Abstract
The integrity of the genome is constantly challenged by intrinsic and extrinsic genotoxic stresses that damage DNA. The cellular responses to DNA damage are orchestrated by DNA damage signaling pathways, also known as DNA damage checkpoints. These signaling pathways play crucial roles in detecting DNA damage, regulating DNA repair, and coordinating DNA repair with other cellular processes. In vertebrates, the ATM- and Rad3-related (ATR) kinase plays a key role in the response to a broad spectrum of DNA damage and DNA replication stress. In this article, we will discuss the recent findings on how ATR is activated by DNA damage, and how it protects the genome against interference with DNA replication.
A model for ATR activation
The DNA damage signaling pathway regulated by the ATR–ATRIP kinase (Box 1 and Table 1) responds to several types of DNA damage and replication interference, such as DNA breaks, adducts, crosslinks, and inhibition of DNA polymerases. The versatility of ATR suggests that it is activated by certain DNA and/or protein structures commonly induced by DNA damage and replication problems. In the past decade, studies using yeast, Xenopus laevis, and mammalian systems have provided strong evidence that single-stranded DNA (ssDNA) is the key structure triggering ATR activation. ssDNA is a DNA replication and repair intermediate commonly induced by stressed DNA replication forks and multiple DNA repair pathways. Once generated in cells, ssDNA is rapidly coated by the ssDNA-binding protein complex RPA (replication protein A) (Fig. 1). ATRIP directly binds RPA-coated ssDNA and provides a crucial means to localize the ATR–ATRIP complex to sites of DNA damage and stressed replication forks [6] (Fig. 1).
BOX 1. The ATR–ATRIP kinase complex.
The ATM- and Rad3-related (ATR) kinase is a member of the phosphoinositide 3-kinase (PI3K)-like protein kinase (PIKK) family [1]. Other members of the PIKK family include ataxia telangiectasia mutated (ATM), DNA dependent protein kinase (DNA-PK), homolog of Caenorhabditis elegans SMG-1 (SMG1), and mammalian target of rapamycin (mTOR; also known as FRAP). All PIKKs display similar domain structures. The kinase domains are located near the C terminus, and are flanked by the conserved FRAP/ATM/TRRAP (FAT) and FAT C terminal (FATC) domains. The N terminal regions of PIKKs contain multiple α-helical HEAT (Huntingtin, elongation factor 3, protein phosphatase 2A, and yeast TOR1) repeats. The similar domain structures of PIKKs suggest that they might be regulated by analogous mechanisms. Several PIKKs are known to associate with distinct regulatory partners. In human cells, ATR associates with ATRIP (ATR-interacting protein)[2]. ATRIP is required for the stability and functions of ATR, suggesting that ATR and ATRIP exist and function as a complex. ATRIP plays a critical role in the localization and stimulation of the ATR–ATRIP kinase complex. In addition to ATRIP, ATR also interacts with TEL2 (telomere maintenance 2; also called HCLK2) [3]. Unlike ATRIP, however, TEL2 binds all PIKKs and is important for the stability of these kinases. Recent studies showed that a large complex containing TEL2, the TEL2-interacting proteins TTI1 and TTI2, and HSP90 preferentially binds newly synthesized ATR, and promotes formation of the ATR–ATRIP complex [4, 5].
Table 1.
