Abstract
Background
Unlike in HIV-1 infection, the majority of HIV-2 patients produce broadly reactive neutralizing antibodies, control viral replication and survive as elite controllers. The identification of the molecular, structural and evolutionary footprints underlying these very distinct immunological and clinical outcomes may lead to the development of new strategies for the prevention and treatment of HIV infection.
Methodology/Principal Findings
We performed a side-by-side molecular, evolutionary and structural comparison of the C2, V3 and C3 envelope regions from HIV-1 and HIV-2. These regions contain major antigenic targets and are important for receptor binding. In HIV-2, these regions also have immune modulatory properties. We found that these regions are significantly more variable in HIV-1 than in HIV-2. Within each virus, C3 is the most entropic region followed by either C2 (HIV-2) or V3 (HIV-1). The C3 region is well exposed in the HIV-2 envelope and is under strong diversifying selection suggesting that, like in HIV-1, it may harbour neutralizing epitopes. Notably, however, extreme diversification of C2 and C3 seems to be deleterious for HIV-2 and prevent its transmission. Computer modelling simulations showed that in HIV-2 the V3 loop is much less exposed than C2 and C3 and has a retractile conformation due to a physical interaction with both C2 and C3. The concealed and conserved nature of V3 in the HIV-2 is consistent with its lack of immunodominancy in vivo and with its role in preventing immune activation. In contrast, HIV-1 had an extended and accessible V3 loop that is consistent with its immunodominant and neutralizing nature.
Conclusions/Significance
We identify significant structural and functional constrains to the diversification and evolution of C2, V3 and C3 in the HIV-2 envelope but not in HIV-1. These studies highlight fundamental differences in the biology and infection of HIV-1 and HIV-2 and in their mode of interaction with the human immune system and may inform new vaccine and therapeutic interventions against these viruses.
Introduction
Human Immunodeficiency Virus type 1 (HIV-1) infection affects more than 40 million individuals throughout the world. It is caused mainly by isolates belonging to group M. Within this group there are nine different subtypes named A to H, six subsubtypes (F1, F2, A1–A4) and at least thirty six recombinant forms named CRF01 up to CRF36 [1]. In contrast to the HIV-1 pandemic, HIV-2 is only prevalent in West Africa where it seems to have been present since the 1940s [2]. In Europe infection with HIV-2 remains rare (2–3% of all AIDS cases), being observed mainly in France and Portugal [3], [4], [5]. Eight different HIV-2 groups named A through H have been reported but only groups A and B cause human epidemics [6], [7], [8], [9]. Isolates from group A are, however, responsible for the vast majority of HIV-2 infections worldwide [10].
For reasons that are still not clear, HIV-1 and HIV-2 infections lead to very different immunological and clinical outcomes. In contrast to HIV-1 infected patients, the majority of HIV-2-infected individuals have reduced general immune activation, normal CD4+ T cell counts, low or absent viremia and absence of clinical disease [11], [12], [13], [14]. This may be related with a more effective immune response produced against HIV-2. In fact, most HIV-2 infected individuals have strong cytotoxic responses to Env and Gag proteins and raise autologous and heterologous neutralizing antibodies [3], [15], [16], [17], [18]. The attenuated course of HIV-2 infection compared to HIV-1 has also been associated to a lower state of immune activation, which may be related to the immunosuppressive activity of the C2-V3-C3 envelope region [19], [20], [21]. Similar immunosuppressive activity has not been found in the homologous C2-V3-C3 region in the HIV-1 envelope [19]. Finally, the transmission rate of HIV-2 is also significantly lower than that of HIV-1 and this has been associated with the low or absent viremia found in most HIV-2 patients [22], [23].
The HIV-1 Env glycoprotein is a trimer on the virion surface with extensive N-linked glycosylation that effectively shields many conserved epitopes from antibody recognition [24]. It is composed of trimers of a surface (SU) glycoprotein with a molecular weight of 120–125 kDa (gp120–125) that is bound to a transmembrane (TM) glycoprotein with 36–41 kDa (gp36–41). SU can be divided into five hipervariable regions, named V1 to V5, bordered by five conserved regions, named C1 to C5. The C2 and C3 regions associate to form the CD4 binding site such that mutations in amino acid at positions 267Q in C2 and 368R in C3 abrogate gp120 binding to CD4 [25], [26]. In HIV-1, V3 is one of the most important determinants of viral tropism and co-receptor usage [27], [28]. This region also contains major antigenic and neutralizing epitopes in HIV-1 which are well exposed upon CD4-binding [29], [30], [31], [32], [33], [34], [35]. Although still debatable, the V3 region in HIV-2 may also contain broadly neutralizing epitopes [36], [37], [38], [39], [40], [41], [42]. However, in contrast to HIV-1, the V3 and flanking C2 and C3 regions are not immunodominant in HIV-2 infected patients [43], [44], [45], [46]. Moreover, it remains to be determined whether these regions are exposed or concealed in the envelope complex of primary isolates of HIV-2.
In HIV-1 infection escape from antibody neutralization occurs frequently and is the major driving force of the molecular evolution of the envelope glycoproteins [47], [48]. Not surprisingly, codons under diversifying selection (positive selection) seem to be clustered mostly in the hypervariable V1/V2 and V3 regions that contain important and accessible neutralizing targets [49], [50]. The impact of the neutralizing antibody response in the in vivo evolution of the HIV-2 Env is currently unknown.
The present study was designed to identify molecular and evolutionary features of the C2, V3 and C3 regions in HIV-1 and HIV-2 infected patients that could be related with their different immunological and clinical outcomes. We describe some potentially important differences in the genetic constitution, molecular evolution and conformation of the C2, V3 and C3 regions in HIV-1 and HIV-2 that provide new insights into their function and may inform the design of HIV vaccines.
Results
HIV-1 is significantly more variable in the envelope C2, V3 and C3 regions than HIV-2
We compared the inter-patient genetic diversity of HIV-1 and HIV-2 in two different datasets: HIV-1 group M (all subtypes) and HIV-2 group A sequences from all over the world (Control dataset composed of reference sequences) and newly derived HIV-1 and HIV-2 sequences obtained from Portuguese (PT) patients. Phylogenetic analysis showed that HIV-1 sequences circulating in Portugal belong to different subtypes and recombinant forms (Figure S1A). Forty five sequences were subtype B and six belonged to the recombinant form CRF14_BG. Subtypes G (4 sequences) and C (2), sub-subtype F1 (2), and CRF02_AG (1) were also found. Regarding HIV-2, all sequences from Portugal clustered together within group A (Figure S1B). Collectively, these results are consistent with previous studies showing a highly complex HIV epidemics in Portugal caused exclusively by HIV-2 group A and different subtypes of HIV-1 group M [51], [52], [53], [54]. Nucleotide diversity between HIV-1 viruses found in Portugal was significantly higher compared to HIV-2 (mean number of substitutions per site, 0.336, 95%CI [0.329; 0.342] vs 0.239, [0.236; 0.243], P<0.0001). Similar results were found for the HIV-1 and HIV-2 Control datasets (Table S1). Hence, we conclude that HIV-1 is genetically more diverse than HIV-2 in the envelope region comprising C2, V3 and C3.
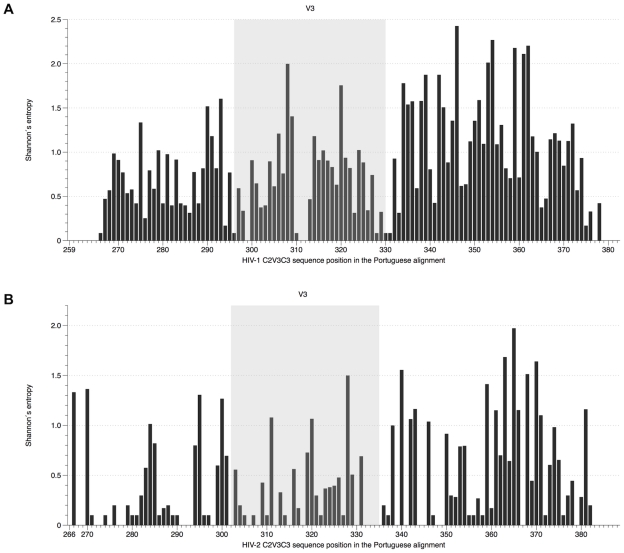
Amino acid diversity in the C2, V3 and C3 regions of HIV-1 and HIV-2 were compared by calculating Shannon's entropy [55]. Mean entropy values for the three regions were significantly higher in HIV-1 than in HIV-2 both in PT (0.794 vs 0.409, P<0.0001) and Control datasets (0.702 vs 0.353, P<0.0001) confirming that these regions are more variable in HIV-1 than in HIV-2. Entropy was also significantly higher in HIV-1 than in HIV-2 in each separate region (C2, P<0.05; V3, P<0.005; C3, P<0.0005) of PT sequences. The region with higher mean entropy was C3 in both viruses (1.031, 95%CI [0.845, 1.217] for HIV-1 vs 0.534, 95%CI [0.378, 0.689] for HIV-2, P<0.0005) followed by V3 (0.674, [0.506, 0.841]) and C2 (0.574, [0.427, 0.721]) in HIV-1 and C2 (0.326, [0.175, 0.477]) and V3 (0.304, [0.176, 0.433]) in HIV-2 (Figure 1). Comparable results were obtained for the Control datasets but in this case V3 was the least entropic region both in HIV-1 and HIV-2 (Table S1 and Figure S2). Not surprisingly, amino acids with higher entropy (values above 1) were primarily located in the C3 region of both viruses and there were more highly entropic amino acids in C3 in HIV-1 than in HIV-2 both in the PT and Control datasets (PT dataset: 51.9% in HIV-1 vs 24.5% in HIV-2; Control dataset: 35.3% in HIV-1 vs 20.8% in HIV-2). Notably, the amino acids in V3 that are related with co-receptor usage, positions 11/25 in HIV-1 (codons 306/320) [56], [57] and possibly positions 18/19/27 in HIV-2 (codons 319/320/328) [17], [58], had a high entropy score in both viruses.
