Abstract
Optogenetic modules offer cell biologists unprecedented new ways to poke and prod cells. The combination of these precision perturbative tools with observational tools, such as fluorescent proteins, may dramatically accelerate our ability to understand the inner workings of the cell.
Biology has always been primarily an observational science, and in the modern era, the development of genetically encoded fluorescent proteins such as GFP has given us the unprecedented ability to peer into the living cell and to observe its inner workings. We can now study individual cells in culture or in the context of a whole organism and directly observe where proteins are localized, their dynamics and their variability in expression level. More than ever, we now appreciate that the cell is not a bag of molecules but an anisotropic structure with highly complex spatial organization. We can see examples of how this organization shifts in dynamic processes, ranging from cell-shape changes to signal transduction propagated from the plasma membrane to the nucleus.
But what are the mechanisms that underlie and orchestrate these complex behaviors? Sadly, our ability to systematically perturb and interrogate the intracellular networks that control cell behavior (and thus our ability to understand their mechanism) has lagged behind our ability to observe these behaviors. The standard genetic perturbation techniques—knockdown, overexpression and mutation—are extremely effective at identifying the proteins involved in a phenotype, but are less effective at extracting mechanism. These perturbations are slow in timescale and broad in effect and, except in lucky circumstances, are more likely to destroy rather than modulate specific spatiotemporal features of the network's response. Pharmacological perturbations have been extremely useful tools—small molecules that target or block specific molecules give the investigator the ability to rapidly switch off the function of a target protein. But these approaches do not allow spatial control, and in most cases good fortune or considerable engineering1 is required to obtain highly specific inhibitors.
Light-gated protein modules provide a potentially transformative solution to the problem of dissecting cellular network function. There is currently an explosion of new light-controlled modules that can, in principle, be used to control the function and localization of diverse proteins. Such general new tools could usher in a new era of perturbative biology that would transform our ability to interrogate, dissect and understand the mechanisms of complex biological systems. Such light-gated modules might serve as the workhorse perturbative tool that complements GFP as an analytical tool. Here we briefly review optogenetic tools that have emerged over the last few years and discuss how they may be applied to cell biology in the near future.
The toolbox: linking light to protein activity
A growing suite of light-controllable tools are already available to the biologist. In the last few years, one of the biggest advances has been the proliferation of genetically encoded light-control modules. Unlike classical photocaging, these elements do not involve chemical modification ex vivo followed by injection or other means of reintroducing the molecule into the cell. Genetically encoded elements also have the convenience and flexibility that has been so powerful for GFP (versus chemical labeling with a fluorophore). A second advance has been the development of light-control modules that are generic in function and can be used with a broad range of signaling currencies. For neuro-optogenetics, some of the workhorse tools have been light-gated ion channels that can activate or inhibit neuronal signaling2,3. A broader range of optogenetically controlled cell signals will be necessary to access the wide spectrum of cellular functions that do not involve ion flux.
Here we focus on the new generation of light-control modules that are genetically encoded and generic (or modular—that is, can be used to control diverse functions in a cell). Nearly all of these modules are borrowed from organisms that have sophisticated light-sensing systems. Generally these are protein modules that contain photoisomerizable chromophores, which, when activated by the proper wavelengths of light, will cause a conformational change in the protein. Broadly, these tools use two general mechanisms to link photoswitching to generic protein activities.
The first strategy is to allosterically link photoactivation to protein activity (Fig. 1a). For example, one can genetically insert a light-responsive light, oxygen, voltage (LOV) domain into a protein of interest such that this domain, in one conformation, will sterically block or perturb protein function. Photoisomerization of the LOV domain releases the allosteric block on protein function. This approach has been used for light-gated control of the GTPase Rac4. This modular mechanism is conceptually similar to chemical (or, more recently, genetic) photocaging strategies5, but has the advantage of reversibility. Photoisomerized LOV domain variants revert to the inactivating state at timescales ranging from seconds to hours.
figure 1.
