Abstract
Background
The Beijing family is a successful group of M. tuberculosis strains, often associated with drug resistance and widely distributed throughout the world. Polymorphic genetic markers have been used to type particular M. tuberculosis strains. We recently identified a group of polymorphic DNA repair replication and recombination (3R) genes. It was shown that evolution of M. tuberculosis complex strains can be studied using 3R SNPs and a high-resolution tool for strain discrimination was developed. Here we investigated the genetic diversity and propose a phylogeny for Beijing strains by analyzing polymorphisms in 3R genes.
Methodology/Principal Findings
A group of 3R genes was sequenced in a collection of Beijing strains from different geographic origins. Sequence analysis and comparison with the ones of non-Beijing strains identified several SNPs. These SNPs were used to type a larger collection of Beijing strains and allowed identification of 26 different sequence types for which a phylogeny was constructed. Phylogenetic relationships established by sequence types were in agreement with evolutionary pathways suggested by other genetic markers, such as Large Sequence Polymorphisms (LSPs). A recent Beijing genotype (Bmyc10), which included 60% of strains from distinct parts of the world, appeared to be predominant.
Conclusions/Significance
We found SNPs in 3R genes associated with the Beijing family, which enabled discrimination of different groups and the proposal of a phylogeny. The Beijing family can be divided into different groups characterized by particular genetic polymorphisms that may reflect pathogenic features. These SNPs are new, potential genetic markers that may contribute to better understand the success of the Beijing family.
Introduction
Mycobacterium tuberculosis is one of the most successful human pathogens, infecting nearly one third of the world's population. Despite efforts to combat the disease, tuberculosis (TB) remains a major public health problem, causing over 9 million new cases and 1.7 million deaths each year [1]. Polymorphic genetic markers have been used to discriminate and subtype M. tuberculosis strains to identify outbreaks. IS6110 restriction fragment length polymorphism typing is one of the most widely used methods, however, this technique is time consuming, technically demanding and insufficiently discriminatory for isolates containing less than five copies of IS6110. This has led to the development of other methods based on the polymorphism of repetitive sequences, either the direct repeat (DR) region (spoligotyping) or mini satellites (variable numbers of tandem repeats (VNTR) typing) [2]. Various M. tuberculosis families, such as the Beijing family, have been defined using these typing techniques [3]. The Beijing family represents a global threat to TB control. It is estimated that more than a quarter of worldwide TB cases are caused by Beijing strains [3], [4]. These strains have frequently been associated with drug resistance and their emergence and wide distribution suggests they have selective advantages over other M. tuberculosis strains [4], [5]. Beijing strains have a characteristic spoligotype pattern [3], [5] and VNTRs have been frequently used to type these strains, exhibiting differing discriminatory abilities per VNTR locus [6], [7].
The availability of whole-genome sequences has enabled comparative genomic analysis to identify single nucleotide polymorphisms (SNPs). SNPs have been used to differentiate between clinical isolates and are preferred over the use of repeats for the construction of phylogenetic trees, because recombination events that could occur independently at the level of repetitive sequences are avoided [8]. Large numbers of SNPs have been identified and used to genotype worldwide strain collections. This supported the grouping of M. tuberculosis into major families and provided useful information about the evolutionary history of this monomorphic bacteria [9], [10], [11]. As an example, the phylogeny of M. tuberculosis was recently established by sequencing 89 genes [12]. Nevertheless, detailed phylogenies about the various M. tuberculosis lineages are still lacking.
We recently identified a group of highly polymorphic genes involved in DNA replication, recombination and repair (3R) in a set of geographically diverse M. tuberculosis strains. We showed that the evolution of M. tuberculosis could be studied using SNPs in 3R genes and a potential, new, high-resolution tool for strain discrimination was developed [13]. Here we investigated the genetic diversity among Beijing family strains and searched for new polymorphisms in this family by sequencing 3R genes in a collection of Beijing strains from different geographic origins in order to disclose the phylogeny of the Beijing family.
Results and Discussion
A collection of 58 clinical isolates with a Beijing spoligotype [3] was used to search for variations in 3R genes. These isolates had different geographic origins: Madagascar (19), USA (18), The Netherlands (6), South Korea (2), South Africa (2), China (3), Malaysia (1), Mongolia (2), Thailand (2), Philippines (1), Singapore (1) and Russia (1) (Table S1). These Beijing isolates included the four different sublineages defined by large sequence polymorphisms (LSPs) previously described [14] (Figure 1B). Two non-Beijing M. tuberculosis strains, designated Myc1, which corresponds to the laboratory strain H37Rv, and Myc2 a clinical strain that belongs to Gutacker's cluster VI [11], were also included in this study.