Conserved proteins in the ATR checkpoint pathway
| Saccharomyces cerevisiae | Xenopus laevis | Human | |
|---|---|---|---|
| PIKK | Mec1 | Xatr | ATR |
| PIKK partner | Ddc2 (Lcd1) | Xatrip | ATRIP |
| RPA complex | Rfa1 | Xrpa1 | RPA1 (RPA70) |
| Rfa2 | Xrpa2 | RPA2 (RPA32) | |
| Rfa3 | Xrpa3 | RPA3 (RPA14) | |
| RAD17-RFC complex | Rad24 | Xrad17 | RAD17 |
| Rfc2 | Xrfc2 | RFC2 | |
| Rfc3 | Xrfc3 | RFC3 | |
| Rfc4 | Xrfc4 | RFC4 | |
| Rfc5 | Xrfc5 | RFC5 | |
| 9-1-1 complex | Ddc1 | Xrad9 | RAD9 |
| Rad17 | Xrad1 | RAD1 | |
| Mec3 | Xhus1 | HUS1 | |
| ATR activator | Dpb11 | XtopBP1 (Xcut5) | TOPBP1 |
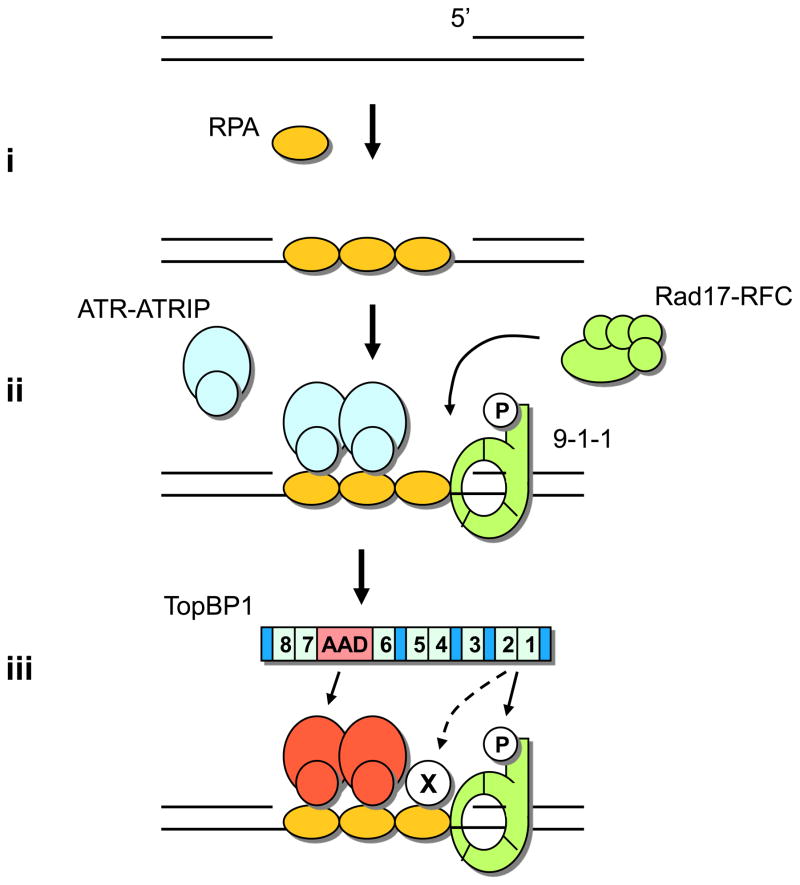
Figure 1. A model for ATR-ATRIP activation by DNA damage.
ssDNA and junctions of ssDNA and dsDNA are the basic DNA structural elements that trigger ATR–ATRIP activation. (i) ssDNA is recognized and coated by RPA (yellow ovals). (ii) ssDNA coated by RPA (RPA–ssDNA) recruits the ATR–ATRIP kinase complex (blue ovals) to sites of DNA damage. In the presence of ssDNA–dsDNA junctions and RPA, the Rad17–RFC complex (green ovals) recruits the 9-1-1 complex (green circle) onto DNA. (iii) RAD9 in the 9-1-1 complex is constitutively phosphorylated at Ser 387. TOPBP1 (blue bar), the stimulator of the ATR–ATRIP kinase, contains 8 BRCT domains (light blue boxes 1–8 in TOPBP1) and an ATR-activation domain (AAD; red box in TOPBP1). The TOPBP1 BRCT domains 1–2 interact with the phosphorylated Ser387 of RAD9, and the AAD domain interacts with ATR–ATRIP. In addition to RAD9 and ATR–ATRIP, TOPBP1 might interact with an unknown factor (label as X) on damaged DNA. Interactions with proteins on damaged DNA recruit TOPBP1 to sites of DNA damage and enable it to stimulate the ATR–ATRIP kinase complex (red ovals indicate stimulated ATR-ATRIP).
