | 0.000069 | 0.002452 | 0.50 | 0.35 | 0.31 | 0.40 | 0.32 | 0.30 | 0.28 | 0.30 |
| 10 | rs3740434 | 72192195 | ADAMTS14 | T/C | 0.000808 | 0.000076 | 0.193978 | 0.000073 | 0.000226 | 0.55 | 0.37 | 0.36 | 0.46 | 0.35 | 0.31 | 0.29 | 0.41 |
| 10 | rs11591930 | 72198570 | ADAMTS14 | A/T | 0.000224 | 0.000026 | 0.152853 | 0.000025 | 0.000037 | 0.55 | 0.37 | 0.35 | 0.46 | 0.35 | 0.29 | 0.27 | 0.26 |
| 12 | rs1358652 | 126947387 | - | C/T | 0.001398 | 0.000014 | 0.148832 | 0.000011 | 0.000139 | 0.18 | 0.07 | 0.08 | 0.16 | 0.04 | 0.06 | 0.06 | 0.14 |
| 12 | rs764377 | 126947603 | - | A/G | 0.001194 | 0.000013 | 0.134585 | 0.000010 | 0.000092 | 0.18 | 0.07 | 0.08 | 0.16 | 0.04 | 0.06 | 0.07 | 0.14 |
| 12 | rs1463670a | 126948493 | - | C/A | 0.000692 | 0.000025 | 0.210417 | 0.000024 | 0.000288 | 0.20 | 0.07 | 0.08 | 0.18 | 0.04 | 0.08 | 0.08 | 0.14 |
| 12 | rs1463669a | 126948655 | - | G/A | 0.000707 | 0.000026 | 0.211419 | 0.000025 | 0.000289 | 0.20 | 0.07 | 0.08 | 0.18 | 0.04 | 0.08 | 0.08 | 0.14 |
| 12 | rs1463666 | 126949325 | - | C/T | 0.001194 | 0.000013 | 0.134585 | 0.000010 | 0.000092 | 0.18 | 0.07 | 0.08 | 0.16 | 0.04 | 0.06 | 0.07 | 0.14 |
| 13 | rs2503340 | 21708964 | - | G/T | 0.002605 | 0.000023 | 0.000112 | 0.000019 | 0.000531 | 0.49 | 0.36 | 0.33 | 0.44 | 0.31 | 0.29 | 0.29 | 0.17 |
| 14 | rs7146962 | 53215067 | - | G/A | 0.003006 | 0.000090 | 0.000017 | 0.000077 | 0.000907 | 0.64 | 0.44 | 0.47 | 0.63 | 0.40 | 0.43 | 0.43 | 0.24 |
| 22 | rs12167903a | 29096466 | CCDC157 | T/C | 0.000450 | 0.000056 | 0.000021 | 0.000054 | 0.000001 | 0.15 | 0.07 | 0.05 | 0.13 | 0.05 | 0.02 | 0.02 | 0.00 |
Naïve and BLUE allele frequencies and are shown.
aROADTRIPS gives a warning for Rχ.
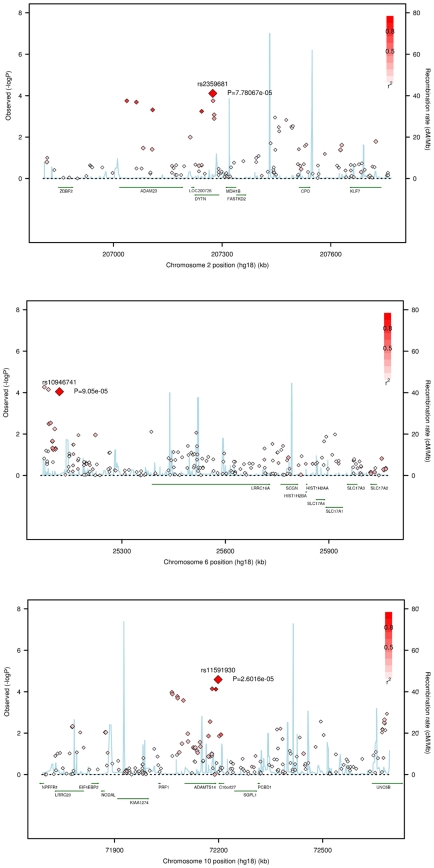
On chromosome 2p, different SNPs in LD which each other showed Rχ below the 1E-4 threshold. These SNPs and the top SNP (rs11125301) are located in introns of the NRXN1 gene.
Two other SNPs at different locations on 2q, rs1864466 and rs2359681, were identified with both Rχ and RM. The former, rs1864466, located in the 3′ of the ALS2CR8 gene, and the latter, rs2359681, is located in an intron of DYTN, and it is in LD with other SNPs located in the nearby ADAM23 gene, for which suggestive association is observed with the Rχ test (p-value = 0.00018, Figure 3 and Table S2).
Figure 3. Regional association plots for the Rχ test.
SNAP plots [45] for Rχ show the strength of association, −log10(p-values), versus chromosomal position (kb) for all SNPs across 1 Mb regions. P-values are plotted with diamonds for all SNPs, shaded white to red by the degree of LD (r2; see inset), estimated from the Talana sample, with the associated SNP (larger red diamond). Talana specific linkage disequilibrium (LD) between SNPs was computed using Haploview [46] from 179 more distantly related individuals selected from the whole sample of genotyped subjects, so that the kinship between any pairs of individuals did not exceed 0.125. Local recombination rates estimated from HapMap CEU (cM/Mb, blue line) are plotted against the secondary y axis, showing recombination hotspots across the region. Labeled green arrows below the plots indicate genes and their orientations.