Figure 1. Shannon's entropy of individual amino acids in the C2, V3 and C3 envelope regions in HIV-1 and HIV-2.
(A) HIV-1 alignment (PT dataset), sites were numbered according to codon env position of HIV-1 HXB2 reference strain; (B) HIV-2 alignment (PT dataset), sites were numbered according to codon env position of HIV-2 ALI reference strain.
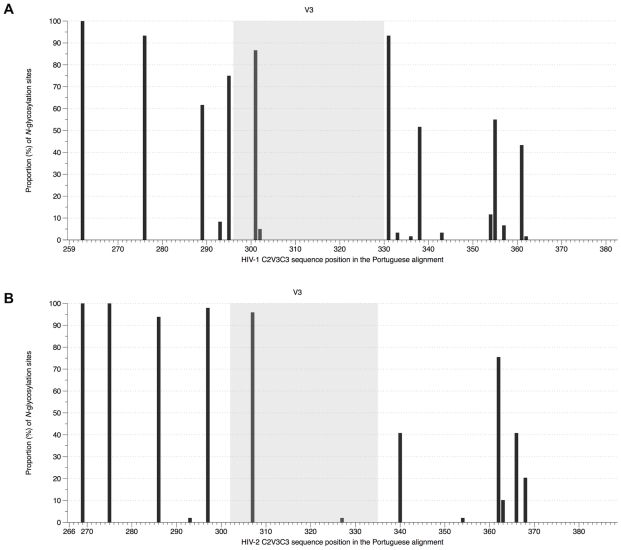
The mean number of potential N-linked glycosylation sites both in HIV-1 and HIV-2 sequences from Portugal was 7 (range: 4–9 in HIV-1; 5–9 in HIV-2). The most conserved glycosylation sites were located in C2 in both viruses (Figure 2). Nonetheless, in this region, there were four highly conserved glycosylation sites in HIV-2 (present in ≥80% of strains) and only two such sites in HIV-1. With the exception of the highly conserved site located in the beginning of C3 in HIV-1, glycosylation sites found in C3 varied from strain to strain in number and location, this being more evident in HIV-1 than in HIV-2. In V3 there were two highly conserved glycosylation sites in both viruses. Similar observations were made for HIV-1 and HIV-2 sequences in the Control datasets (Table S1 and Figure S3).
Figure 2. Frequency of N-glycosylation sites in the C2, V3 and C3 envelope regions in HIV-1 and HIV-2.
(A) HIV-1 alignment (PT dataset), sites were numbered according to codon env position of HIV-1 HXB2 reference strain; (B) HIV-2 alignment (PT dataset), sites were numbered according to codon env position of HIV-2 ALI reference strain.
Selective pressures act differently in HIV-1 and HIV-2
We have recently found that HIV-2 displays a faster evolutionary rate in the envelope gp125 and C2-V3-C3 region than HIV-1 in patients with chronic and advanced disease [52], [59]. The faster evolutionary rate in HIV-2 was more pronounced in synonymous sites than in non-synonymous sites suggesting a weaker positive selection in HIV-2 than in HIV-1. To investigate this possibility, we analysed diversifying selection in the C2-V3-C3 region of both viruses using codon-based models of molecular evolution. Firstly, we estimated the ratio of non-synonymous and synonymous substitution rates (dN/dS ratio) averaged over all sites. For HIV-1 sequences from Portugal dN/dS ratio was 0.703, 95%CI [0.668, 0.740]; for HIV-2 it was 0.451, [0.419, 0.484]. Similar values were obtained for the Control alignments (Table S1). These results are consistent with the higher degree of genetic conservation of the C2, V3 and C3 regions in HIV-2.
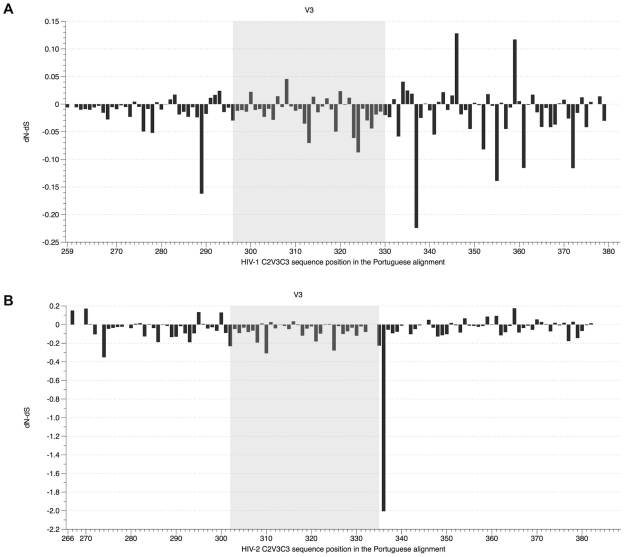
Site-by-site analysis revealed that diversifying selection is unevenly distributed along the studied region between the two viruses (PT, P<0.001; Controls, P<0.001) (Figures 3 and S4). For HIV-2 sequences from the PT dataset, there were between 7 and 9 positively selected (PS) sites depending on the method that was used (SLAC/FEL/REL) while for HIV-1 the number of sites ranged from 7 to 17 (Table 1). Taking into account only sites that were selected by at least two methods, HIV-2 had a total of 7 PS sites whereas in HIV-1 there were 9 sites. The sites were distributed as follows: in C2 there were 3 sites in HIV-2 and 2 in HIV-1; in V3 there were 2 sites in HIV-1, and no sites in HIV-2; in C3 there were 4 sites in HIV-2 and 5 in HIV-1, including one codon within the CD4 binding site (codon 378 in HIV-1) and two in the α2-helix (codons 343 and 346) [60]. In Control data sets the number of PS sites was slightly lower but they were similarly distributed, with the exception of the V3: 1 PS site in HIV-2, but no sites in HIV-1 (Tables S1 and S2). Importantly, we found that when compared to HIV-1, positive selection was stronger in HIV-2 in most sites (Tables 1 and S2).
Figure 3. Positive selection in the C2, V3 and C3 envelope regions in HIV-1 and HIV-2.
dN-dS values were estimated by FEL and scaled by the total codon tree length. (A) HIV-1 alignment (PT dataset), sites were numbered according to codon env position of HIV-1 HXB2 reference strain; (B) HIV-2 alignment (PT dataset), sites were numbered according to codon env position of HIV-2 ALI reference strain.