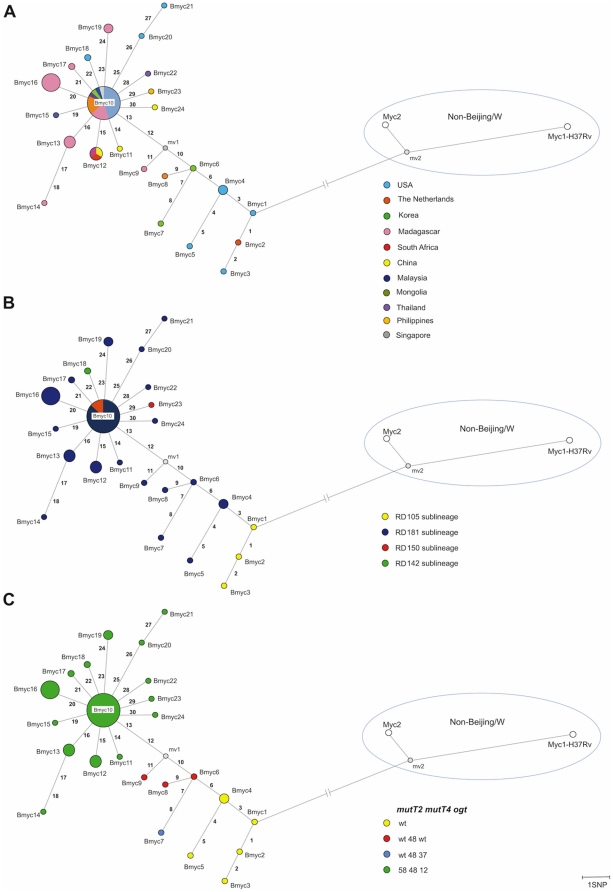
Figure 1. Phylogenetic network based on SNPs discovered in the collection of 58 Beijing isolates.
This phylogenetic network was constructed using the median-joining algorithm with the final set of 48 SNPs characterized by sequencing 22 3R genes in 58 Beijing isolates plus one non-Beijing isolate (Myc2). Isolates are color coded according to their geographic origin (A), large sequence polymorphisms (LSPs) (B) and, variations in mutT2 mutT4 and ogt genes (C). The reference strain M. tuberculosis H37Rv (Myc1) was also included. The numbers in each branch correspond to SNPs (Table 1) that enabled discrimination of sequence types. Node sizes are proportional to the number of isolates belonging to the same sequence type: Bmyc4 node (2); Bmyc12 node (3); Bmyc13 node (3); Bmyc19 (2); Bmyc16 node (7); Bmyc10 node (23). See Table S1 for details about strains belonging to each node. Mv represents a median vector created by the software and can be interpreted as possibly extant unsampled sequences or extinct ancestral sequences.
Of the 56 described genes encoding 3R components [13], 22 were previously demonstrated to be polymorphic among Beijing strains [13], [15]. These 22 genes (Table S2) were sequenced for each of the 58 Beijing isolates and the non-Beijing strain Myc2, resulting in approximately 1,6 Mbp of sequence data. Comparative analysis with the M. tuberculosis H37Rv (Myc1) genome sequence identified 48 SNPs (Table S2). Forty-one (85%) SNPs appeared to be specific for Beijing strains, as these were absent from the non-Beijing strain included in this study (Myc2) (Table S2), and also from the 86 non-Beijing M. tuberculosis strains included in a previous study [13]. Nineteen (46%) of these SNPs corresponded to new variations, not previously described in Beijing strains [12], [13], [15]. Thirty of the 41 Beijing specific SNPs (Table 1) enabled discrimination of 24 different sequence types for which a phylogenetic network was constructed using the Network software [16] (Figure 1A). Based on the inferred proteins, the number of non-synonymous SNPs (nsSNPs) was twice the number of synonymous SNPs (sSNPs) (Table 1). Phylogenetic relationships established by sequence types were in agreement with evolutionary pathways suggested by LSPs [14] and by SNPs in the putative DNA repair genes mutT2, muT4 and ogt [15] (Figure 1B and 1C). However, sequencing of the 22 genes was more discriminatory than LSPs; 24 sequence types versus four sublineages defined by the LSPs.