On 6p three SNPs were associated with a Rχ p-value<1E-4, but only one, rs10946741, had also a RM p-value <1E-2 (p-value = 0.00026). Other SNPs in the region and in LD with rs10946741 showed marginal association. The highest association is found in a region near the 5′ of the LRRC16A gene, found to be associated in a large meta-analysis with serum uric acid levels [29], although different SNPs located in introns of LRRC16A or in introns in flanking genes (SLC17A4 and SLC17A1) showed association with either the Rχ or the RM statistics (Figure 3 and Table S3). Evidence of association through the RM test was observed at different SNPs located in introns of LRRC16A, and at SLC17A4, where a nonsense SNP provided a p-value = 0.00354. Strong LD is observed at SNPs in the SLC17A2 gene, but no evidence of association is observed with any of the test statistics.
On chromosome 8, the Rχ test resulted in the most significant p-value, 8.95E-08 over the genome (genome-wide significant after Bonferroni correction, p-value corrected = 0.03), at rs12707927. The closest gene to rs12707927, PVT1, lies 90kb upstream, and neighbouring SNPs located within the gene were only showing marginal significance. Also, this SNP was tagged by a warning message that the minor allele count was small, and as a result the p-value, which is calculated based on a null distribution assumption of a χ2 with 1 degree of freedom, may not be exact. Indeed the allele frequency estimated with the BLUE was 0.082 in cases and 0.038 in controls for the A allele (allele frequency is 0.062 in Hapmap-CEU). Note that this SNP was not removed in the QC stage because the estimated naïve allele frequency was 0.054, and therefore slightly above the set MAF threshold of 5%.
In a region on 10q, three SNPs in strong LD showed association (rs12784847, rs3740434, and rs11591930) in all statistics, with a lowest Rχ p-value of 0.00003 at rs11591930 (Figure 3 and Table S4). One of this SNP, rs12784847, is located in an intron of the ADAMTS14 gene. Further, different SNPs in LD with rs11591930, and located either in introns or in the 5′UTR of the gene showed nominal association (Rχ p-value<0.05), and one SNP (rs10999500) was a synonymous coding variant of the gene. Further, rs11591930 is tagging additional SNPs located in introns of the LRRC20 gene which showed marginal association (best Rχ p-value = 0.00469, and RM p-value = 0.00552), and in introns of the UNC5B gene (best Rχ p-value = 0.00116, and RM p-value = 0.00034).
Finally, a SNP located on chromosome 22, rs12167903, was a missense variant of the CCDC157 gene. ROADTRIPS gave the warning that Rχ p-value might not be correct because of the low MAF (which is 0.024 in the whole sample estimated by BLUE). This variant is indeed rare in the literature (2%), and resulted in a BLUE estimated frequency in our cases of 0.125 (SD = 0.060).
Other regions identified in the initial GWAS did not contain genes with a direct role in stones formation or were regions devoid of known genes. These regions were not examined further adding tSNPs in the replication set, but only the top SNPs obtained in the initial GWAS were typed in the independent sample.
Case-controls study in the replication set
The results of the analysis carried out for the 96 SNPs in the continental Italian, Sardinian, and merged sets (for a total of 282 individuals, 125 cases and 157 controls) are shown in Table 4. When considering the Sardinian and the continental Italian sample separately and merged together, using the CMH tests allowing for two strata, we observed association to UAN in the chromosome 2 and 6 regions.
Table 4. Results of the replication study.
| Continental Italy + Sardinia | Continental Italy | Sardinia | |||||||||||||||||
| ch | SNP | position | location | GENE | alleles | ref | cases | controls | CMH-P | OR | BD-P | cases | controls | P | OR | cases | controls | P | OR |
| 2 | rs11125301 | 50452354 | intron | NRXN1 | T,A | T | 0.55 | 0.48 | 0.09835 | 1.33 | NS | 0.59 | 0.48 | 0.03890 | 1.59 | 0.49 | 0.48 | NS | - |
| 2 | rs16838282 | 207036840 | intron | ADAM23 | G,A | G | 0.15 | 0.18 | NS | - | - | 0.12 | 0.20 | 0.04393 | 0.53 | 0.20 | 0.16 | NS | - |
| 2 | rs11891267 | 207063312 | intron | ADAM23 | A,G | A | 0.15 | 0.18 | NS | - | - | 0.11 | 0.20 | 0.02732 | 0.49 | 0.20 | 0.16 | NS | - |
| 2 | rs10194632 | 207066922 | intron | ADAM23 | A,G | A | 0.08 | 0.04 | 0.09925 | 1.80 | NS | 0.07 | 0.04 | NS | - | 0.10 | 0.06 | NS | - |
| 2 | rs3755224 | 207110679 | intron | ADAM23 | G,A | G | 0.15 | 0.18 | NS | - | - | 0.12 | 0.20 | 0.04393 | 0.53 | 0.20 | 0.16 | NS | - |
| 2 | rs4085933 | 207179129 | intron | ADAM23 | A,G | A | 0.51 | 0.47 | NS | - | - | 0.54 | 0.46 | NS | - | 0.48 | 0.47 | NS | - |