Table 1. Positively selected sites detected by SLAC, FEL, REL and/or IFEL in HIV-1 and HIV-2 env C2, V3 and C3 regions1.
| HIV-1 | HIV-2 | ||||||||||||||||||
| Region | Codon | SLAC | FEL | REL | IFEL | Region | Codon | SLAC | FEL | REL | IFEL | ||||||||
| C2 | 283 | 0.249 | (0.083) | 0.017 | (0.027) | 0.282 | (0.950) | −0.007 | (0.237) | C2 | 267 | 1.805 | (<0.001) | 0.151 | (<0.001) | 1.007 | (1.000) | 0.147 | (0.004) |
| 291 | 0.252 | (0.096) | 0.011 | (0.334) | 0.005 | (0.277) | 0.008 | (0.648) | 270 | 1.561 | (0.003) | 0.171 | (<0.001) | 0.892 | (1.000) | 0.179 | (0.010) | ||
| 292 | 0.269 | (0.063) | 0.017 | (0.167) | 0.547 | (<0.001) | 0.014 | (0.419) | 295 | 1.049 | (0.051) | 0.134 | (0.095) | 0.070 | (0.910) | 0.109 | (0.316) | ||
| 293 | 0.401 | (0.066) | 0.024 | (0.230) | 0.924 | (0.972) | 0.063 | (0.050) | 300 | 0.787 | (0.210) | 0.130 | (0.077) | 0.714 | (<0.001) | 0.131 | (0.109) | ||
| V3 | 300 | 0.335 | (0.079) | 0.022 | (0.022) | 0.219 | (0.846) | 0.016 | (0.093) | V3 | 331 | −0.312 | (0.859) | 0.020 | (0.591) | 0.193 | (0.987) | 0.069 | (0.390) |
| 306 | 0.312 | (0.106) | 0.014 | (0.541) | 0.947 | (0.984) | −0.004 | (0.867) | |||||||||||
| 308 | 0.619 | (0.008) | 0.046 | (0.065) | 0.869 | (0.973) | 0.094 | (0.012) | |||||||||||
| 314 | 0.314 | (0.052) | 0.014 | (0.291) | 0.163 | (0.178) | −0.001 | (0.971) | |||||||||||
| 317 | 0.301 | (0.057) | 0.011 | (0.401) | 0.192 | (0.140) | 0.005 | (0.749) | |||||||||||
| C3 | 332 | 0.124 | (0.267) | 0.009 | (0.341) | −0.432 | (<0.001) | 0.031 | (0.093) | C3 | 346 | 0.478 | (0.236) | 0.051 | (0.173) | 1.008 | (1.000) | 0.050 | (0.397) |
| 334 | 0.543 | (0.004) | 0.041 | (0.027) | 1.142 | (0.997) | 0.065 | (0.024) | 351 | 0.207 | (0.298) | 0.018 | (0.087) | 0.114 | (<0.001) | 0.000 | (1.000) | ||
| 335 | 0.458 | (0.010) | 0.025 | (0.109) | 0.893 | (0.936) | 0.035 | (0.147) | 354 | 0.689 | (0.047) | 0.067 | (0.016) | 0.988 | (1.000) | 0.000 | (1.000) | ||
| 336 | 0.452 | (0.058) | 0.019 | (0.583) | 0.817 | (0.907) | 0.011 | (0.743) | 361 | 0.887 | (0.035) | 0.093 | (0.011) | 0.988 | (1.000) | 0.016 | (0.693) | ||
| 343 | 0.405 | (0.060) | 0.022 | (0.109) | 0.885 | (0.989) | 0.017 | (0.370) | 364 | −0.085 | (0.704) | 0.014 | (0.690) | 0.130 | (0.983) | −0.069 | (0.072) | ||
| 345 | 0.392 | (0.024) | 0.016 | (0.330) | 0.492 | (0.657) | 0.070 | (0.018) | 365 | 1.074 | (0.089) | 0.175 | (0.070) | 0.463 | (0.561) | −0.056 | (0.626) | ||
| 346 | 1.080 | (<0.001) | 0.128 | (<0.001) | 0.945 | (0.982) | 0.281 | (<0.001) | 378 | 0.415 | (0.088) | 0.030 | (0.043) | 0.116 | (<0.001) | 0.029 | (0.119) | ||
| 348 | 0.270 | (0.096) | 0.011 | (0.664) | 1.183 | (<0.001) | −0.005 | (0.899) | |||||||||||
| 353 | 0.319 | (0.143) | 0.018 | (0.476) | −0.118 | (0.347) | 0.120 | (0.035) | |||||||||||
| 359 | 0.558 | (0.022) | 0.117 | (<0.001) | 0.882 | (1.000) | 0.214 | (0.001) | |||||||||||
| 363 | 0.169 | (0.295) | 0.017 | (0.279) | 0.860 | (0.995) | 0.003 | (0.900) | |||||||||||
| 378 | 0.244 | (0.021) | 0.014 | (0.020) | 0.217 | (<0.001) | 0.000 | (1.000) | |||||||||||
PT dataset.
Codon – codons selected under 10% level of significance (SLAC, FEL and IFEL) or above a Bayes Factor of 50 (REL) and numbered according to codon env position of HIV-1 HXB2 for HIV-1 dataset or of HIV-2 ALI for HIV-2 dataset. Codons selected simultaneously by SLAC, FEL and REL methods are bold and underlined.
SLAC, FEL and IFEL – the first numbers are the dN-dS difference for each site scaled by the total codon tree length, the numbers in parenthesis show P-values for corresponding test of non-synonymous rate being superior to synonymous rate; REL - the first numbers are the expected posterior dN-dS difference for each site scaled to the total codon tree length, the number in parenthesis show the posterior probability of non-synonymous rate being superior to synonymous rate; Bold dN-dS differences correspond to significant P-values or posterior probabilities.
The comparison of diversifying selection between terminal and internal branches of the phylogenetic trees revealed two distinct profiles for HIV-1 and HIV-2. Firstly, non-synonymous substitution rates were significantly different between the internal nodes and the tips of the tree in all datasets: PT, P = 0.002 for HIV-2 and P = 0.011 for HIV-1; Controls, P<0.001 and P = 0.004 (data not shown). Stronger selection was in general found at codons selected simultaneously at the tips and the external branches of the HIV-1 and HIV-2 trees. Importantly, however, only 2 of the 7 sites (29%) detected in terminal branches of PT HIV-2 tree were also under positive selection along the internal branches (codons 267 and 270 in C2). In contrast, for HIV-1 most positively selected sites (6/9, 67%) were present both in the internal and the terminal branches. In Control datasets these percentages were 43% for HIV-2 and 71% for HIV-1 (Table S2). These results suggest that natural selection affects less the transmission fitness of HIV-1 than HIV-2.
Structure and solvent accessibility of V3 differ in HIV-1 and HIV-2
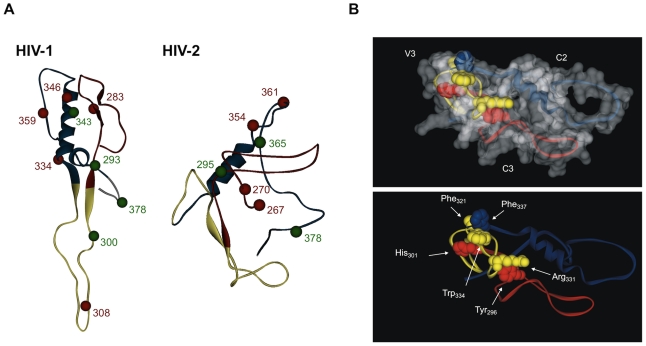
A model of the structure of the C2-V3-C3 region was built for HIV-1 and HIV-2 based on the atomic coordinates of the HIV-1 gp120 and SIV gp120 using consensus sequences from both the PT and Control HIV-1 and HIV-2 alignments. For HIV-1, the structures of PT and Control sequences were almost identical having only a slight difference in V3, which presents less regular secondary structure in the PT sequence (Figure S5). For HIV-2, the structures of PT and Control sequences were identical. The structure of the C2-V3-C3 region was however markedly different between HIV-1 and HIV-2, the most striking differences being the significant retraction of the V3 loop in HIV-2 and its potential interaction both with C2 and C3 (Figure 4A). Identical results were obtained when comparing the HIV-1 and HIV-2 control sequences (Figure S6). The predicted non-covalent interaction between V3, C3 and C3 in HIV-2 involves residues Tyr296 and His301 in C2 binding, respectively, to Arg331 and Trp334 in V3, and Phe337 in C3 binding to Phe321 in V3 (Figure 4B).
Figure 4. Conformational structure of C2, V3 and C3 envelope regions in HIV-1 and HIV-2.
The conformational structure of consensus amino acid sequences derived from the PT datasets was obtained by homology modeling as indicated in Material and Methods. In the schematics, C2 is shown in red, V3 in yellow and C3 in blue. (A) Balls represent the amino acids under positive selection. The red balls represent codons selected simultaneously by SLAC, FEL and REL methods, while green balls stand for codons selected by at least two of these methods; (B) Model structure showing the predicted interactions between V3, C2 and C3 in HIV-2 gp125. The non-covalent interaction involves residues Tyr296 and His301 in C2 binding, respectively, to Arg331 and Trp334 in V3, and Phe337 in C3 binding to Phe321 in V3.