Table 1. Description of SNPs that enabled discrimination of the 26 sequence types among 305 M. tuberculosis Beijing strains (Figures 1 and 2).
| SNP number | Gene | Codon position | SNP type |
| 1* | ligD | 580 (CTG>TTG) | Synonymous |
| 2 | ligD | 162 (GAT>GCT) | Non-synonymous |
| 3* | recR | 44 (GGT>TGT) | Non-synonymous |
| 4 | ligD | 346 (GGC>GGT) | Synonymous |
| 5 | uvrC | 388 (CGG>CGC) | Synonymous |
| 6* | mutT4 | 48 (CGG>GGG) | Non-synonymous |
| 7* | ogt | 37 (CGC>CTC) | Non-synonymous |
| 8 | uvrC | 166 (CAG>AAG) | Non-synonymous |
| 9 | recX | 8 (CCG>CTG) | Non-synonymous |
| 10* | recX | 59 (GTT>CTT) | Non-synonymous |
| 11 | recG | 285 (CCT>TCT) | Non-synonymous |
| 12* | muT2 | 58 (GGA>CGA) | Non-synonymous |
| 13* | ogt | 12 (GGG>GGA) | Synonymous |
| 14 | recR | 89 (GAC>GAT) | Synonymous |
| 15* | recF | 269 (GGG>GGT) | Synonymous |
| 16* | uvrD1 | 462 (GGC>AGC) | Non-synonymous |
| 17 | ligB | 77 (GTC>GCC) | Non-synonymous |
| 18 | dnaQ | 161 (TTC>TTT) | Synonymous |
| 19 | nth | 122 (TTG>TGG) | Non-synonymous |
| 20* | dnaZX | 92 (CTG>TTG) | Synonymous |
| 21 | nth | 34 (GAG>GCG) | Non-synonymous |
| 22 | alkA | 11 (GCG>ACG) | Non-synonymous |
| 23* | mutT4 | 99 (TCG>TCA) | Synonymous |
| 24* | tagA | 129 (GCG>ACG) | Non-synonymous |
| 25* | recX | 153 (GGC>GAC) | Non-synonymous |
| 26* | radA | 276 (ATC>ACC) | Non-synonymous |
| 27 | recD | 139 (GTA>TTA) | Non-synonymous |
| 28 | recD | 277 (ACG>ACA) | Non-synonymous |
| 29 | radA | 186 (GTC>GCC) | Non-synonymous |
| 30 | tagA | 179 (GTC>GTT) | Synonymous |
*Most informative SNPs observed in this study.
Next we investigated the set of 30 polymorphic SNPs (Table 1), discovered by sequence analysis of the 3R genes, in a larger collection of Beijing strains including 192 Beijing clinical isolates from China and 55 Beijing strains isolated in South Africa (Table S1). The M. tuberculosis Beijing strain, GC 1237, responsible for a tuberculosis epidemic in Gran Canaria, Spain [17] was also included.
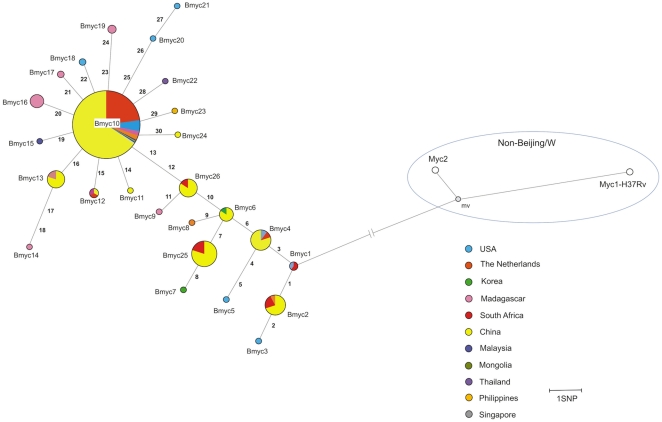
A phylogenetic network was constructed from this larger set of isolates (Figure 2). Certain SNPs that were previously found in a single isolate, were confirmed with this larger sample. Overall, fourteen SNPs were found in more than one isolate and were therefore informative (Table 1). Two new sequence types (Bmyc25 and Bmyc26) were identified (Figure 2).
Figure 2. Phylogenetic network based on SNPs charaterized in the entire collection of 305 Beijing isolates.