| 2 | rs6704787 | 207263993 | intron | DYTN | G,A | G | 0.05 | 0.03 | 0.08968 | 2.12 | NS | 0.07 | 0.03 | 0.04583 | 2.92 | 0.03 | 0.03 | NS | - |
| 6 | rs10946741 | 25118978 | flanking_5UTR | LRRC16A | A,G | A | 0.55 | 0.46 | 0.03393 | 1.44 | NS | 0.53 | 0.44 | NS | - | 0.58 | 0.48 | NS | - |
| 6 | rs12665174 | 25152787 | flanking_5UTR | LRRC16A | G,C | G | 0.14 | 0.24 | 0.00146 | 0.49 | NS | 0.13 | 0.20 | 0.08020 | 0.59 | 0.14 | 0.30 | 0.00502 | 0.40 |
| 6 | rs458664 | 25163144 | flanking_5UTR | LRRC16A | A,C | A | 0.41 | 0.34 | 0.07173 | 1.37 | NS | 0.39 | 0.35 | NS | - | 0.44 | 0.32 | 0.07511 | 1.63 |
| 6 | rs302966 | 25177342 | flanking_5UTR | LRRC16A | A,G | A | 0.18 | 0.14 | NS | - | NS | 0.19 | 0.16 | NS | - | 0.17 | 0.09 | 0.08541 | 1.99 |
| 6 | rs1002539 | 25199319 | flanking_5UTR | LRRC16A | G,A | G | 0.22 | 0.28 | 0.08344 | 0.71 | NS | 0.21 | 0.26 | NS | - | 0.22 | 0.31 | NS | - |
| 6 | rs3792970 | 25208104 | flanking_5UTR | LRRC16A | G,A | G | 0.27 | 0.21 | 0.08196 | 1.42 | NS | 0.26 | 0.23 | NS | - | 0.29 | 0.18 | 0.05244 | 1.85 |
| 6 | rs3812105 | 25212233 | flanking_5UTR | LRRC16A | C,A | C | 0.36 | 0.29 | 0.07306 | 1.39 | NS | 0.36 | 0.33 | NS | - | 0.36 | 0.23 | 0.03228 | 1.87 |
| 6 | rs302976 | 25226413 | flanking_5UTR | LRRC16A | A,G | A | 0.26 | 0.19 | 0.05305 | 1.49 | NS | 0.25 | 0.19 | NS | - | 0.27 | 0.18 | NS | - |
| 6 | rs302970 | 25236412 | flanking_5UTR | LRRC16A | A,G | A | 0.35 | 0.27 | 0.03079 | 1.49 | NS | 0.36 | 0.30 | NS | - | 0.34 | 0.23 | 0.06291 | 1.73 |
| 6 | rs2149228 | 25502652 | intron | LRRC16 | T,A | T | 0.07 | 0.12 | 0.03741 | 0.53 | NS | 0.07 | 0.12 | NS | - | 0.06 | 0.12 | NS | - |
| 8 | rs12707927 | 129274206 | flanking_5UTR | PVT1 | A,G | A | 0.04 | 0.06 | NS | - | - | 0.07 | 0.04 | NS | - | 0.02 | 0.08 | 0.02260 | 0.20 |
The table shows only SNPs with a p-value<0.05 in at least one of the sample (p-values<0.01 are bold and underlined; p-values<0.05 are bold; p-values>0.1 are indicated with NS). The Cochran-Mantel-Haenszel tests for stratified tables (CMH-P), and the Breslow-Day (BD-P) homogeneity of odds ratio tests are shown. Bold SNPs are those analyzed in the initial GWAS. Naïve allele frequencies are shown. Talana - All samples: results with ROADTRIPS considering all related cases and controls; Talana - Sub-samples: results with PLINK considering distantly related cases and controls. The latter samples were used in the Continental Italy + Sardinia + Talana merged set.
In the replication set on 2q33.3 no significant association was observed when considering the merged sample, but nominal significance was obtained at three different SNPs in the continental Italian sample. The SNPs are located in the introns of ADAM23, with the highest evidence at rs11891267 (p-value = 0.02732). The same SNPs were also found to be associated in the Talana sample, but the allele frequencies were oppositely distributed in cases and controls. In the whole Talana sample (Table S5) two SNPs intronic to ADAM23 (rs1025077 and rs3755224, Rχ p-value = 0.01884, and Rχ p-value = 0.00069, respectively), and a SNP in the intron region of DYTN (rs2163033, Rχ p-value = 0.00818) were additionally found to be associated to UAN.
In the region encompassing the LRRC16 gene on 6p22.2–p21.3, different SNPs showed marginal significance in the upstream region of the gene with peak evidence at rs12665174 with a CMH p-value of 0.00146. One SNP (rs2149228) located in the intronic region of the gene also showed nominal significance in the merged replication set. All the SNPs but rs10946741 (identified in the initial study and not found significant in the replication set, i.e. CMH p-value of 0.09925) had the same allele more frequent in cases compared to controls, both within each strata (Sardinia and continental Italy samples) and in the Talana cohort. Therefore, when considering the distantly related cases and controls from Talana as an additional strata in a merged dataset, evidence for association in the region increased for all SNPs with the highest evidence at rs12665174 (CMH p-value = 0.00085). When analyzing the continental Italy and Sardinian samples separately, significant evidence for association was observed only in the Sardinian sample, with the highest evidence at rs12665174 (p-value = 0.00502). When looking at the results obtained in the chromosome 6 region with ROADTRIPS in the whole Talana sample (Table S5), two additional SNPs showed nominal significance in the LRRC16 region (rs9461102, located upstream of LRRC16, and rs880226, located in an intron of the gene), and one additional SNP showed a Rχ p-value of 0.015627 at SLC17A1. None of these SNPs showed evidence for association in the replication set.