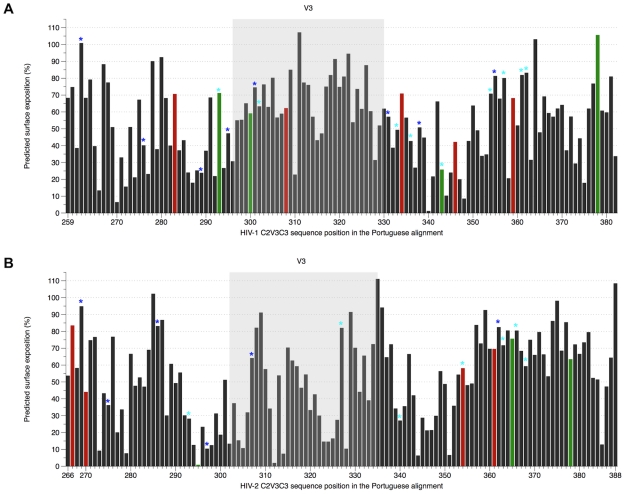
The solvent accessibilities of amino acid residues were also calculated for both models (Figure 5). As expected, both in HIV-1 and HIV-2 most PS sites and N-glycans had at least 50% surface exposure. In HIV-2, 8 out of 37 (22%) amino acids in C2, 8/34 (24%) in V3 and 19/53 (36%) in C3 were highly exposed (≥70% solvent accessibility) whereas in HIV-1 these were 9/37 (24%), 15/35 (43%) and 10/52 (19%), respectively. Consistent with the high exposure of the V3 region in HIV-1, the two amino acids at positions 306 and 320 involved in binding to co-receptors were well exposed (≥50% solvent accessibility). In contrast, in HIV-2, among amino acids 319/320 and 328 in V3 loop potentially involved in co-receptor binding, only 319 was relatively well exposed. Despite the potential interaction between V3 and C3 (Figure 4B), the overall exposition of C3 was higher in HIV-2 than in HIV-1. Thus, for instance, 42% (5/12) of the residues in C3 that may contribute for the formation of the CD4-binding site (positions 377–388) in HIV-2 showed high solvent accessibility. In HIV-1 only 3 out of 16 (19%) amino acids with similar function (positions 367–382) were highly exposed. Similar results were obtained when comparing the HIV-1 and HIV-2 control sequences (Figure S7).
Figure 5. Solvent accessible surface area, positive selection and potential N-glycosylation sites in C2-V3-C3 region.
(A) HIV-1 alignment (PT dataset), sites were numbered according to codon env position of HIV-1 HXB2 reference strain; (B) HIV-2 alignment (PT dataset), sites were numbered according to codon env position of HIV-2 ALI reference strain. Coloured bars represent the amino acids under positive selection and have the same colours (red and green) as the corresponding positions (balls) highlighted in Figure 4A. The dark blue stars over the bars correspond to potential N-glycosylation sites conserved along the alignment (present in ≥50% of strains), whereas the light blue stars represent sites only present in less than 50% of sequences.
Discussion
To investigate the molecular and structural features underlying the differences in HIV-1 and HIV-2 biology and human infection, we have analysed the C2-V3-C3 envelope region from a significant number of HIV-1 and HIV-2 infected patients living in Portugal and worldwide. HIV-2 sequences from Portugal belonged to group A and the majority of HIV-1 sequences belonged to subtype B (75%) followed by subtypes G, C and F, CRF02_AG and CRF14_BG. Collectively, these results are consistent with previous studies showing a highly complex HIV epidemic in Portugal caused by HIV-2 group A and different subtypes of HIV-1 group M [44], [51], [52], [53], [54], [61], [62].
Genetic distances and amino acid diversity between HIV-1 viruses were significantly higher compared to HIV-2. This was surprising since at the individual level HIV-2 displays a similar [52] or even faster evolutionary rate than HIV-1 in the C2-V3-C3 region [59]. The more pronounced evolutionary rate in synonymous sites than in non-synonymous sites in HIV-2 [59] together with the rare escape of this virus from autologous neutralizing antibodies [17], suggested that the lower amino acid diversity in HIV-2 could be related with a weaker positive selection or even with negative selection [62]. This was not the case however since most sites in C2 and C3 were under stronger positive selection in HIV-2 than in HIV-1. The C3 region of HIV-1 is antibody accessible [63] and is subject to diversifying selection because it is a major neutralizing target [64], [65], [66], [67]. Therefore, the high level of positive selection detected in C3 together with its high solvent exposure strongly suggests that this region is also antibody accessible in HIV-2 and might be a major neutralizing domain.
Strength of selection was significantly different between internal and external branches of the HIV-1 and HIV-2 phylogenetic trees. This is expected in populations of highly variable RNA viruses and implies that non-synonymous substitutions can be highly deleterious [68], [69]. In HIV-1, most of the codons selected in the tips of the tree were also under selection along the internal branches, indicating that adaptation in these sites is occurring at the host and population levels [68]. In contrast, most adaptive mutations in HIV-2 were only found in the tips of the tree indicating that they are recent maladaptive substitutions that are transitory at the population level [68], [70]. Thus, in contrast to HIV-1, diversification of C2 and C3 in HIV-2 seems to have a dominant negative effect on viral fitness and transmission. This data suggests that one possible consequence of the unexpectedly high evolutionary rate of HIV-2 at the patient level can be the frequent accumulation of deleterious mutations and production of defective viruses [52], [59], [71]. A high frequency of defective viruses in HIV-2 infected individuals could explain the poor replication of this virus in vivo as well as its very low transmissibility.
Unlike in HIV-1, the V3 loop in HIV-2 always presented the lower amino acid diversity. This result might be a consequence of significant structural and conformational constraints due to its role in preventing chronic and disruptive immune activation [20] and in co-receptor binding [58]. On the other hand, these results imply that the V3 loop is not well exposed in the HIV-2 envelope complex in vivo. Indeed, by computer modelling simulations we show that in HIV-2 the V3 loop is much less exposed than C2 and C3 and likely has a retractile conformation due to non-covalent interaction both with C2 and C3. In contrast, HIV-1 had, as previously found, an extended and highly accessible V3 loop [66], [67], [72]. Such conformation is entirely consistent with its immunodominant and neutralizing nature and with its crucial role in HIV-1 co-receptor binding and tropism [33], [34], [35], [73], [74], [75]. Conversely, the concealed nature of V3 in the HIV-2 envelope complex implies that this region may not be immunodominant in HIV-2 infection. Indeed, a significant number of HIV-2 patients do not raise antibodies against the V3 loop [43] or against a polypeptide comprising the C2, V3 and C3 regions [45]. Thus, the occlusion of V3 in the HIV-2 envelope complex may prevent it from over immune recognition and associated sequence changes thereby preserving its crucial functions in viral entry. It has been shown that removal or antigenic dampening of the HIV-1 V3 loop redirects the neutralizing immune response to other epitopes of the Env protein that otherwise would be non-neutralizing or non-antibody responsive [33], [76], [77], [78]. In this context, the occluded nature of the V3 region in the HIV-2 envelope complex might favour a more effective production of broadly neutralizing antibodies targeting other regions in gp125 such as the C2, V1, V2, V4 and C5 regions [37], [38], [39], [79].
In conclusion, the C2 and C3 regions are well exposed in the HIV-2 envelope complex and are under strong diversifying selection suggesting that, like in HIV-1, they may harbour neutralizing epitopes. However, extreme diversification of C2 and C3 in HIV-2 seems to be deleterious for the virus and prevent its transmission. On the other hand, V3 is highly conserved in HIV-2 and is concealed within the envelope complex, possibly due to a physical interaction with C2 and C3. In contrast, V3 is highly exposed and variable in HIV-1 which is consistent with its immunodominant and neutralizing properties. Collectively, we identify significant structural and functional constrains to the diversification and evolution of C2, V3 and C3 in the HIV-2 envelope but not in HIV-1. These studies highlight fundamental differences in the biology and infection of HIV-1 and HIV-2 and in their mode of interaction with the human immune system and may inform new vaccine and therapeutic interventions against these viruses.
Materials and Methods
Amplification, cloning and sequencing of HIV-1 and HIV-2 viruses from Portugal
Portuguese (PT) samples were collected from HIV infected patients, followed in hospitals in the North and South of Portugal and presenting different clinical stages of infection and CD4+ T-cell counts. HIV-2 samples were collected between 1997 and 2005 from 49 patients, some of whom were infected in late-1970s [52], [80]. HIV-1 samples were collected from 60 patients between 1993 and 1998.
Proviral DNA was extracted from uncultured PBMCs, or viral genomic RNA was extracted from plasma and reverse transcribed. A nested PCR technique was used to amplify a 373 bp HIV-2 C2-V3-C3 env gene region and a 409 pb HIV-1 C2-V3-C3 env region as described elsewhere [62], [81]. PCR products were sequenced using the BigDye Terminator Cycle sequencing kit (Applied Biosystems) and an automated capillary sequencer (ABI PRISM 310, Applied Biosystems). Newly derived HIV-1 sequences from Portugal have been assigned GenBank accession numbers: EU335962 - EU335903. Newly derived HIV-2 sequences from Portugal have been assigned GenBank accession numbers: AY913773-AY913794, AY649545-AY649554 and GU591163.
Additionally, 16 HIV-2 consensus sequences from a previous publication [52] were also included in this study. The samples used to obtain these consensus sequences were: 03PTHCC1, 03PTHCC2, 03PTHCC4, 03PTHCC5, 03PTHCC7, 03PTHCC8, 03PTHCC12, 05PTHCC13, 03PTHCC14, 03PTHCC17, 03PTHCC19, 03PTHSM2, 05PTHSM3, 03PTHSM7, 03PTHSM9 and 03PTHSM10.