In addition to ssDNA, the double-stranded DNA (dsDNA) adjacent to ssDNA also has an important role in ATR activation. In vitro studies using Xenopus egg extracts and purified yeast proteins show that ssDNA annealed with primers, but not ssDNA alone, triggers the activation of Xatr (“X” refers to Xenopus) [SC1]and its yeast homolog Mec1 (Table 1) [7, 8]. The junctions of ssDNA and dsDNA are recognized by two key regulators of ATR, the Rad17–RFC complex and the Rad9–Rad1–Hus1 (9-1-1) complex (Table 1). In the presence of ssDNA–dsDNA junctions and RPA, the Rad17–RFC complex functions as a “loader” to recruit the clamp-shaped 9-1-1 complex onto DNA [9, 10] (Fig. 1). The recruitment of ATR–ATRIP by RPA–ssDNA and the recruitment of Rad17–RFC and 9-1-1 by ssDNA–dsDNA junctions are largely independent of each other [11], and they provide the key mechanisms to bring ATR and its regulators together (Fig. 1). In yeast, tethering Mec1 and Ddc1, the homologue of human RAD9, to a tandem array of artificial binding sites leads to activation of the Mec1 pathway in the absence of DNA damage [12]. This finding suggests that the colocalization of ATR and 9-1-1 at sites of DNA damage and stressed replication forks might be a critical event for the activation of ATR and the phosphorylation of ATR substrates.
In human cells, the full activation of ATR requires not only RPA, RAD17–RFC, 9-1-1, but also topoisomerase (DNA) II binding protein 1 (TOPBP1). TOPBP1 functions in both the initiation of DNA replication and activation of the checkpoint. Interestingly, TOPBP1 contains 8 BRCT domains, and is known to interact with a number of proteins involved in the DNA damage response including RAD9 [13, 14] (Fig. 1). In vitro, TOPBP1 directly stimulates the kinase activity of ATR–ATRIP in the absence of other proteins and DNA [15]. How TOPBP1 is regulated by DNA damage is still not fully understood. The localization of ATR–ATRIP, Rad17, and 9-1-1 complexes have been recently reviewed (see [16–18]). Here, we focus on recent findings related to how the DNA structures that activate ATR–ATRIP are generated, how ATR–ATRIP is activated by its regulators on DNA, and how activated ATR regulates its downstream cellular processes to maintain genomic stability.
The DNA structures that trigger the ATR checkpoint
To understand the DNA structure specificity of the ATR pathway, it is important to first understand how ATR and ATM differ. In response to double-stranded DNA breaks (DSBs), ATM and ATR are sequentially activated, and ATM is required for the nuclease-mediated resection of DNA ends and subsequent ATR activation [19, 20]. A recent study showed that whereas dsDNA with blunt ends or short ssDNA overhangs efficiently activate ATM, the ability of dsDNA to activate ATM is attenuated by long ssDNA overhangs [21]. By contrast, ATR activation by dsDNA is enhanced by resection of DNA ends and lengthening of ssDNA overhangs. These results suggest that in the context of DSBs, the length of ssDNA overhangs is the key factor that determinates whether ATM or ATR is efficiently activated.
The length of ssDNA is also critical for the activation of ATR at stressed replication forks. In Xenopus egg extracts, aphidicolin inhibits DNA polymerase α at replication forks, leading to extensive DNA unwinding by the minichromosome maintenance (MCM) helicase and robust activation of Xatr [22, 23]. In this assay, the extent of checkpoint kinase 1 (Xchk1) phosphorylation correlates with the extent of DNA unwinding. Like aphidicolin, ultraviolet light (UV)-induced DNA lesions stall DNA polymerases but not the replicative helicase (Fig. 2). If replication forks collapse at sites of DNA damage, “one-ended” DSBs with ssDNA overhangs might be generated (Fig. 2).
Figure 2. Generation of ssDNA and ssDNA–dsDNA junctions at stressed DNA replication forks.