No other SNP identified in the initial study showed nominal significance in the merged replication sample, but two SNPs showed evidence for association in specific sub-samples: rs11125301 located in an intron of NRXN1 on chromosome 2 was associated in the Italian sample only, and with a different allele more frequent in cases compared to Talana; and rs12707927 on chromosome 8, that provided the highest evidence in the initial study, and for which a different allele was associated only at a nominal level in the Sardinian replication set (p-value = 0.02260), but with allele frequencies in cases and controls in the opposite direction compared to Talana. For this SNP, in the analysis with ROADTRIPS without considering the unknown population controls, the RM and the Rχ tests were both tagged with a warning message due to the low MAF of the SNP, and were less significant than in the initial scan RM p-value = 0.00020 and Rχ p-value = 0.00079.
On chromosome 10 only one SNP (rs826460) showed marginal evidence for Rχ in the flanking 5UTR region of ADAMTS14 in the whole Talana sample with ROADTRIPS, whereas none of the replication sets, nor the merged sample showed any evidence in this region.
Finally, in the initial scan, we identified a missense variant of the CCDC157 gene, located on chromosome 22, whose frequency estimated with BLUE was increased in affected cases (12.5%), compared to either unaffected controls (5.0%) or population controls (2.2%). The population control frequency is comparable to the 2% frequency reported in the literature. This variant is too rare to be identified in the relatively small replication set, and we did observed any significant results, nor in the merged Italian samples, not considering the two distinct geographical origins.
Discussion
In the present case-control GWAS including 73 stones formers and 92 controls, all related to each other, and deriving from an isolated Sardinian village, we identified different SNPs that showed suggestive associations with UAN.
We applied a recently proposed method [20], ROADTRIPS, that allows for the analysis of the complex type of data we have, and we showed the improvement of the method in this founder population over previously proposed methods implemented in the MQLS software [18]. Indeed, providing the pair-wise kinship for all pairs of cases and controls was not sufficient to control for spurious association in our dataset using the MQLS test, as additional structure was still present. The statistics implemented in the MQLS software do not use an empirical structure matrix, and, in the presence of additional cryptic relatedness or unknown population structure, we observed inflated type 1 error. The remaining population structure was accounted for in the RM test implemented in the ROADTRIPS software by using an empirical covariance matrix calculated using genome-wide data, while also incorporating known genealogical information about the cases and controls into the analysis. In contrast, both χ2 corrected statistics (either corrected on pedigree or on genome data) showed similar results, indicating that with a sufficiently well-characterized genealogy data, the corrected χ2 test as implemented in the MQLS software shows less inflation of type 1 errors over the genome.
A deviation from the χ2 null distribution was observed throughout the genome for both RM and MQLS for SNPs with rarer alleles (MAF<0.1), which is an artifact of the small number of samples in our study. Furthermore, we observed that for a number of SNPs, the difference between RM and Rχ was largely being driven by the complex pedigree structure in the sample and the small number of founders (see Text S2). For samples like the Talana sample (as well as samples from founder populations like the Hutterites) with only a small number of founders, it actually is not clear at this time if a reasonable assessment of p-values can be obtained in the extreme tail of the χ2 distribution (e.g., genome-wide significance p-values <1E-8), and this is future research to be conducted.
A small sample is expected and unavoidable when focusing on small, isolated villages like Talana with only 1,200 inhabitants. Nevertheless, we were able to identify suggestive candidate genes for UAN in the initial GWAS, and to validate some of them in an independent Italian sample of well characterize cases and controls. Based on the associated SNPs in the initial scan, and their tagged SNPs (basing on LD pattern in Talana), we identified candidate genes on 2q33.3, 6p22.2–p21.3 and 10q22.1, that were particularly interesting for UAN due to their physiological function. These regions were also investigated in the independent samples by typing additional tSNPs. Since the geographical origin of the replication samples were either continental Italy or Sardinia, we considered these two distinct groups in the statistical analysis using the CMH test and tested the homogeneity of odds ratio by the BD test.
The 6p22.2 region contains the LRRC16A, SLC17A1, SLC17A4 genes, and was identified in the initial scan with a significance at LRRC16A of Rχ p-value = 0.00863, and RM p-value = 0.00306, at SLC17A1 of Rχ p-value = 0.01048, and at SLC17A4 for RM p-value = 0.00354 at a nonsense SNP (rs2328894). Interesting, Kolz and colleagues [29], in a meta-analysis of 14 GWAs including a total of 28,141 participants, identified the same genes (except SLC17A4) significantly associated with serum UA levels. Therefore, peak SNPs in the region (Table S3) and additional 16 tSNPs for LRRC16A and 6 tSNPs for SLC17A4-SLC17A1 were typed in the replication set. Interestingly, different SNPs showed significant association in the upstream and intronic regions of the LRRC16 gene in the merged replication sample, with the highest evidence at rs12665174 (CMH p-value = 0.00146). Most of the associated SNPs in the region showed the same allele more frequent in cases compared to controls, both within each strata (Sardinia and Italy samples) and in the Talana cohort. Therefore, when considering the distantly related cases and controls from Talana as an additional strata in a merged sample, evidence for association in the region increased with the highest evidence of CMH p-value = 0.00085 at rs12665174. When analyzing the Italian and Sardinian samples separately, significant evidence for association was observed only in the Sardinian sample, with the highest evidence at rs12665174 (p-value = 0.00502). When looking at the results obtained in the chromosome 6 region with ROADTRIPS in the whole Talana sample (Table S5), 2 additional SNPs showed nominal significance in the LRRC16 region (rs9461102, located in the upstream region of LRRC16, and rs880226, located in an intron of the gene), and one additional SNP showed a Rχ p-value of 0.01563 at rs1165208, located in the intronic region of SLC17A1 (Table S5). None of these SNPs showed evidence for association in the replication set, and no association was observed in the replication set in the SLC17A4-SLC17A1 region.