Control datasets
As Control datasets to this study, HIV-1 group M (all subtypes) reference sequence alignment (94 sequences) was obtained from the Los Alamos HIV database (http://www.hiv.lanl.gov/). HIV-2 group A reference sequence alignment was also obtained from the Los Alamos HIV database. Additional C2-V3-C3 sequences derived from group A primary isolates were retrieved from the Los Alamos Database adding to a total of 59 HIV-2 Control sequences. Both control alignments are available as supplementary information (Alignment S1 and S2).
Molecular and phylogenetic analysis
Nucleotide sequences were aligned using ClustalX 1.8 [82]. Maximum likelihood analyses were performed using the best-fit models of molecular evolution estimated by Modeltest [83]. These were GTR+G+I [84] for the PT HIV-2 dataset and TVM+G+I for PT HIV-1 and for HIV-1 and HIV-2 Control datasets [85].
Evolutionary distances were estimated under these models using PAUP version 4.0 [86]. Tree searches were also conducted in PAUP version 4.0 using either nearest-neighbor interchange (NNI) or subtree pruning-regrafting (SPR) heuristic strategies, with bootstrap resampling. All positions containing gaps and missing data were eliminated from the dataset. In the final datasets there were a total of 369 nucleotide positions in PT HIV-2 and 372 positions in PT HIV-1 alignments, and 369 positions in HIV-2 and HIV-1 Control alignments. Both alignments were tested for recombination with the Single Breakpoint Recombination (SBP) tool [87] in the DATAMONKEY web-server [88]; evidence for recombination, inferred by the small sample AIC score, was only found for HIV-1 Control dataset. Thus, when appropriate, a multiple partition dataset was used for HIV-1 Control analysis. Detection of N-linked glycosylation sites was performed with Glycosite [89]. The entropy at each position in protein alignment was measured with Shannon's entropy [55].
Tests for codon selection
Selection pressures over the HIV-1 and HIV-2 C2-V3-C3 regions were examined with the HYPHY software package [90] and the DATAMONKEY web-server [88]. All estimations were performed using the MG94 codon substitution model [91] crossed with the nucleotide substitution model previously selected with Modeltest, GTR for PT HIV-2 and TVM for PT HIV-1 and Control alignments. To understand if selection pressure within a host is different from selection for transmission among hosts, non-synonymous substitutions were compared between terminal and internal branches of the phylogenetic tree, with the TestBranchDNDS.bf batch file in HyPHy, as described elsewhere [92].
Four different approaches were used to identify codons under selection: single-likelihood ancestor counting (SLAC), fixed-effects likelihood (FEL), internal fixed effects likelihood (IFEL) and relaxed-effects likelihood (REL) methods [68], [93]. While SLAC, FEL and REL detect sites under selection at the external branches of the phylogenetic three, IFEL identifies such sites only along the internal branches. To classify a site as positively or negatively selected the cut-off P-value was 10% for SLAC, FEL and IFEL. For REL, codons under selection were detected with a cut-off value for the Bayes factor of 50. Since SLAC, FEL and IFEL can estimate site-specific ratios of non-synonymous and synonymous substitutions rates (dN/dS ratios) as undefined or infinite due to dS = 0, we reported dN-dS values instead, which were scaled by the total codon tree length to allow a better comparison between the two datasets. A multiple partition dataset was used for the identification of codons under selection in HIV-1 Control analysis. Site-by-site variation of synonymous substitution rates can bias estimations of codon's diversifying selection [94]. Although all four methods described above model for this variation, variation of synonymous rates from codon to codon in each dataset was tested with the dNdSRateAnalysis.bf batch file in HyPHy, as described elsewhere [92]. Finally, comparison of the dN/dS distribution rates and the strength of selection between the HIV-1 and HIV-2 alignments, was performed with dNdSDistributionComparison.bf batch file also in HyPHy, as described elsewhere [92].
Molecular modelling and calculation of solvent accessible surfaces
Consensus amino acid sequences were derived for the different HIV-1 and HIV-2 datasets. Structural models of HIV-1 and HIV-2 C2-V3-C3 were produced with SWISS-MODEL homology modelling server in project mode resorting to Swiss-Pdb Viewer (DeepView) version 4.0, using PDB file 2B4C (from HIV-1 JR-FL gp120) for HIV-1, and PDB file 2BF1 (from SIV gp120) for HIV-2 as templates [95], [96], [97], [98]. Accelrys Discovery Studio Visualizer 2.5 [99] was used to produce three dimensional images of the models obtained. Solvent accessible surface area in Å2 was calculated by Gerstein's calc-surface software on UCSF Chimera [100], [101] with a probe size of 1.4 Å. All atoms in the input PDB file were included in the calculation. The solvent accessible surface data was normalized dividing each amino acid residue solvent accessible surface value added by the solvent accessible surface value of the corresponding amino acid residue (X) in the tripeptide Gly-X-Gly. The inter-chain H-Bonds formed by HIV-2 V3 with C2 and C3 were calculated with H-Bond Finder software on UCSF Chimera [100], [101] with a probe size of 1.4 Å. All atoms in the input PDB file were included in the calculation.
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 4.0c for Macintosh (GraphPad Software, 2005, San Diego, California, USA, www.graphpad.com) with a level of significance of 5%. Non-parametric Mann-Whitney U test was used to compare Shannon's entropy values and nucleotide distances.
Supporting Information
Genotyping HIV-1(A) and HIV-2 (B) by maximum-likelihood phylogenetic analysis. The phylogenetic trees were constructed using the SPR heuristic search strategy and 1000 bootstrap replications, with reference sequences from HIV-1, under the TVM+G+I evolutionary model (A) and with reference sequences from HIV-2, under the GTR+G+I evolutionary model (B). The bootstrap values (above 50%) supporting the internal branches are shown. The scale bar represents evolutionary distances in substitutions per site.
(0.21 MB PDF)
Shannon's entropy of individual amino acids in the C2, V3 and C3 envelope regions in HIV-1 and HIV-2. (A) HIV-1 alignment (Control dataset), sites were numbered according to codon env position of HIV-1 HXB2 reference strain; (B) HIV-2 alignment (Control dataset), sites were numbered according to codon env position of HIV-2 ALI reference strain.
(0.96 MB TIF)
Frequency of N-glycosylation sites in the C2, V3 and C3 envelope regions in HIV-1 and HIV-2. (A) HIV-1 alignment (Control dataset). Sites were numbered according to codon env position of HIV-1 HXB2 reference strain. (B) HIV-2 alignment (Control dataset). Sites were numbered according to codon env position of HIV-2 ALI reference strain.
(0.58 MB TIF)
Positive selection in the C2, V3 and C3 envelope regions in HIV-1 and HIV-2. dN-dS values were estimated by FEL and scaled by the total codon tree length. (A) HIV-1 alignment (Control dataset). Sites were numbered according to codon env position of HIV-1 HXB2 reference strain. (B) HIV-2 alignment (Control dataset). Sites were numbered according to codon env position of HIV-2 ALI reference strain.
(0.53 MB TIF)
Superimposition of the conformational structures generated by homology modelling of Portuguese and Control C2, V3 and C3 regions of HIV-1 and HIV-2. In the schematics, Portuguese structures are represented in red, and Control structures are in blue.
(0.78 MB TIF)
Conformational structure of C2, V3 and C3 envelope regions in HIV-1 and HIV-2. The conformational structure of consensus amino acid sequences derived from the Control datasets was obtained by homology modeling as indicated in “Materials and Methods.” In the schematics, C2 is shown in red, V3 in yellow, and C3 in blue. Balls represent the amino acids under positive selection. (A) The red balls represent codons selected simultaneously by SLAC, FEL and REL methods, while green balls stand for codons selected by at least two of these methods. (B) Model structure showing the predicted interactions between V3, C2 and C3 in HIV-2 gp125. The non-covalent interaction involves residues Tyr296 and His301 in C2 binding, respectively, to Arg331 and Trp334 in V3, and Phe337 in C3 binding to Phe321 in V3.
(0.88 MB TIF)
Solvent accessible surface area, positive selection and potential N-glycosylation sites in C2-V3-C3 region. (A) HIV-1 alignment (Control dataset). Sites were numbered according to codon env position of HIV-1 HXB2 reference strain. (B) HIV-2 alignment (Control dataset). Sites were numbered according to codon env position of HIV-2 ALI reference strain. Coloured bars represent the amino acids under positive selection and have the same colours (red and green) as the corresponding positions (balls) highlighted in Figure S6. The dark blue stars over the bars correspond to potential N-glycosylation sites conserved along the alignment (present in ≥50% of strains), whereas the light blue stars represent sites only present in less than 50% of sequences.
(1.60 MB TIF)
Summary of results for phylogenetic, codon selection and solvent accessibility analysis for C2, V3 and C3 regions of HIV-1 and HIV-2 Control datasets.