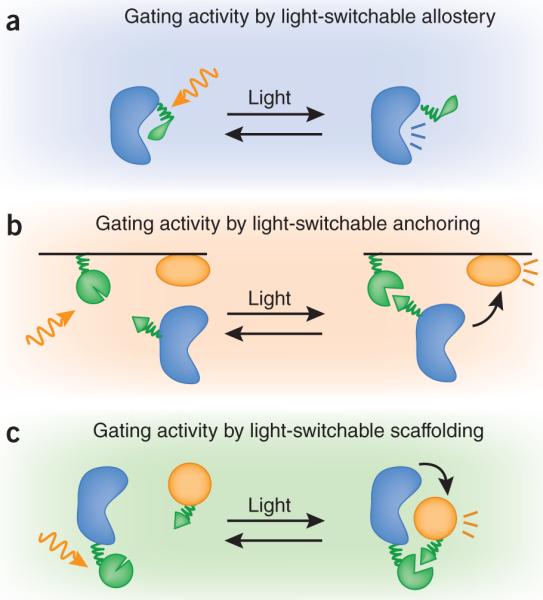
Modes of light-regulated biochemistry. (a) Protein activity can be put directly under light control by fusion to light-responsive domains or residues (green). Upon stimulation with light (gold arrow), allosteric inhibition is removed, leading to activation. (b,c) Protein activity can be indirectly controlled using light-dependent anchoring to a subcellular compartment (b) or scaffolding (c).
Another strategy for regulating cell signaling is through light controlled protein-protein interaction. Throughout cell biology, we know that recruitment of proteins to new locations and new complexes is frequently used to gate their function. Harnessing this property, researchers have used chemically gated protein dimerization to regulate many signaling pathways6. Similarly, photoactivated interaction pairs, such as the phytochrome-PIF interaction pair from Arabidopsis thaliana7 can be used to regulate diverse cell functions. These interaction pairs can be used to control protein subcellular localization (Fig. 1b), as has been demonstrated by the light-gated localization of a Rac guanine nucleotide exchange factor to the membrane, which is sufficient to allow it to activate Rac and lead to subsequent actin polymerization7. In principle, a similar inverse strategy could also be used to recruit proteins away from their site of action, thereby turning them off8. An alternative way to use interaction pairs is to link split portions of proteins that must associate to function. Examples of this strategy include a light-controlled yeast two-hybrid transcriptional switch9 or a light-controlled activation of a split Cre recombinase10. Light interactions could be used to directly recruit partners in a signaling cascade together (for example, kinase and substrate), acting as a scaffold to promote pathway activation (Fig. 1c). Notably, the development of new light-gated interaction pairs that are activated at different wavelengths, such as the cryptochrome-CIB1 system10, suggests a future in which the spectrum of control-lable modules matches that of fluorescent proteins as observational modules.
Variable inputs: opening the black box of cell signaling
In the coming years, researchers will undoubtedly discover more light-switchable modules as well as new ways in which these modules can be used to control molecular functions. But what remains less certain is how exactly these tools might be used to push forward our understanding of cell biology. Here we focus on some possible research applications.
As with any emerging technology, we cannot predict exactly which biological investigations will be transformed by optogenetic tools. However, these techniques are likely to be immediately useful for systems whose proper function requires activation that is transient or spatially restricted. Perhaps the most important issue is that the cell biologist who uses these tools may need to rethink their paradigm for experimental design to move away from simply manipulating the system in `natural' ways.
Highly controlled perturbative tools are traditionally used in engineering to deconstruct and decode the internal mechanism and workings of, for example, a complex electronic device. Similarly in biochemistry and biophysics, systems are reconstituted with diverse compositions and concentrations, even in regimes that are far from those observed in vivo because one can mechanistically distinguish distinct models by moving to these regimes. In molecular mechanical systems (for example, motors), exposure to nonnatural forces is useful to determine their global physical and energetic properties and thus their underlying mechanism. These approaches are unified by the logic of interrogating a system by probing it with variable inputs, thereby learning about its inner workings by observing the ways in which it responds.
In an analogous way, cell biologists can take advantage of light-controlled tools to perturb spatial signaling in diverse but systematic ways. In many cellular and developmental processes (for example, cell polarization, migration and developmental patterning) the spatial dynamics of intracellular signals are likely to be critical for function. Imagine if optogenetic tools could be used to paint on arbitrary and diverse spatial distribution functions of inputs. These approaches could be incredibly helpful in discriminating between models for how molecular circuits interpret these signals. Although some microfluidic systems have been used to create diverse input patterns, these have largely been limited to diffusible, extracellular inputs. Optogenetics has the potential to create arbitrary input patterns at almost any level in a network, potentially even in a developing organism.