This phylogenetic network was constructed using the median-joining algorithm with the set of SNPs identified in the 3R genes analyzed on the final collection of 305 Beijing isolates. Isolates are color coded according to their geographic origin. M. tuberculosis strains Myc1 (H37Rv) and Myc2 are included as non-Beijing strains. The numbers in each branch correspond to SNPs (Table 1) that enabled discrimination of SNP types. Node sizes are proportional to the number of isolates belonging to the same SNP type: Bmyc1 node (2); Bmyc2 node (14); Bmyc4 node (13); Bmyc6 node (7); Bmyc25 node (28); Bmyc26 node (13); Bmyc12 node (3); Bmyc13 node (13); Bmyc16 node (7); Bmyc19 node (2); Bmyc10 node (188). See Table S1 for details about strains belonging to each node. Mv represents a median vector created by the software and can be interpreted as possibly extant unsampled sequences or extinct ancestral sequences. The relative proportion of isolates in each node, of a given geographic origin, may not reflect the population structure of the Beijing family of that geographic region.
The Beijing family can be divided into different groups characterized by particular SNPs. However, a recent sequence type, represented by the Bmyc10 node, appeared to be predominant in this family (Figure 2). Sixty-two percent of the isolates belonged to this group. This sequence type was found not only in China, where the Beijing family is highly prevalent, but also in other countries, where the Beijing family is less prevalent, such as Madagascar, The Netherlands and South Africa. In a recent study, a group of Beijing strains characterized by RD181 deletion and polymorphisms in mutT4 and mutT2 appear to be predominant in a collection of strains isolated in Italy [18]. Strains belonging to the Bmyc10 node also had the RD181 deletion and the same SNPs in mutT4 and mutT2 genes (SNP6 and SNP12). This suggests that this might indeed be a prevalent group of Beijing strains which can be found in different parts of the world. The effect on enzyme characteristics of the variation in the mutT2 gene (a characteristic of all isolates found in the R1 node, (SNP12, Figure 2)) has been investigated [19]. The results revealed significant changes in enzyme properties caused by a single amino acid substitution that leads to protein destabilization. It was suggested that this altered MutT2 enzyme may contribute to the success of strains due to an increase in nucleotide-dependent reactions. This suggests that the SNPs that we have discovered may have an effect on protein function and consequently confer advantageous phenotypes. Considering the high percentage of nsSNPs found (Table 1) it may be informative to investigate which of these variants might have a functional effect. They may confer advantageous phenotypes on certain Beijing genotypes, and play an important role in the evolution of the family. Our results showed that the Bmyc25 group might represent another predominant group of Beijing strains. This includes the Gran Canaria TB outbreak strain GC 1237 [17]. These observations suggest that several Beijing subtypes may be the result of the resurgence of tuberculosis in different regions.
When compared to other pathogens, M. tuberculosis complex strains are highly clonal, sharing 99% similarity at the nucleotide level [20]. In recent years, SNPs have been identified and used in order to get a more detailed insight into the evolutionary history of this organism [9], [10], [11], [12]. SNP analysis is a simple and relatively fast way to compare organisms and trace back the evolutionary history of strains, as some SNPs are highly informative. The increasing number of genome sequencing projects is making SNP analyses more and more attractive. This will provide important data, particularly relevant to understanding the genetic basis for strain differences in pathogenesis. Allelic variation in 3R genes seems to be an important mechanism in evolution and adaptation of microorganisms. Therefore, defective 3R systems could potentially increase genomic variability due to higher mutation rates. Strains with higher mutation rates (mutators) may, under certain conditions, have a selective advantage. For example, a strain may acquire mutations that induce antibiotic resistance or facilitate evasion of the host immune response [21]. The evolutionary history of a collection of 305 Beijing isolates was investigated by analyzing polymorphisms in 3R genes. We found SNPs in 3R genes associated with the Beijing family. These SNPs enabled discrimination of 26 different groups enabling a phylogenetic network to be constructed. The Beijing family can be divided into different groups presenting specific polymorphisms that may reflect pathogenic features. These new SNPs are potential genetic markers for Beijing strains that may contribute to a better understanding of the role of the Beijing family in the worldwide epidemic of tuberculosis.