(i–ii) Stalling of DNA polymerases on either the leading or lagging strand of DNA replication forks can increase the length of ssDNA at the forks. If the lesions (red triangles) are not removed or bypassed, ssDNA gaps can be generated behind the forks. (iii) DNA replication inhibitors such as aphidicolin can inhibit DNA polymerases on the lagging strand or both the leading and lagging strands. Compromised DNA polymerase function leads to increased amounts of ssDNA and an accumulation of short DNA primers at the forks. (iv) DNA lesions such as single-stranded DNA breaks can lead to collapse of DNA replication forks and generate one-ended double-stranded breaks. The resulting DNA breaks can contain ssDNA overhangs and might subsequently be resected by nucleases. The ssDNA generated at stressed replication forks not only contributes to ATR–ATRIP activation, but also participates in the DNA recombination events that facilitate the recovery of stressed forks.
The ssDNA generated at stressed replication forks is a dynamic DNA structure. The BACH1 (also known as the Fanconia anemia (FA) protein FANCJ) helicase is recruited onto chromatin through an interaction with TOPBP1 in response to replication stress [24]. When BACH1 recruitment is disrupted, RPA fails to accumulate on chromatin and CHK1 activation is compromised. Furthermore, SMARCAL1 (also called HARP), an annealing helicase that rewinds complementary ssDNA into dsDNA, is recruited to replication forks by RPA [25–29]. In the absence of SMARCAL1, increased amounts of RPA are recruited onto chromatin in response to replication stress, and the association of RPA with chromatin persists for a longer time [27]. Thus, the length of ssDNA at stressed replication forks is not only determined by the initial replication stress, but also by the ssDNA-processing factors recruited to the forks.
In Xenopus extracts, aphidicolin and UV induce the accumulation of DNA polymerase α on chromatin [30, 31]. When DNA replication is partially inhibited by aphidicolin, continuous synthesis of short DNA primers occurs [32] (Fig. 2). The accumulation of DNA primers, which is dependent upon proliferating cell nuclear antigen (Xpcna) and DNA polymerases α, δ, and ε, correlates with Xatr activation. It is possible that the accumulation of short DNA primers at stressed replication forks increases the number of ssDNA–dsDNA junctions and enhances ATR activation.
Although the DNA damage interfering with DNA replication induces ssDNA at replication forks, the resulting ssDNA does not always colocalize with the forks. In yeast cells treated with UV, ssDNA is detected not only at replication forks, but also many kilobases behind the forks [33] (Fig. 2). In the absence of translesion DNA polymerases, DNA replication forks lose the ability to bypass UV-induced DNA lesions, leaving an increased number of ssDNA gaps behind the forks [33]. In a fission yeast mutant lacking the translesion DNA polymerases, UV-induced DNA damage does not reduce the rate of DNA replication, but prolongs activation of the checkpoint [34]. Thus, in this context, the ssDNA gaps left behind the forks probably are the signals for prolonged checkpoint activation.
Together, these recent studies provide further support to the model in which ssDNA and ssDNA–dsDNA junctions are the key DNA structures that activate the ATR pathway. Furthermore, the length of ssDNA, the number of ssDNA–dsDNA junctions, and whether or not these DNA structures associate with replication forks can influence the strength and timing of ATR activation.
Activation of the ATR–ATRIP kinase
The full activation of the ATR pathway requires not only the localization of ATR–ATRIP to sites of DNA damage and stressed replication forks, but also the stimulation of the ATR–ATRIP kinase complex by its regulators. Studies using budding yeast suggested that the Ddc1–Rad17–Mec3 complex and Dpb11, the yeast homologues of 9-1-1 and TOPBP1 (Table 1), are activators of the Mec1 kinase. In G1 cells, Mec1 activation is dependent upon Ddc1, whereas in G2 cells, both Ddc1 and Dpb11 contribute to Mec1 activation [35]. In vitro, both Ddc1 and Dpb11 can directly stimulate Mec1–Ddc2 (the yeast homologue of ATR–ATRIP) kinase activity [7, 36, 37]. In a reconstituted Mec1 activation assay using purified proteins and primed ssDNA, the stimulation of Mec1–Ddc2 by the Ddc1–Rad17–Mec3 complex depends on the loading of this complex onto DNA, as well as a bipartite Mec1-activating domain in Ddc1 [7, 35]. Furthermore, the Ddc1–Rad17–Mec3 complex and Dpb11 function synergistically to stimulate Mec1–Ddc2 in the presence of primed ssDNA [36]. These results suggest that in budding yeast, there is more than one way to stimulate the kinase activity of Mec1–Ddc2 on damaged DNA.