The LRRC16A gene is, for the larger part, located in an LD block encompassing also SCGN. In this study the coverage obtained by adding tSNPs in this region was only 51% of the total variation with an r2 of at least 0.8, therefore further studies are needed to validate the involvement of these genes to UAN.
On 2q33.3, different SNPs showed association in the initial scan, with a peak at rs2359681 identified with both Rχ and RM (p-value = 0.00008 and p-value = 0.00254, respectively). The SNP is located in an intron of DYTN, and it is in LD with other associated SNPs located in the nearby ADAM23 gene, for which suggestive association is observed (Figure 3 and Table S2). In the replication set no significant association was observed when considering the Italian and Sardinia samples together, but nominal significance was obtained at different SNPs located in the introns of ADAM23, with the highest evidence at rs11891267 (p-value of 0.02732) in the Italian sample alone. The allele frequencies were oppositely distributed in cases and controls compared to Talana, suggesting that putative causal variant/s at the gene implicated in UAN etiology are in LD with different alleles at the SNPs examined. In the whole Talana sample (Table S5) two tSNPs intronic to ADAM23 (rs1025077 and rs3755224, Rχ p-value = 0.01884, and p-value = 0.00069, respectively) and a SNP in the intron region of DYTN (rs2163033, Rχ p-value = 0.00818) were additionally found to be associated in the replication study.
The other interesting region identified in the initial study and investigated further in the replication set was on 10q22.1, with the highest association evidence observed in the initial study at the ADAMTS14 gene. (Rχ p-value = 0.00003; RM p-value = 0.00022). Two other genes were found to be associated in the region on 10q, namely LRRC20 and UNC5B (most significant RM p-value = 0.00552, and RM p-value = 0.00034, respectively). Interestingly, the SNPs identified on chromosome 10 are located within the critical region identified through linkage analysis in Talana [14]. In the previous study we performed a genome-wide linkage search in 14 closely-related affected individuals using 382 microsatellites, and followed up suggestive regions on 37 individuals more distantly-related affecteds. The original linkage region spanned approximately 9 Mb, with the second highest peak at D10S537 (position ∼72,065 kb), located in the upstream region of ADAMTS14. In the replication set we did not observe any signal of association in either the ADAMTS14 region or in the UNC5B region, although by typing additional tSNPs. In the Talana sample a SNP located in the upstream region of ADAMTS14 showed marginal evidence of association (Rχ p-value = 0.04647 at rs826460, Table S5), but the SNPs that showed association in the original scan when also including the unknown phenotype controls in the analysis, were not found to be significantly associated in the replication study. Further analyses are needed to evaluate the role of this region in UAN etiology.
Among the remaining top SNPs identified in the initial GWAS only one showed marginal evidence for association in a specific sub-sample: rs11125301 located in an intron of NRXN1 on chromosome 2 was only found to be associated in the Italian sample and with a different allele that is more frequent in cases compared to the Talana sample.
In conclusion, we obtained evidence for association to UAN for some interesting genes in this study, whereas further investigation is needed to validate the involvement of other genes/regions identified in the initial GWAS. In particular, LRRC16A, already associated to serum UA levels from previous studies, encodes for CARMIL protein, an inhibitor of actin capping protein (CP) and has profound effects on cell behavior. Removal of CP may be a means to harness actin polymerization for processes such as cell movement and endocytosis and plays important roles in intracellular transport (the movement of vesicles and organelles). It is interesting that this protein showed the highest expression in kidney and other epithelial tissues [30]. The mechanism by which variants at this gene regulate UA remains unclear. We can envisage that this gene may be involved with the kidney, for example, in podocytes that are glomerular cells with an actin-based contractile apparatus and they are insulin sensitive [31]. The insulin response of the podocytes occurs via the facilitative glucose transporters GLUT1 and GLUT4, and this process is dependent on the filamentous actin cytoskeleton [32]. Insulin responsiveness in this key structural component of the glomerular filtration barrier may have a central role in the establishment of states of insulin resistance. Different studies have emphasized the increasing importance of insulin resistance in the pathogenesis of UA stones and insulin resistance is strongly correlated with low urine pH [33]. Numerous epidemiologic studies have shown a significant association between nephrolithiasis, obesity, glucose intolerance, type 2 diabetes mellitus, hypertension and chronic kidney disease [10], [34]–[37]. There are likely still many unrecognized renal manifestations of the metabolic syndrome. UAN, secondary to low urine pH, might only be the tip of the iceberg. Nevertheless, UA stone formers may have yet undisclosed mechanisms leading to unduly low urinary pH that are not entirely accounted for by insulin resistance [33]. Similarly, we can envisage that the F-actin reorganization is important also in tubular cells of kidney for proteins sorting directly involved in metabolism of UA. For the different endophenotypes we examined (Table S6), we observed normal serum parameters and not significant differences between cases and controls (after correcting for age and sex). Indeed in Talana we observed a general low urinary pH (Figure S3), significantly lower than the distribution in the general population (95%CI = [5.4;5.7]), that could explain the high proportion of UAN cases among renal stones formers.