(0.04 MB DOC)
Positively selected sites detected by SLAC, FEL, REL and/or IFEL in Control HIV-1 and HIV-2 env C2, V3 and C3 regions.
(0.10 MB DOC)
Alignment of HIV-1 reference sequences used as a Control for the Portuguese HIV-1 dataset. Each sequence is identified by the corresponding GenBank accession number.
(0.04 MB TXT)
Alignment of HIV-2 sequences used as a Control for the Portuguese HIV-2 dataset. Each sequence is identified by the corresponding GenBank accession number.
(0.02 MB TXT)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by projects PTDCSAU-FCF6767/2006 from Fundo para a Ciencia e Tecnologia, Portugal and by Collaborative HIV and Anti-HIV Drug Resistance Network (CHAIN), from the European Union. PB and IB are supported by PhD grants from Fundo para a Ciencia e Tecnologia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhang M, Foley B, Schultz AK, Macke JP, Bulla I, et al. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology. 2010;7:25. doi: 10.1186/1742-4690-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemey P, Pybus OG, Wang B, Saksena NK, Salemi M, et al. Tracing the origin and history of the HIV-2 epidemic. Proc Natl Acad Sci U S A. 2003;100:6588–6592. doi: 10.1073/pnas.0936469100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Silva TI, Cotten M, Rowland-Jones SL. HIV-2: the forgotten AIDS virus. Trends Microbiol. 2008;16:588–595. doi: 10.1016/j.tim.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Semaille C, Barin F, Cazein F, Pillonel J, Lot F, et al. Monitoring the dynamics of the HIV epidemic using assays for recent infection and serotyping among new HIV diagnoses: experience after 2 years in France. J Infect Dis. 2007;196:377–383. doi: 10.1086/519387. [DOI] [PubMed] [Google Scholar]
- 5.Valadas E, Franca L, Sousa S, Antunes F. 20 years of HIV-2 infection in Portugal: trends and changes in epidemiology. Clin Infect Dis. 2009;48:1166–1167. doi: 10.1086/597504. [DOI] [PubMed] [Google Scholar]
- 6.Gao F, Yue L, Robertson DL, Hill SC, Hui H, et al. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J Virol. 1994;68:7433–7447. doi: 10.1128/jvi.68.11.7433-7447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Luckay A, Sodora DL, Telfer P, Reed P, et al. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J Virol. 1997;71:3953–3960. doi: 10.1128/jvi.71.5.3953-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi J, Devare SG, Brennan CA. Identification of a new HIV-2 subtype based on phylogenetic analysis of full-length genomic sequence. AIDS Res Hum Retroviruses. 2000;16:925–930. doi: 10.1089/08892220050042864. [DOI] [PubMed] [Google Scholar]
- 9.Damond F, Worobey M, Campa P, Farfara I, Colin G, et al. Identification of a highly divergent HIV type 2 and proposal for a change in HIV type 2 classification. AIDS Res Hum Retroviruses. 2004;20:666–672. doi: 10.1089/0889222041217392. [DOI] [PubMed] [Google Scholar]
- 10.Rowland-Jones S. Protective immunity against HIV infection: lessons from HIV-2 infection. Future Microbiol. 2006;1:427–433. doi: 10.2217/17460913.1.4.427. [DOI] [PubMed] [Google Scholar]
- 11.Berry N, Ariyoshi K, Jaffar S, Sabally S, Corrah T, et al. Low peripheral blood viral HIV-2 RNA in individuals with high CD4 percentage differentiates HIV-2 from HIV-1 infection. J Hum Virol. 1998;1:457–468. [PubMed] [Google Scholar]
- 12.Soares R, Foxall R, Albuquerque A, Cortesao C, Garcia M, et al. Increased frequency of circulating CCR5+ CD4+ T cells in human immunodeficiency virus type 2 infection. J Virol. 2006;80:12425–12429. doi: 10.1128/JVI.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marlink R, Kanki P, Thior I, Travers K, Eisen G, et al. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science. 1994;265:1587–1590. doi: 10.1126/science.7915856. [DOI] [PubMed] [Google Scholar]
- 14.Drylewicz J, Matheron S, Lazaro E, Damond F, Bonnet F, et al. Comparison of viro-immunological marker changes between HIV-1 and HIV-2-infected patients in France. AIDS. 2008;22:457–468. doi: 10.1097/QAD.0b013e3282f4ddfc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez SK, Sarr AD, MacNeil A, Thakore-Meloni S, Gueye-Ndiaye A, et al. Comparison of heterologous neutralizing antibody responses of human immunodeficiency virus type 1 (HIV-1)- and HIV-2-infected Senegalese patients: distinct patterns of breadth and magnitude distinguish HIV-1 and HIV-2 infections. J Virol. 2007;81:5331–5338. doi: 10.1128/JVI.02789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lizeng Q, Skott P, Sourial S, Nilsson C, Andersson SS, et al. Serum immunoglobulin A (IgA)-mediated immunity in human immunodeficiency virus type 2 (HIV-2) infection. Virology. 2003;308:225–232. doi: 10.1016/s0042-6822(02)00088-0. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y, Brandin E, Vincic E, Jansson M, Blaxhult A, et al. Evolution of human immunodeficiency virus type 2 coreceptor usage, autologous neutralization, envelope sequence and glycosylation. J Gen Virol. 2005;86:3385–3396. doi: 10.1099/vir.0.81259-0. [DOI] [PubMed] [Google Scholar]
- 18.Bjorling E, Scarlatti G, von Gegerfelt A, Albert J, Biberfeld G, et al. Autologous neutralizing antibodies prevail in HIV-2 but not in HIV-1 infection. Virology. 1993;193:528–530. doi: 10.1006/viro.1993.1160. [DOI] [PubMed] [Google Scholar]
- 19.Cavaleiro R, Sousa AE, Loureiro A, Victorino RM. Marked immunosuppressive effects of the HIV-2 envelope protein in spite of the lower HIV-2 pathogenicity. AIDS. 2000;14:2679–2686. doi: 10.1097/00002030-200012010-00007. [DOI] [PubMed] [Google Scholar]
- 20.Cavaleiro R, Brunn GJ, Albuquerque AS, Victorino RM, Platt JL, et al. Monocyte-mediated T cell suppression by HIV-2 envelope proteins. Eur J Immunol. 2007;37:3435–3444. doi: 10.1002/eji.200737511. [DOI] [PubMed] [Google Scholar]
- 21.Grossman Z, Meier-Schellersheim M, Sousa AE, Victorino RM, Paul WE. CD4+ T-cell depletion in HIV infection: are we closer to understanding the cause? Nat Med. 2002;8:319–323. doi: 10.1038/nm0402-319. [DOI] [PubMed] [Google Scholar]
- 22.Blaak H, van der Ende ME, Boers PH, Schuitemaker H, Osterhaus AD. In vitro replication capacity of HIV-2 variants from long-term aviremic individuals. Virology. 2006;353:144–154. doi: 10.1016/j.virol.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 23.MacNeil A, Sarr AD, Sankale JL, Meloni ST, Mboup S, et al. Direct evidence of lower viral replication rates in vivo in human immunodeficiency virus type 2 (HIV-2) infection than in HIV-1 infection. J Virol. 2007;81:5325–5330. doi: 10.1128/JVI.02625-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 25.Douagi I, Forsell MN, Sundling C, O'Dell S, Feng Y, et al. Influence of Novel CD4 binding-defective HIV-1 Envelope Glycoprotein Immunogens on Neutralizing Antibody and T Cell Responses in Non-human Primates. J Virol. 2009 doi: 10.1128/JVI.01896-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore JP, Willey RL, Lewis GK, Robinson J, Sodroski J. Immunological evidence for interactions between the first, second, and fifth conserved domains of the gp120 surface glycoprotein of human immunodeficiency virus type 1. J Virol. 1994;68:6836–6847. doi: 10.1128/jvi.68.11.6836-6847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang SS, Boyle TJ, Lyerly HK, Cullen BR. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 28.Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, et al. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 29.Javaherian K, Langlois AJ, McDanal C, Ross KL, Eckler LI, et al. Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc Natl Acad Sci U S A. 1989;86:6768–6772. doi: 10.1073/pnas.86.17.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Javaherian K, Langlois AJ, LaRosa GJ, Profy AT, Bolognesi DP, et al. Broadly neutralizing antibodies elicited by the hypervariable neutralizing determinant of HIV-1. Science. 1990;250:1590–1593. doi: 10.1126/science.1703322. [DOI] [PubMed] [Google Scholar]
- 31.Krachmarov CP, Honnen WJ, Kayman SC, Gorny MK, Zolla-Pazner S, et al. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. Journal of Virology. 2006;80:7127–7135. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis KL, Bibollet-Ruche F, Li H, Decker JM, Kutsch O, et al. Human immunodeficiency virus type 2 (HIV-2)/HIV-1 envelope chimeras detect high titers of broadly reactive HIV-1 V3-specific antibodies in human plasma. J Virol. 2009;83:1240–1259. doi: 10.1128/JVI.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forsell MN Schief WR, Wyatt RT. Immunogenicity of HIV-1 envelope glycoprotein oligomers. Curr Opin HIV AIDS. 2009;4:380–387. doi: 10.1097/COH.0b013e32832edc19. [DOI] [PubMed] [Google Scholar]