Materials and Methods
M. tuberculosis Beijing clinical isolates included in this study are listed in Table S1. DNA from the 58 Beijing isolates, used to search for variations in 3R genes, was provided by the Madagascar Pasteur Institute (MG), RIVM, The Netherlands (NL), Scientific Institute of Public Health, Belgium (BE) and was used to amplify the 22 3R genes with primers listed in Table 2. These fragments were sequenced by the dideoxy chain-termination method using the Big Dye Terminator v3.1 cycle sequencing Kit (Perkin Elmer Applied Biosystems, Courtaboeuf, France) according to the manufacter's instructions. Sequencing products were run on an ABI prism 3100 Genetic Analyser (Applied Biosystems). Sequencing was also performed for SNP analysis of the non-beijing strain (myc2), the Bejing isolates from South Africa (ZA) and the GC 1237 strain (DNA provided from NRF Centre of Excellence in Biomedical Tuberculosis Research/MRC Centre for Molecular and Cellular Biology, South Africa and available in our laboratory).
Table 2. List of oligonucleotides (5′-3′) used in this study.
| Primer name | Sequence | Primer name (mismatch) | Sequence |
| ligD_f | GTCACGGCGAAATTCCACGCGATATTTGA | ligD580_f | CGGGCATTGGCGGAGGATCT |
| ligD_r | CCCGACCAGATCCAGCAACGACACGTC | ligD580_wt-f | TGCGTTAGCTAGGGTTTCGAGCAG |
| ligD_2 | TCACCAGCGGCAGCAAGGGATTGCAT | ligD580_mt-r | TGCGTTAGCTAGGGTTTCGAGCAA |
| ligD_3 | GATACACACCGAGGACCACCCGCTGGAATA | ligD162_f | CGACGACCTATCCGATCATCG |
| ligB_f | CCACATAGCCCCCAGGCGGTATTGGTA | ligD162_wt-r | GGAAGGTGACCAACCCGATAT |
| ligB_r | CGCTTGGTCGACGAGCGTGAATCTG | ligD162_mt-r | GGAAGGTGACCAACCCGATAG |
| ligB_2 | GGCACTCTACCGGGCAAAGGGTCTCAG | rccR44_f | AAGCGCCCCGCCCAGGACGTG |
| ligC_f | ACCCCAGCTTCGGGAAATACATCCTGT | rccR44_wt-r | CGGACATCGACCGGCTGACCG |
| ligC_r | TCGCCACACAGACGACAAGTCCCAA | rccR44_mt-r | CGGACATCGACCGGCTGACCT |
| dnaZX_f | CGCCGAAATCACGCCGAACGTTCA | ligD346_f | ACCACCATCGCGCCGTACTCA |
| dnaZX_r | CGAACGAAACAACCTGCAGCTACATCACG | ligD346_wt-r | ACCGCCCACGAGACCAGCACG |
| dnaZX_2 | AACACCTGATCTTCATATTCGCCACCA | ligD346_mt-r | ACCGCCCACGAGACCAGCACA |
| dnaZX_3 | CTGCTGCTGGAAGTGGTTTGCG | uvrC388_r | GGATCCCGAAGTGGCGGTAGT |
| recD_f | GGTGTGTTCACCTGGAACCCGCCCA | uvrC388_wt-f | CAACAACACAAGCTGAAGCGG |
| recD_r | GTCGCCGTGCTGTTCGTGTATGCGATGT | uvrC388_mt-f | CAACAACACAAGCTGAAGCGC |
| recD_2 | TCTCGCAAGGTGTTACGGTGTTGACTGG | mutT4-48_r | CGCATCAAATAATGGTGGACG |
| recG_f | CATGTGCACGACCACCATCCAGGCAC | mutT4-48_wt-f | CGGCAACGGCGAAGCGGTCCC |
| recG_r | CGATGATCCCAGCGTCTGATACGCGA | mutT4-48_mt-f | CGGCAACGGCGAAGCGGTCCG |
| recG_2 | CAGCACAAAAGTGCAGAGCTGGGACATCTT | ogt37_f | AGCTGGGCCTGCCTGCACAAC |
| recG_3 | GATGACGGCAGGGCAGAAGAAGCAAGTTC | ogt37_wt-r | GGTCGGGTGTCCAGTGTGTGC |
| recX_f | CCGACGTGGCTGACGAGATCGAGAAGAA | ogt37_mt-r | GGTCGGGTGTCCAGTGTGTGA |