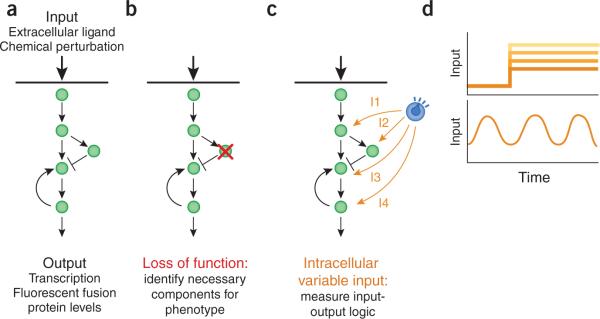
Optogenetic tools also have the potential to fulfill the promise of `in vivo biochemistry'. Whereas fluorescent proteins allow us to quantitatively measure concentrations and distributions of molecules in vivo, any good biochemist knows that uncovering mechanism requires systematic variation of concentrations and other system parameters. Traditional tools for dissecting signaling pathways in vivo excel at identifying the components required for signal propagation and the signs of interaction (activating or inhibiting) between components (Fig. 2a,b).
figure 2.
In vivo biochemistry: from component lists to signal processing. (a) Cell regulatory networks are comprised of cascades of interacting proteins as well as feedback and feed-forward loops. Typically, they are stimulated by extracellular ligands or pharmacological agents and observed by following protein levels or pathway activity (such as a transcriptional response). (b) Classical chemical and genetic perturbations block or enhance individual nodes to identify phenotype changes. (c,d) Light-gated inputs can be used to specifically and precisely perturb activation at distinct nodes in a pathway (inputs 1–4, labeled I1 to I4) (c), using a rich set of temporal inputs including fixed levels of activation and frequency-modulated signals (d).
More and more, crucial questions go beyond identifying pathway components to ask how collections of components operate together to perform their function. Light control presents an opportunity to dial in the activity or local concentration of intracellular components through modulation of the activating light intensity. It may be possible to clamp concentrations of some active intermediate at fixed levels, something akin to voltage clamping of channels or positional clamping in optical tweezer experiments. In the complex dynamical systems in cells, these types of experiments may yield a wealth of important mechanistic information.
In cell signaling, there is much interest in understanding how a signal is transmitted, changed and interpreted as it passes down a cascade or through a network. The flexibility of optogenetic control promises to enable one to insert a light-controlled dial into many steps of a pathway, including intracellular steps (Fig. 2c). Switching extracellular stimuli on and off has been a standard technique for studying these circuits, but switchable intracellular perturbations have typically been inaccessible. By walking down a pathway and systematically varying input (while also observing output at various steps in the pathway), one can directly observe how signals are processed at each step. This mode of analysis could be used to uncover detailed information about signaling networks such as the critical nodes for feedback control or ultrasensitivity11.
The speed and control of optogenetics also offers the tantalizing possibility of probing nearly any cell-signaling system with arbitrary time-variant or oscillatory-input patterns (Fig. 2d). This would be obviously important for systems that appear to use frequency variation to encode information (for example, calcium signaling and frequency-modulated nuclear import)12,13. But even for signaling systems that do not normally interpret changing input frequencies, presenting a cell with variable time inputs could prove to be very informative14. Currently we have very poor capabilities to map or identify feedback control in cellular networks even though we postulate that feedback control has a central role in cellular signal processing. There is a long history in electronics of using time-varying input stimulation to identify modes of feedback present in a circuit. Similar approaches, now controlled by light oscillations, could be used to uncover feedback linkages in living cells.
Challenges facing optogenetic cell biology methods
To make optogenetic systems widely accessible and easy to use by the broad scientific community, both the technologies of light delivery and analysis, and the properties of the molecular components themselves must be improved. In the case of fluorescent proteins, technological developments in microscopy as well as improvements in the properties of the proteins themselves greatly improve their utility.
Fluorescent proteins are ideal observational tools because they report spatial and temporal information, and are highly modular (they can be flexibly fused to almost any target protein). Similarly, to realize their full potential, light-based perturbative tools must meet a set of core criteria: they must be reversible, rapid and modular, and must permit the direct readout of light-induced activity. Without reversible activation, it is impossible to maintain high-resolution spatial inputs, even if the system is exposed to patterned light inputs (Fig. 3). This arises because once a mobile light-gated molecule is activated in a lit region, it would be free to travel to an unlit region and maintain its activity, blurring the spatial pattern of activity. Reversibility is also required for time-varying stimuli implementing decreases in activity, and could prove useful for fixing activation at intermediate levels.
figure 3.