- 34.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 35.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Traincard F, Rey-Cuille MA, Huon I, Dartevelle S, Mazie JC, et al. Characterization of monoclonal antibodies to human immunodeficiency virus type 2 envelope glycoproteins. AIDS Res Hum Retroviruses. 1994;10:1659–1667. doi: 10.1089/aid.1994.10.1659. [DOI] [PubMed] [Google Scholar]
- 37.Bjorling E, Broliden K, Bernardi D, Utter G, Thorstensson R, et al. Hyperimmune antisera against synthetic peptides representing the glycoprotein of human immunodeficiency virus type 2 can mediate neutralization and antibody-dependent cytotoxic activity. Proc Natl Acad Sci U S A. 1991;88:6082–6086. doi: 10.1073/pnas.88.14.6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bjorling E, Chiodi F, Utter G, Norrby E. Two neutralizing domains in the V3 region in the envelope glycoprotein gp125 of HIV type 2. J Immunol. 1994;152:1952–1959. [PubMed] [Google Scholar]
- 39.McKnight A, Shotton C, Cordell J, Jones I, Simmons G, et al. Location, exposure, and conservation of neutralizing and nonneutralizing epitopes on human immunodeficiency virus type 2 SU glycoprotein. J Virol. 1996;70:4598–4606. doi: 10.1128/jvi.70.7.4598-4606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morner A, Achour A, Norin M, Thorstensson R, Bjorling E. Fine characterization of a V3-region neutralizing epitope in human immunodeficiency virus type 2. Virus Res. 1999;59:49–60. doi: 10.1016/s0168-1702(98)00133-6. [DOI] [PubMed] [Google Scholar]
- 41.Matsushita S, Matsumi S, Yoshimura K, Morikita T, Murakami T, et al. Neutralizing monoclonal antibodies against human immunodeficiency virus type 2 gp120. J Virol. 1995;69:3333–3340. doi: 10.1128/jvi.69.6.3333-3340.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcelino JM, Borrego P, Rocha C, Barroso H, Quintas A, et al. Potent and broadly reactive HIV-2 neutralizing antibodies elicited by a vaccinia virus vector prime-C2V3C3 polypeptide boost immunization strategy. J Virol. 2010;84:12429–12436. doi: 10.1128/JVI.01102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plantier JC, Damond F, Souquieres S, Brun-Vezinet F, Simon F, et al. V3 serological subtyping of human immunodeficiency virus type 2 infection is not relevant. J Clin Microbiol. 2001;39:3803–3807. doi: 10.1128/JCM.39.10.3803-3807.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcelino JM, Nilsson C, Barroso H, Gomes P, Borrego P, et al. Envelope-specific antibody response in HIV-2 infection: C2V3C3-specific IgG response is associated with disease progression. AIDS. 2008;22:2257–2265. doi: 10.1097/QAD.0b013e3283155546. [DOI] [PubMed] [Google Scholar]
- 45.Marcelino JM, Barroso H, Goncalves F, Silva SM, Novo C, et al. Use of a new dual-antigen enzyme-linked immunosorbent assay to detect and characterize the human antibody response to the human immunodeficiency virus type 2 envelope gp125 and gp36 glycoproteins. J Clin Microbiol. 2006;44:607–611. doi: 10.1128/JCM.44.2.607-611.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang ML, Essex M, Lee TH. Localization of immunogenic domains in the human immunodeficiency virus type 2 envelope. J Virol. 1991;65:5073–5079. doi: 10.1128/jvi.65.9.5073-5079.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frost SD, Wrin T, Smith DM, Kosakovsky Pond SL, Liu Y, et al. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci U S A. 2005;102:18514–18519. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemey P, Rambaut A, Pybus OG. HIV evolutionary dynamics within and among hosts. AIDS Rev. 2006;8:125–140. [PubMed] [Google Scholar]
- 49.Liang B, Luo M, Ball TB, Plummer FA. QUASI analysis of the HIV-1 envelope sequences in the Los Alamos National Laboratory HIV sequence database: pattern and distribution of positive selection sites and their frequencies over years. Biochem Cell Biol. 2007;85:259–264. doi: 10.1139/o06-143. [DOI] [PubMed] [Google Scholar]
- 50.Canducci F, Marinozzi MC, Sampaolo M, Berre S, Bagnarelli P, et al. Dynamic features of the selective pressure on the human immunodeficiency virus type 1 (HIV-1) gp120 CD4-binding site in a group of long term non progressor (LTNP) subjects. Retrovirology. 2009;6:4. doi: 10.1186/1742-4690-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antunes R, Figueiredo S, Bartolo I, Pinheiro M, Rosado L, et al. Evaluation of the clinical sensitivities of three viral load assays with plasma samples from a pediatric population predominantly infected with human immunodeficiency virus type 1 subtype G and BG recombinant forms. J Clin Microbiol. 2003;41:3361–3367. doi: 10.1128/JCM.41.7.3361-3367.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borrego P, Marcelino JM, Rocha C, Doroana M, Antunes F, et al. The role of the humoral immune response in the molecular evolution of the envelope C2, V3 and C3 regions in chronically HIV-2 infected patients. Retrovirology. 2008;5:78. doi: 10.1186/1742-4690-5-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Esteves A, Parreira R, Venenno T, Franco M, Piedade J, et al. Molecular epidemiology of HIV type 1 infection in Portugal: high prevalence of non-B subtypes. AIDS Res Hum Retroviruses. 2002;18:313–325. doi: 10.1089/088922202753519089. [DOI] [PubMed] [Google Scholar]
- 54.Palma AC, Araujo F, Duque V, Borges F, Paixao MT, et al. Molecular epidemiology and prevalence of drug resistance-associated mutations in newly diagnosed HIV-1 patients in Portugal. Infect Genet Evol. 2007;7:391–398. doi: 10.1016/j.meegid.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 55.Korber BT, Kunstman KJ, Patterson BK, Furtado M, McEvilly MM, et al. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Jong JJ, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Resch W, Hoffman N, Swanstrom R. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology. 2001;288:51–62. doi: 10.1006/viro.2001.1087. [DOI] [PubMed] [Google Scholar]
- 58.Isaka Y, Sato A, Miki S, Kawauchi S, Sakaida H, et al. Small amino acid changes in the V3 loop of human immunodeficiency virus type 2 determines the coreceptor usage for CXCR4 and CCR5. Virology. 1999;264:237–243. doi: 10.1006/viro.1999.0006. [DOI] [PubMed] [Google Scholar]
- 59.Skar H, Borrego P, Wallstrom TC, Mild M, Marcelino JM, et al. HIV-2 genetic evolution in patients with advanced disease is faster than in matched HIV-1 patients. J Virol. 2010;84:7412–7415. doi: 10.1128/JVI.02548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soriano V, Gomes P, Heneine W, Holguin A, Doruana M, et al. Human immunodeficiency virus type 2 (HIV-2) in Portugal: clinical spectrum, circulating subtypes, virus isolation, and plasma viral load. J Med Virol. 2000;61:111–116. [PubMed] [Google Scholar]
- 62.Barroso H, Taveira N. Evidence for negative selective pressure in HIV-2 evolution in vivo. Infect Genet Evol. 2005;5:239–246. doi: 10.1016/j.meegid.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 63.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore PL, Gray ES, Choge IA, Ranchobe N, Mlisana K, et al. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J Virol. 2008;82:1860–1869. doi: 10.1128/JVI.02187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rong R, Gnanakaran S, Decker JM, Bibollet-Ruche F, Taylor J, et al. Unique mutational patterns in the envelope alpha 2 amphipathic helix and acquisition of length in gp120 hypervariable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J Virol. 2007;81:5658–5668. doi: 10.1128/JVI.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaschen B, Taylor J, Yusim K, Foley B, Gao F, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 67.Choisy M, Woelk CH, Guegan JF, Robertson DL. Comparative study of adaptive molecular evolution in different human immunodeficiency virus groups and subtypes. J Virol. 2004;78:1962–1970. doi: 10.1128/JVI.78.4.1962-1970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pond SL, Frost SD, Grossman Z, Gravenor MB, Richman DD, et al. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput Biol. 2006;2:e62. doi: 10.1371/journal.pcbi.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pybus OG, Rambaut A, Belshaw R, Freckleton RP, Drummond AJ, et al. Phylogenetic evidence for deleterious mutation load in RNA viruses and its contribution to viral evolution. Mol Biol Evol. 2007;24:845–852. doi: 10.1093/molbev/msm001. [DOI] [PubMed] [Google Scholar]
- 70.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 71.Lemey P, Kosakovsky Pond SL, Drummond AJ, Pybus OG, Shapiro B, et al. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput Biol. 2007;3:e29. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pinter A. Roles of HIV-1 Env variable regions in viral neutralization and vaccine development. Curr HIV Res. 2007;5:542–553. doi: 10.2174/157016207782418470. [DOI] [PubMed] [Google Scholar]