| recX_r | CCGCCATCAAGTCGAGGTAAATTCGTTCA | uvrC166_f | CCGCTACCGCGACGACAAGTC |
| ruvB_f | GATACGGTGCTGGCCGCCAACCAT | uvrC166_wt-r | GCAGGCATGGACGATCGATCTG |
| ruvB_r | GGGGTCATTGCCAACGGCTCCTTTG | uvrC166_mt-r | GCAGGCATGGACGATCGATCTT |
| uvrC_f | CAATGCACCCGACCAACAGTGGGATAGC | recX8_r | GGCCGAGTTCGACATCCTCTA |
| uvrC_r | CCGGACAGCCCGGTTACCAAGACGA | recX8_wt-f | CTTCGCGCTCAGAAGTCGACG |
| uvrC_2 | TACATCGACAAATGTTCCGCGCCGTGT | recX8_mt-f | CTTCGCGCTCAGAAGTCGACA |
| uvrC_3 | CGGTGCACCGAAACGCAGAAGATGC | recX59_wt-f | GGTGTCATCCACCAGGCCAAC |
| recR_f | AAGATGGCGCAGGAACGGCTGGGT | recX59_mt-f | GGTGTCATCCACCAGGCCAAG |
| recR_r | GAGATCAACATTTTGCAGGCAAGGTGCG | recX59-r | CTCGGCCAGGGCAAGGAGAAT |
| nei_f | TCTGGTCGAGCGGGCCGACGGCAT | recG285_r | CGTGCGGCAGGTGCTCGATGT |
| nei_r | GGTGGCAGGCAATATCTGCCCAAGGCGG | recG285_wt-f | TCCCGCCGTCAGCTCAAAAGG |
| nth_f | ATGACACAAGGAGAGTAAACATGGC | recG285_mt-f | TCCCGCCGTCAGCTCAAAAGA |
| nth_r | AATAGTCATGCAGTTGGGCAACCA | mutT2-58_f | CCGGCCATAAACGTCGGAAAC |
| rv2979_f | GTTCGAAGGTCCACAGGGCCAGAACG | mutT2-58_wt-r | GAGGTCGGCGACCTCGAGTCC |
| rv2979_r | TCCAGTTGTATGCCTTGCGACGAGCA | mutT2-58_mt-r | GAGGTCGGCGACCTCGAGTCG |
| tagA_f | TGAGCTCGAGGCGCTACGCTCTCAGC | ogt12_f | CCGCAGGAGAAGATCGCAT |
| tagA_r | CCCCGCCATTGGATTTCCAGCCATA | ogt12_wt-r | GCCCGGCCAGGGTTAATAGC |
| uvrD1_f | CCCGCAAAAACTTGGCGGGAAAAGTG | ogt12_mt-r | GCCCGGCCAGGGTTAATAGT |
| uvrD1_r | GGACTTAGCGTCGGCAATTACACCGGTTGA | recR89_f | CGGACGCGATCCGTGTGACGG |
| uvrD1_2 | CAACCTGAAGAACGAGTTGATCGACCC | recR89_wt-r | GTGCATTGTCGAGGAACCCAAAGAC |
| uvrD1_3 | CGAGGGTAGCGAGATCACCTACAACGAT | recR89_mt-r | GTGCATTGTCGAGGAACCCAAAGAT |
| dnaQ_f | CGGGTGGTTACCACCCGGGCAGTTTAC | recF269_f | GGCGGAGCACGGGGCTGAACT |
| dnaQ_r | TCTCGCAAGGTGTTACGGTGTTGACTGG | recF269_wt-r | CGGTGCGGACCAACTAGACAAACC |
| radA_f | TAATGGTGCCGATCTCGGCCGGATT | recF269_mt-r | CGGTGCGGACCAACTAGACAAACA |
| radA_r | GTTGCTGCATAGCGGACATCGAGGGAGAA | uvrD1-462_f | TCCGCGCCGGTATTCCGTACA |
| radA_2 | GAGATCTACCTCGCCGCACAGTCCGA | uvrD1-462_wt-r | GACGAGCGCGTCACCGAAGCC |
| recF_f | GGAGCGAGTGTCTTTCGGGTTTACGACTGC | uvrD1-462_mt-r | GACGAGCGCGTCACCGAAGCT |
| recF_r | CGCCCTCGACCGGCGTCTTGTCC | ligB77_r | TGTCGGCGAGACATGCCAAGCT |
| mutT2_f | CTGCCAGCCGTTGAGGTCGT | ligB77_wt-f | GGGTGGCGTCGACACCGGTGA |
| mutT2_f | CGGGCATGCAAACCCAAGTTA | ligB77_mt-f | GGGTGGCGTCGACACCGGTGG |
| mutT4_f | TCGAAGGTGGGCAAATCGTG | dnaQ161_f | GGACCAGCGGGCGGCCCTGGA |
| mutT4_r | TGGGGTTCGCTGGAAGTGG | dnaQ161_wt-r | CAACGGCCGCACGATGCATTC |
| ogt_f | CAGCGCTCGCTGGCGCC | dnaQ161_mt-r | CAACGGCCGCACGATGCATTT |
| ogt_r | GACTCAGCCGCTCGCGA | nth122_f | CCCGCCGTCCGTGAAAGATCA |
| alkA_f | AGCCGCGTAGGTAACCT | nth122_wt-r | GCCGGCCACCATGGACAAGTT |
| alkA_r | TGCTCGAGCATCCGCAG | nth122_mt-r | GCCGGCCACCATGGACAAGTG |