The ADAM23 gene, for which nominal significance was observed in the Italian replication sample, encodes a member of the ADAM (a disintegrin and metalloprotease domain) family. ADAMs are membrane-anchored cell surface proteins with putative roles in cell–cell and/or cell–matrix interactions and in protease activities [38]. Members of this family have a unique structural organization including metalloprotease, desintegrin, cystein-rich, epidermal growth factor-like, transmembrane and cytoplasmatic domains [38]. The available data indicate that three of the ADAM family members are expressed at high levels in normal brain (ADAMs 11, 22, and 23) while other members are either expressed in the testis or are ubiquitous. More recently Ru et al. [39] detected the ADAM23 protein in Human urine samples. ADAM23 exhibits the typical structure of the ADAM family members; however, the metalloproteinase domain is inactive, suggesting that it is exclusively involved in cell adhesion. The disintegrin and cysteine-rich domain of ADAMs have been shown to interact with cell adhesion molecules including the receptors of the extracellular matrix, integrins [40], as well as proteoglycans (e.g.syndecans) [41]. It is interesting that the proteoglycans (GAGs) are inhibitors of crystallization and appear to be involved in kidney stone formation. In a previous study we showed that the lower excretion of GAGs in stone formers could impair their inhibitory activity on UA stone formation, and, as a consequence, it may represent a risk factor for this form of urolithiasis [42]. Furthermore, a proteoglycan like Syndecan-4 was up-regulated in proliferative renal disease and mice deficient in syndecan-4 were more susceptible to κ-carrageenan induced renal damage indicating that syndecan-4 plays an important role in renal diseases [43]. Finally, Hwang et al. [44] reported a strong association with ADAM23 for urinary albumin excretion, that is a marker of kidney function.
Due to the small sample of affected subjects used in the initial scan, statistical power was consequently relatively low in this study, and indeed the significance of the evidence for association with the identified SNPs is lower than genome-wide significance considering Bonferroni correction. On the other hand, we have the advantage of using a homogenous cohort of individuals, sharing a very similar life style and dietary habits, and with an increased genetic homogeneity, as a consequence of a strong founder effect and of genetic drift deriving from isolation that endured for centuries. A consequence of association studies in founder populations can be lower statistical power due to having small sample sizes. A compelling advantage, however, for such samples is increased homogeneity in terms of both environmental and genetic factors involved in disease etiology, which can ultimately improve the power to detect association. Although our sample was relatively small, we were nevertheless able to identify different candidate genes with a potential role in UAN, and to provide evidence for association in an independent sample for the gene LRRC16A on 6p, already found to be associated to serum UA levels in a large meta-analysis of 14 GWAS and possibly for ADAM23 on 2q.
To our knowledge this GWAS is the first one carried out for UAN. It is also the first application of ROADTRIPS to a founder population. The original application of ROADTRIPS [20] was to both simulated and real data in samples from outbred populations. The sample sizes of the cases and/or the controls in the previous applications used to evaluate the method were also more than five times the sample we analyzed in this study. We were able to evaluate the performance of the method using real data from a small sample in a genetic isolate, which likely has different properties and complexities than the data sets previously used to evaluate the type 1 error and power of ROADTRIPS.
Supporting Information
MAF corresponding to warnings in ROADTRIPS. Box-plots for the MAF (naïve estimates) for the statistics tagged by a warning message for each specific case/control cohort. Box-plots are shown for the 500K and 50K sets separately, as a different number of individuals were genotyped for the two sets (829 and 514 subjects, respectively).
(0.08 MB TIF)
Q-Q plots for RM and Rχ stratified by warning messages in ROADTRIPS. For the Rχ statistic warning messages entailing the case, controls or both samples were considered together.
(0.06 MB TIF)
pH distribution in Talana (N = 218). Red bars indicate pH in affected cases (N = 43). The red line indicates the pH level ( = 6) in the general population.
(0.20 MB TIF)
Tagged regions for the replication study and resulting coverage based on r2 >0.8.
(0.03 MB DOC)
Region 2q.
(0.07 MB DOC)
Region 6p.
(0.09 MB DOC)
Region 10q.
(0.11 MB DOC)
Results obtained with ROADTRIPS for the whole Talana sample in the replication study for the 96 SNPs.
(0.10 MB DOC)
Phenotypic characteristics of the case/control sample used in this analysis.
(0.04 MB DOC)
Quality control of genome-wide SNP data in Talana.
(0.31 MB DOC)
Investigating SNPs in the Talana sample.