- 74.Binley J. Specificities of broadly neutralizing anti-HIV-1 sera. Curr Opin HIV AIDS. 2009;4:364–372. doi: 10.1097/COH.0b013e32832e06fe. [DOI] [PubMed] [Google Scholar]
- 75.Tomaras GD, Haynes BF. HIV-1-specific antibody responses during acute and chronic HIV-1 infection. Curr Opin HIV AIDS. 2009;4:373–379. doi: 10.1097/COH.0b013e32832f00c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Selvarajah S, Puffer BA, Lee FH, Zhu P, Li Y, et al. Focused dampening of antibody response to the immunodominant variable loops by engineered soluble gp140. AIDS Res Hum Retroviruses. 2008;24:301–314. doi: 10.1089/aid.2007.0158. [DOI] [PubMed] [Google Scholar]
- 77.Garrity RR, Rimmelzwaan G, Minassian A, Tsai WP, Lin G, et al. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J Immunol. 1997;159:279–289. [PubMed] [Google Scholar]
- 78.Kim YB, Han DP, Cao C, Cho MW. Immunogenicity and ability of variable loop-deleted human immunodeficiency virus type 1 envelope glycoproteins to elicit neutralizing antibodies. Virology. 2003;305:124–137. doi: 10.1006/viro.2002.1727. [DOI] [PubMed] [Google Scholar]
- 79.Skott P, Achour A, Norin M, Thorstensson R, Bjorling E. Characterization of neutralizing sites in the second variable and fourth variable region in gp125 and a conserved region in gp36 of human immunodeficiency virus type 2. Viral Immunol. 1999;12:79–88. doi: 10.1089/vim.1999.12.79. [DOI] [PubMed] [Google Scholar]
- 80.Mota-Miranda A, Gomes H, Lima-Alves C, Araujo F, Cunha-Ribeiro LM, et al. Perinatally acquired HIV-2 infection diagnosed at 15 and 24 years of age. AIDS. 2001;15:2460–2461. doi: 10.1097/00002030-200112070-00018. [DOI] [PubMed] [Google Scholar]
- 81.Leitner T, Escanilla D, Marquina S, Wahlberg J, Brostrom C, et al. Biological and molecular characterization of subtype D, G, and A/D recombinant HIV-1 transmissions in Sweden. Virology. 1995;209:136–146. doi: 10.1006/viro.1995.1237. [DOI] [PubMed] [Google Scholar]
- 82.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 84.Rodriguez F, Oliver JL, Marin A, Medina JR. The general stochastic model of nucleotide substitution. J Theor Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- 85.Posada D. Using Modeltest and PAUP* to select a model of nucleotide substitution. In: A. D. Baxevanis DBD, R. D. M. Page et al, editors. Current Protocols in Bioinformatics. Chichester, UK: John Wiley & Sons, Inc; 2003. pp. 6.5.1–6.5.14. [DOI] [PubMed] [Google Scholar]
- 86.Swofford DL. Sunderland, MA: Sinauer-Associates; 1998. PAUP*. Phylogenetic Analysis using Parsimony (*and other Methods). Version 4. Version 4 ed. [Google Scholar]
- 87.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 88.Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 89.Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, et al. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 90.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 91.Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol. 1994;11:715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- 92.Kosakovsky Pond SL, Poon AWF, Frost SDW. Estimating selection pressures on aligments of coding sequences: practice. In: Lemey P, Salemi M, Vandamme A, editors. The Phylogenetic Handbook: A Paratical Approach to Phylogenetic Analysis and Hypothesis Testing, 2nd ed. Cambridge, UK: Cambridge University Press; 2009. pp. 452–490. [Google Scholar]
- 93.Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 94.Pond SK, Muse SV. Site-to-site variation of synonymous substitution rates. Mol Biol Evol. 2005;22:2375–2385. doi: 10.1093/molbev/msi232. [DOI] [PubMed] [Google Scholar]
- 95.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 96.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peitsch MC. Protein Modeling by E-mail. Nature Biotechnology. 1995;13:658–660. [Google Scholar]
- 98.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 99.Accelrys Inc. San Diego, CA: Accelrys Inc.; 2008. Discovery Studio 2.1. [Google Scholar]
- 100.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera, a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 101.Gerstein M. A Resolution-Sensitive Procedure for Comparing Protein Surfaces and its Application to the Comparison of Antigen-Combining Sites. Acta Cryst. 1992;A48:271–276. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotyping HIV-1(A) and HIV-2 (B) by maximum-likelihood phylogenetic analysis. The phylogenetic trees were constructed using the SPR heuristic search strategy and 1000 bootstrap replications, with reference sequences from HIV-1, under the TVM+G+I evolutionary model (A) and with reference sequences from HIV-2, under the GTR+G+I evolutionary model (B). The bootstrap values (above 50%) supporting the internal branches are shown. The scale bar represents evolutionary distances in substitutions per site.
(0.21 MB PDF)
Shannon's entropy of individual amino acids in the C2, V3 and C3 envelope regions in HIV-1 and HIV-2. (A) HIV-1 alignment (Control dataset), sites were numbered according to codon env position of HIV-1 HXB2 reference strain; (B) HIV-2 alignment (Control dataset), sites were numbered according to codon env position of HIV-2 ALI reference strain.
(0.96 MB TIF)
Frequency of N-glycosylation sites in the C2, V3 and C3 envelope regions in HIV-1 and HIV-2. (A) HIV-1 alignment (Control dataset). Sites were numbered according to codon env position of HIV-1 HXB2 reference strain. (B) HIV-2 alignment (Control dataset). Sites were numbered according to codon env position of HIV-2 ALI reference strain.
(0.58 MB TIF)
Positive selection in the C2, V3 and C3 envelope regions in HIV-1 and HIV-2. dN-dS values were estimated by FEL and scaled by the total codon tree length. (A) HIV-1 alignment (Control dataset). Sites were numbered according to codon env position of HIV-1 HXB2 reference strain. (B) HIV-2 alignment (Control dataset). Sites were numbered according to codon env position of HIV-2 ALI reference strain.
(0.53 MB TIF)
Superimposition of the conformational structures generated by homology modelling of Portuguese and Control C2, V3 and C3 regions of HIV-1 and HIV-2. In the schematics, Portuguese structures are represented in red, and Control structures are in blue.
(0.78 MB TIF)
Conformational structure of C2, V3 and C3 envelope regions in HIV-1 and HIV-2. The conformational structure of consensus amino acid sequences derived from the Control datasets was obtained by homology modeling as indicated in “Materials and Methods.” In the schematics, C2 is shown in red, V3 in yellow, and C3 in blue. Balls represent the amino acids under positive selection. (A) The red balls represent codons selected simultaneously by SLAC, FEL and REL methods, while green balls stand for codons selected by at least two of these methods. (B) Model structure showing the predicted interactions between V3, C2 and C3 in HIV-2 gp125. The non-covalent interaction involves residues Tyr296 and His301 in C2 binding, respectively, to Arg331 and Trp334 in V3, and Phe337 in C3 binding to Phe321 in V3.
(0.88 MB TIF)
Solvent accessible surface area, positive selection and potential N-glycosylation sites in C2-V3-C3 region. (A) HIV-1 alignment (Control dataset). Sites were numbered according to codon env position of HIV-1 HXB2 reference strain. (B) HIV-2 alignment (Control dataset). Sites were numbered according to codon env position of HIV-2 ALI reference strain. Coloured bars represent the amino acids under positive selection and have the same colours (red and green) as the corresponding positions (balls) highlighted in Figure S6. The dark blue stars over the bars correspond to potential N-glycosylation sites conserved along the alignment (present in ≥50% of strains), whereas the light blue stars represent sites only present in less than 50% of sequences.
(1.60 MB TIF)
Summary of results for phylogenetic, codon selection and solvent accessibility analysis for C2, V3 and C3 regions of HIV-1 and HIV-2 Control datasets.
(0.04 MB DOC)
Positively selected sites detected by SLAC, FEL, REL and/or IFEL in Control HIV-1 and HIV-2 env C2, V3 and C3 regions.
(0.10 MB DOC)
Alignment of HIV-1 reference sequences used as a Control for the Portuguese HIV-1 dataset. Each sequence is identified by the corresponding GenBank accession number.
(0.04 MB TXT)
Alignment of HIV-2 sequences used as a Control for the Portuguese HIV-2 dataset. Each sequence is identified by the corresponding GenBank accession number.
(0.02 MB TXT)