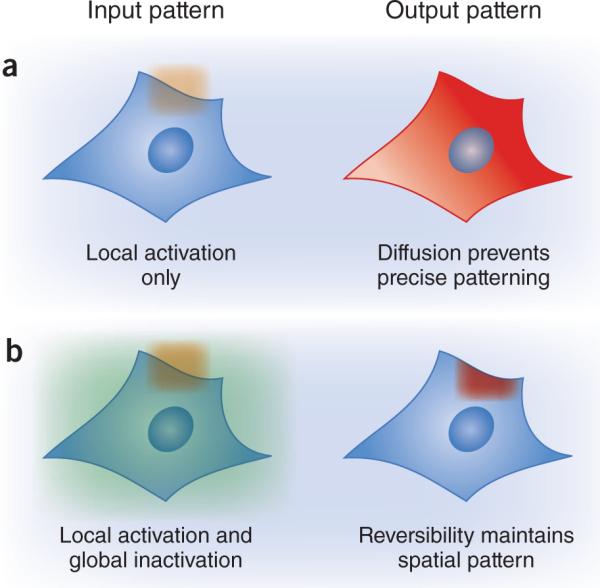
Reversibility and spatial precision. (a) Optogenetics allows the investigator to apply spatially restricted light inputs (orange). However, diffusion of protein activity (red) from the site of activation destroys the applied spatial pattern. (b) Faithful spatial patterns can be maintained by coupling local activation with global inactivation (green), either by implementing an inactivating light wavelength or a short-lived active state.
Finally, for such techniques to be truly quantitative, live cell readouts of the magnitude of the optogenetic input will be essential. Quantitative in vivo biochemistry approaches rely on measuring the response to a known amount of enzyme (or enzymatic activity). However, because concentrations of components vary between cells, the same light dose is unlikely to lead to the same activity increase in different cells. Fortunately, a variety of direct readouts are possible, depending on the optogenetic technique. For light-gated protein-protein interactions, directly reading out complex formation by fluorescence resonance energy transfer or localization changes is appealing. The activation of light-controlled ion channels can be measured by voltage sensors. Doubtless, future developments will suggest additional ingenious solutions to this challenge, especially for unimolecular modes of light-induced activation.
Addressing other challenges is less crucial but could greatly aid the ease with which optogenetic approaches can be used. Some current methods require the addition of an exogenous chromophore, often chemically synthesized or purified from another organism. Although preventing light responsivity until the time of chromophore addition is often useful for experimental design, chromophore addition can be an obstacle. This is especially true in the case of organism or tissue imaging, where accessibility to the added chromophore is limited. In whole organisms, efficient delivery of both the light input and any additional cofactors will be critical. It will be exciting to see how refinement of each technique will enable them to meet each of these challenges.
Optogenetics is one of a growing number of approaches to transform biology from an observational to a generative discipline. In recent years, synthetic biology has been successful both in engineering biological circuits with new function and in rewiring natural pathways to modulate their response. Optogenetics nicely extends and complements these approaches: in addition to determining how natural systems will respond when perturbed at various levels, synthetic biologists can envision varying pathway inputs to characterize engineered biological components, or tune the strength of connections to optimize system function. One might even envision using light as an input for fine-tuning pathway activation over time, with an eye toward controlling cell behaviors in useful or informative ways.
ACKNOWLEDGMENTS
This work was partially supported by the Cancer Research Institute postdoctoral fellowship (to J.E.T.); US National Institutes of Health (NIH) grants EY016546 and AI067699, National Science Foundation (NSF) grants BES-0547637, EEC-0540879 and CBET-0943302, Office of Naval Research grant N00014-10-1-0245 and the NSF Synthetic Biology Engineering Research Center (to C.A.V.); NIH grant GM084040 (to O.D.W.); and NIH grants GM55040, GM62583, GM081879 and EY016546, the Howard Hughes Medical Institute, Packard Foundation and the NSF Synthetic Biology Engineering Research Center (to W.A.L.).
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Bishop AC, et al. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 2.Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Nat. Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunaydin LA, et al. Nat. Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 4.Wu YI, et al. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gautier A, et al. J. Am. Chem. Soc. 2010;132:4086–4088. doi: 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]
- 6.Spencer DM, Graef I, Austin DJ, Schreiber SL, Crabtree GR. Proc. Natl. Acad. Sci. USA. 1995;92:9805–9809. doi: 10.1073/pnas.92.21.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levskaya A, Weiner OD, Lim WA, Voigt CA. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haruki H, Nishikawa J, Laemmli UK. Mol. Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 9.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. Nat. Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy MJ, et al. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cluzel P, Surette M, Leibler S. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 12.Cai L, Dalal CK, Elowitz MB. Nature. 2008;455:485–490. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsien RW, Tsien RY. Annu. Rev. Cell Biol. 1990;6:715–760. doi: 10.1146/annurev.cb.06.110190.003435. [DOI] [PubMed] [Google Scholar]
- 14.Mettetal JT, Muzzey D, Gomez-Uribe C, van Oudenaarden A. Science. 2008;319:482–484. doi: 10.1126/science.1151582. [DOI] [PMC free article] [PubMed] [Google Scholar]