(0.97 MB PDF)
Acknowledgments
The authors would like to thank Marcella Devoto for her helpful comments and suggestions on this study. We thank the Talana population and all the individuals who participated in this study. We are very grateful to the municipal administrators for their collaboration to the project and for economic and logistic support.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by grants from the Italian Ministry of Education, University, and Research (MIUR) n: 5571/DSPAR/2002 and (FIRB) DM n.718/Ric/2005. TT was supported by research grant K01 CA 148958 from the National Cancer Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rivers K, Shetty S, Menon M. When and how to evaluate a patient with nephrolithiasis. Urol Clin North Am. 2000;27:203–213. doi: 10.1016/s0094-0143(05)70251-2. [DOI] [PubMed] [Google Scholar]
- 2.Stamatelou KK, Francis ME, Jones CA, Nyberg LM, Curhan GC. Time trends in reported prevalence of kidney stones in the United States: 1976–1994. Kidney Int. 2003;63:1817–1823. doi: 10.1046/j.1523-1755.2003.00917.x. [DOI] [PubMed] [Google Scholar]
- 3.Soucie JM, Thun MJ, Coates RJ, McClellan W, Austin H. Demographic and geographic variability of kidney stones in the United States. Kidney Int. 1994;46:893–899. doi: 10.1038/ki.1994.347. [DOI] [PubMed] [Google Scholar]
- 4.Pak CY, Poindexter JR, Adams-Huet B, Pearle MS. Predictive value of kidney stone composition in the detection of metabolic abnormalities. Am J Med. 2003;115:26–32. doi: 10.1016/s0002-9343(03)00201-8. [DOI] [PubMed] [Google Scholar]
- 5.Bid HK, Chaudhary H, Mittal RD. Association of vitamin-D and calcitonin receptor gene polymorphism in paediatric nephrolithiasis. Pediatr Nephrol. 2005;20:773–776. doi: 10.1007/s00467-005-1846-4. [DOI] [PubMed] [Google Scholar]
- 6.Onaran M, Yilmaz A, Sen I, Ergun MA, Camtosun A, et al. A HindIII polymorphism of fibronectin gene is associated with nephrolithiasis. Urology. 2009;74:1004–1007. doi: 10.1016/j.urology.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Sayer JA. The genetics of nephrolithiasis. Nephron Exp Nephrol. 2008;110:e37–43. doi: 10.1159/000151730. [DOI] [PubMed] [Google Scholar]
- 8.Brikowski TH, Lotan Y, Pearle MS. Climate-related increase in the prevalence of urolithiasis in the United States. Proc Natl Acad Sci U S A. 2008;105:9841–9846. doi: 10.1073/pnas.0709652105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez-Nemegyei J, Medina-Escobedo M, Villanueva-Jorge S, Vazquez-Mellado J. Prevalence and risk factors for urolithiasis in primary gout: is a reappraisal needed? J Rheumatol. 2005;32:2189–2191. [PubMed] [Google Scholar]
- 10.Sakhaee K. Nephrolithiasis as a systemic disorder. Curr Opin Nephrol Hypertens. 2008;17:304–309. doi: 10.1097/MNH.0b013e3282f8b34d. [DOI] [PubMed] [Google Scholar]
- 11.Pak CY, Sakhaee K, Peterson RD, Poindexter JR, Frawley WH. Biochemical profile of idiopathic uric acid nephrolithiasis. Kidney Int. 2001;60:757–761. doi: 10.1046/j.1523-1755.2001.060002757.x. [DOI] [PubMed] [Google Scholar]
- 12.Angius A, Hyland FC, Persico I, Pirastu N, Woodage T, et al. Patterns of linkage disequilibrium between SNPs in a Sardinian population isolate and the selection of markers for association studies. Hum Hered. 2008;65:9–22. doi: 10.1159/000106058. [DOI] [PubMed] [Google Scholar]
- 13.Fraumene C, Belle EM, Castri L, Sanna S, Mancosu G, et al. High resolution analysis and phylogenetic network construction using complete mtDNA sequences in sardinian genetic isolates. Mol Biol Evol. 2006;23:2101–2111. doi: 10.1093/molbev/msl084. [DOI] [PubMed] [Google Scholar]
- 14.Ombra MN, Forabosco P, Casula S, Angius A, Maestrale G, et al. Identification of a new candidate locus for uric acid nephrolithiasis. Am J Hum Genet. 2001;68:1119–1129. doi: 10.1086/320105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gianfrancesco F, Esposito T, Ombra MN, Forabosco P, Maninchedda G, et al. Identification of a novel gene and a common variant associated with uric acid nephrolithiasis in a Sardinian genetic isolate. Am J Hum Genet. 2003;72:1479–1491. doi: 10.1086/375628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourgain C, Hoffjan S, Nicolae R, Newman D, Steiner L, et al. Novel case-control test in a founder population identifies P-selectin as an atopy-susceptibility locus. Am J Hum Genet. 2003;73:612–626. doi: 10.1086/378208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slager SL, Schaid DJ. Evaluation of candidate genes in case-control studies: a statistical method to account for related subjects. Am J Hum Genet. 2001;68:1457–1462. doi: 10.1086/320608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thornton T, McPeek MS. Case-control association testing with related individuals: a more powerful quasi-likelihood score test. Am J Hum Genet. 2007;81:321–337. doi: 10.1086/519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasky-Su J, Won S, Mick E, Anney RJ, Franke B, et al. On genome-wide association studies for family-based designs: an integrative analysis approach combining ascertained family samples with unselected controls. Am J Hum Genet. 2010;86:573–580. doi: 10.1016/j.ajhg.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thornton T, McPeek MS. ROADTRIPS: case-control association testing with partially or completely unknown population and pedigree structure. Am J Hum Genet. 2010;86:172–184. doi: 10.1016/j.ajhg.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollack HM, Arger PH, Goldberg BB, Mulholland SG. Ultrasonic detection of nonopaque renal calculi. Radiology. 1978;127:233–237. doi: 10.1148/127.1.233. [DOI] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 25.McPeek MS, Wu X, Ober C. Best linear unbiased allele-frequency estimation in complex pedigrees. Biometrics. 2004;60:359–367. doi: 10.1111/j.0006-341X.2004.00180.x. [DOI] [PubMed] [Google Scholar]