| alkA_2 | CGCATGCAGACCGCCCG | nth34_f | TCACCGCCAAACCGCTCAA |
| alkA_3 | CACTGCACGTTGCCGAC | nth34_wt-r | ATTTCCGCACGTATACTGAGA |
| alkA_4 | GCTGACGATGCCGTTGCC | nth_34_mt-r | ATTTCCGCACGTATACTGAGC |
| dnaZX92_f | GCGAGCAACGCCCGCATAGT | ||
| dnaZX92_wt-r | GCAGCATCGACGTGGTAGAGC | ||
| dnaZX92_mt-r | GCAGCATCGACGTGGTAGAGT | ||
| alkA11_f | ATCGCCCGCGCCACGACGTCA | ||
| alkA11_wt-r | ACTTCGAACGCTGCTACCGGG | ||
| alkA11_mt-r | ACTTCGAACGCTGCTACCGGA | ||
| mutT4-99_f | GGCGGCGCGCTGCGGCTACAG | ||
| mutT4-99_wt-r | CAACTCGATGTGCCCCTTGGGTAGC | ||
| mutT4-99_mt-r | CAACTCGATGTGCCCCTTGGGTAGT | ||
| recX153_f | GCGGGCGAACGCAGCAAAGAG | ||
| recX153_wt-r | CCTCGCACGCCAAGGTCTGGC | ||
| recX153_mt-r | CCTCGCACGCCAAGGTCTGGT | ||
| recD277_r | CGGGCCTGGCACCGGGAAGAC | ||
| recD277_wt-f | TCGGCCAGCCGGGCCATCAGC | ||
| recD277_mt-f | TCGGCCAGCCGGGCCATCAGT | ||
| radA186_f | TCCGGACGGCGCGCGCTCTAT | ||
| radA186_wt-r | CCTGCGTGACCCCGCCGGTGA | ||
| radA186_mt-r | CCTGCGTGACCCCGCCGGTGG | ||
| tagA179_f | GTGCGCAACCGCGCCAAGATT | ||
| tagA179_wt-r | CCAGCATGCTTGGATATGGTCGTCG | ||
| tagA179_mt-r | CCAGCATGCTTGGATATGGTCGTCA |
The name of the target gene and position of the oligonucleotide is followed by the oligonucleotide sequence. (f) for forward and (r) for reverse oligonucleotides used for amplification and sequencing reactions. Oligonucleotides whose name finishes in number were used for sequencing reactions. (wt) for wild-type and (mt) for mutant oligonucleotides used for detection of SNPs by mismatched PCR (see materials and methods).
Sequences were analysed using the software Genalys obtained at http://software.cng.fr/. The genome sequences of M.tuberculosis H37Rv were obtained from the Institut Pasteur at http://genolist.pasteur.fr and used for detection of SNPs.
A mismatched PCR method, using one wild-type primer and one containing the SNP which matched/mismatched the template DNA at the 3′-end of the primer (Table 2), was used to detect SNPs in the Beijing isolates from China (CN).
SNPs were concatenated resulting in one character string (nucleotide sequence) for each clinical isolate analyzed. A FASTA file was created to run in the Network software [16] to build a phylogeny based on the median-joining method. This software assumes that there is no recombination between genomes.
Supporting Information
Description of M. tuberculosis Beijing strains belonging to each node found in Fig. 1 and 2 , and respective country of isolation.
(DOC)
Full list of 48 SNPs identified in this study. The first line indicates the gene and the second line indicates the position on that gene where polymorphisms were identified in relation to M. tuberculosis H37Rv strain (bottom). Polymorphisms that characterize and allowed discrimination of the 26 sequence types (Figure 2 and Table 1) are marked in red.