- 26.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 27.Falchi M, Forabosco P, Mocci E, Borlino CC, Picciau A, et al. A genomewide search using an original pairwise sampling approach for large genealogies identifies a new locus for total and low-density lipoprotein cholesterol in two genetically differentiated isolates of Sardinia. Am J Hum Genet. 2004;75:1015–1031. doi: 10.1086/426155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Browning SR, Briley JD, Briley LP, Chandra G, Charnecki JH, et al. Case-control single-marker and haplotypic association analysis of pedigree data. Genet Epidemiol. 2005;28:110–122. doi: 10.1002/gepi.20051. [DOI] [PubMed] [Google Scholar]
- 29.Kolz M, Johnson T, Sanna S, Teumer A, Vitart V, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang C, Pring M, Wear MA, Huang M, Cooper JA, et al. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev Cell. 2005;9:209–221. doi: 10.1016/j.devcel.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faul C, Asanuma K, Yanagida-Asanuma E, Kim K, Mundel P. Actin up: regulation of podocyte structure and function by components of the actin cytoskeleton. Trends Cell Biol. 2007;17:428–437. doi: 10.1016/j.tcb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Coward RJ, Welsh GI, Yang J, Tasman C, Lennon R, et al. The human glomerular podocyte is a novel target for insulin action. Diabetes. 2005;54:3095–3102. doi: 10.2337/diabetes.54.11.3095. [DOI] [PubMed] [Google Scholar]
- 33.Abate N, Chandalia M, Cabo-Chan AV, Jr, Moe OW, Sakhaee K. The metabolic syndrome and uric acid nephrolithiasis: novel features of renal manifestation of insulin resistance. Kidney Int. 2004;65:386–392. doi: 10.1111/j.1523-1755.2004.00386.x. [DOI] [PubMed] [Google Scholar]
- 34.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 35.Deitel M. Overweight and obesity worldwide now estimated to involve 1.7 billion people. Obes Surg. 2003;13:329–330. doi: 10.1381/096089203765887598. [DOI] [PubMed] [Google Scholar]
- 36.Taylor EN, Stampfer MJ, Curhan GC. Obesity, weight gain, and the risk of kidney stones. Jama. 2005;293:455–462. doi: 10.1001/jama.293.4.455. [DOI] [PubMed] [Google Scholar]
- 37.Taylor EN, Stampfer MJ, Curhan GC. Diabetes mellitus and the risk of nephrolithiasis. Kidney Int. 2005;68:1230–1235. doi: 10.1111/j.1523-1755.2005.00516.x. [DOI] [PubMed] [Google Scholar]
- 38.Primakoff P, Myles DG. The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet. 2000;16:83–87. doi: 10.1016/s0168-9525(99)01926-5. [DOI] [PubMed] [Google Scholar]
- 39.Ru QC, Katenhusen RA, Zhu LA, Silberman J, Yang S, et al. Proteomic profiling of human urine using multi-dimensional protein identification technology. J Chromatogr A. 2006;1111:166–174. doi: 10.1016/j.chroma.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 40.White JM. ADAMs: modulators of cell-cell and cell-matrix interactions. Curr Opin Cell Biol. 2003;15:598–606. doi: 10.1016/j.ceb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Thodeti CK, Albrechtsen R, Grauslund M, Asmar M, Larsson C, et al. ADAM12/syndecan-4 signaling promotes beta 1 integrin-dependent cell spreading through protein kinase Calpha and RhoA. J Biol Chem. 2003;278:9576–9584. doi: 10.1074/jbc.M208937200. [DOI] [PubMed] [Google Scholar]
- 42.Ombra MN, Casula S, Biino G, Maestrale G, Cardia F, et al. Urinary glycosaminoglycans as risk factors for uric acid nephrolithiasis: case control study in a Sardinian genetic isolate. Urology. 2003;62:416–420. doi: 10.1016/s0090-4295(03)00473-4. [DOI] [PubMed] [Google Scholar]
- 43.Ishiguro K, Kadomatsu K, Kojima T, Muramatsu H, Matsuo S, et al. Syndecan-4 deficiency increases susceptibility to kappa-carrageenan-induced renal damage. Lab Invest. 2001;81:509–516. doi: 10.1038/labinvest.3780259. [DOI] [PubMed] [Google Scholar]
- 44.Hwang SJ, Yang Q, Meigs JB, Pearce EN, Fox CS. A genome-wide association for kidney function and endocrine-related traits in the NHLBI's Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S10. doi: 10.1186/1471-2350-8-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MAF corresponding to warnings in ROADTRIPS. Box-plots for the MAF (naïve estimates) for the statistics tagged by a warning message for each specific case/control cohort. Box-plots are shown for the 500K and 50K sets separately, as a different number of individuals were genotyped for the two sets (829 and 514 subjects, respectively).
(0.08 MB TIF)
Q-Q plots for RM and Rχ stratified by warning messages in ROADTRIPS. For the Rχ statistic warning messages entailing the case, controls or both samples were considered together.
(0.06 MB TIF)
pH distribution in Talana (N = 218). Red bars indicate pH in affected cases (N = 43). The red line indicates the pH level ( = 6) in the general population.
(0.20 MB TIF)
Tagged regions for the replication study and resulting coverage based on r2 >0.8.
(0.03 MB DOC)
Region 2q.
(0.07 MB DOC)
Region 6p.
(0.09 MB DOC)
Region 10q.
(0.11 MB DOC)
Results obtained with ROADTRIPS for the whole Talana sample in the replication study for the 96 SNPs.
(0.10 MB DOC)
Phenotypic characteristics of the case/control sample used in this analysis.
(0.04 MB DOC)
Quality control of genome-wide SNP data in Talana.
(0.31 MB DOC)
Investigating SNPs in the Talana sample.
(0.97 MB PDF)