(XLS)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work is part of the TB-ADAPT (LSHP-CT-2006-037919) and TB-VIR (Grant agreement n° 200973) projects supported by the European Commission under the Health Cooperation Work Programme of the 6th and 7th Framework Programme, respectively. This work was also supported by the Key Project of Chinese National Programs (2008ZX10003-010). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Anonymous. Geneva, Switzerland: World Health Organization; 2008. WHO report 2008: global tuberculosis control—surveillance, planning, financing. [Google Scholar]
- 2.Van Soolingen D. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J Intern Med. 2001;249:1–26. doi: 10.1046/j.1365-2796.2001.00772.x. [DOI] [PubMed] [Google Scholar]
- 3.Kremer K, Glynn JR, Lillebaek T, Niemann S, Kurepina NE, et al. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J Clin Microbiol. 2004;42:4040–4049. doi: 10.1128/JCM.42.9.4040-4049.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg Infect Dis. 2006:736–743. doi: 10.3201/eid1205.050400. 2006/05/18 ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 2002;10:45–52. doi: 10.1016/s0966-842x(01)02277-6. [DOI] [PubMed] [Google Scholar]
- 6.Kremer K, Au BK, Yip PC, Skuce R, Supply P, et al. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J Clin Microbiol. 2005;43:314–320. doi: 10.1128/JCM.43.1.314-320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, Chen J, Shen X, Gui X, Mei J, et al. Highly polymorphic variable-number tandem repeats loci for differentiating Beijing genotype strains of Mycobacterium tuberculosis in Shanghai, China. FEMS Microbiol Lett. 2008;282:22–31. doi: 10.1111/j.1574-6968.2008.01081.x. [DOI] [PubMed] [Google Scholar]
- 8.Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 9.Baker L, Brown T, Maiden MC, Drobniewski F. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg Infect Dis. 2004;10:1568–1577. doi: 10.3201/eid1009.040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filliol I, Motiwala AS, Cavatore M, Qi W, Hazbon MH, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol. 2006;188:759–772. doi: 10.1128/JB.188.2.759-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutacker MM, Smoot JC, Migliaccio CA, Ricklefs SM, Hua S, et al. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics. 2002;162:1533–1543. doi: 10.1093/genetics/162.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dos Vultos T, Mestre O, Rauzier J, Golec M, Rastogi N, et al. Evolution and diversity of clonal bacteria: the paradigm of Mycobacterium tuberculosis. PLoS One. 2008;3:e1538. doi: 10.1371/journal.pone.0001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsolaki AG, Gagneux S, Pym AS, Goguet de la Salmoniere YO, Kreiswirth BN, et al. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J Clin Microbiol. 2005;43:3185–3191. doi: 10.1128/JCM.43.7.3185-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rad ME, Bifani P, Martin C, Kremer K, Samper S, et al. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg Infect Dis. 2003;9:838–845. doi: 10.3201/eid0907.020803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 17.Caminero JA, Pena MJ, Campos-Herrero MI, Rodriguez JC, Garcia I, et al. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am J Respir Crit Care Med. 2001;164:1165–1170. doi: 10.1164/ajrccm.164.7.2101031. [DOI] [PubMed] [Google Scholar]
- 18.Rindi L, Lari N, Cuccu B, Garzelli C. Evolutionary pathway of the Beijing lineage of Mycobacterium tuberculosis based on genomic deletions and mutT genes polymorphisms. Infect Genet Evol. 2009;9:48–53. doi: 10.1016/j.meegid.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Moreland NJ, Charlier C, Dingley AJ, Baker EN, Lott JS. Making sense of a missense mutation: characterization of MutT2, a Nudix hydrolase from Mycobacterium tuberculosis, and the G58R mutant encoded in W-Beijing strains of M. tuberculosis. Biochemistry. 2009;48:699–708. doi: 10.1021/bi8009554. [DOI] [PubMed] [Google Scholar]
- 20.Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denamur E, Matic I. Evolution of mutation rates in bacteria. Mol Microbiol. 2006;60:820–827. doi: 10.1111/j.1365-2958.2006.05150.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of M. tuberculosis Beijing strains belonging to each node found in Fig. 1 and 2 , and respective country of isolation.
(DOC)
Full list of 48 SNPs identified in this study. The first line indicates the gene and the second line indicates the position on that gene where polymorphisms were identified in relation to M. tuberculosis H37Rv strain (bottom). Polymorphisms that characterize and allowed discrimination of the 26 sequence types (Figure 2 and Table 1) are marked in red.
(XLS)