Compared to the activation of Mec1 in yeast, the activation of ATR–ATRIP in vertebrates is less clear. Like yeast Dpb11, human and Xenopus TOPBP1 can directly stimulate ATR–ATRIP kinase activity; however, it remains unknown whether the 9-1-1 complex is capable of doing so [15]. In vitro, the ATR-activating domain (AAD) of TOPBP1 located between the BRCT domains 6 and 7 is sufficient to stimulate the ATR–ATRIP kinase in the absence of other proteins and DNA [15]. When overexpressed in mouse cells, the TOPBP1 AAD induces some ATR-mediated responses even in the absence of DNA damage [38]. Whereas the direct stimulation of ATR–ATRIP by the TOPBP1 AAD apparently bypasses the regulation of ATR by DNA damage, it provides some mechanistic details on how TOPBP1 interacts with ATR–ATRIP. A recent study showed that the AAD interacts with both ATRIP and the PRD (PIKK regulatory domain) of ATR [39]. Specific mutations in the PRD of ATR reduce the interaction between ATR–ATRIP and TopBP1, and compromise the stimulation of ATR–ATRIP by the AAD.
How the function of TOPBP1 in ATR activation is regulated by DNA damage is an important issue still under investigation. The TOPBP1 BRCT domains 1–2 interact with the RAD9 C-terminus via a phosphorylated residue (Ser 387 of human RAD9 and Ser 373 of Xenopus rad9) [13, 14] (Fig. 1). This constitutive interaction between RAD9 and TOPBP1 might enable the 9-1-1 complex to recruit TOPBP1 to sites of DNA damage and stressed replication forks. Consistent with this model, RAD9–TOPBP1 fusion proteins support ATR activation even when the RAD9 C-terminus and TOPBP1 BRCT domains 1–2 are deleted [14]. However, a recent study using Xenopus extracts showed that whereas the XtopBP1 BRCT domains 1–2 are required for the binding of XtopBP1 to chromatin in response to replication stress, Xrad9 is dispensable [40]. In contrast, another Xenopus study showed that 9-1-1 is required for the binding of XtopBP1 to chromatin, but the Ser373 of Xrad9 is not needed [41]. In addition, purified human TOPBP1 binds RPA–ssDNA in the presence of ATR–ATRIP and the absence of 9-1-1 [42]. Although the discrepancies among these studies remain to be clarified, the recruitment of TOPBP1 to sites of DNA damage appears to involve multiple protein–protein interactions (Fig. 1).
The stimulation of ATR–ATRIP by TOPBP1 on DNA also appears to be more complex than previously thought. A recent Xenopus study suggested that the Ser373 of Xrad9, which interacts with XtopBP1, is not needed for the recruitment of XtopBP1 to chromatin, but is important for activation of the Xatr pathway [41]. This finding suggests that 9-1-1 not only regulates the recruitment of TOPBP1, but also its function on DNA. Furthermore, experiments using purified human proteins showed that the stimulation of ATR–ATRIP by full-length TOPBP1, but not that by the TOPBP1 AAD, is enhanced by RPA–ssDNA in the absence of 9-1-1 [42]. This finding indicates that TOPBP1 might directly interact with the ATR–ATRIP bound to RPA–ssDNA through regions outside of its AAD. Together, these recent studies suggest that the stimulation of ATR–ATRIP by TOPBP1 involves not only the contacts between the TOPBP1 AAD and ATR–ATRIP, but also additional interactions between TOPBP1 and the 9-1-1 and ATR–ATRIP complexes.
The role of ATR in cell cycle arrest and replication fork stabilization
ATR is essential for the sustained survival of proliferating cells [43], indicating that its activity is crucial for cells to cope with intrinsic cellular stresses. Ablation of ATR and its effector kinase CHK1 leads to increased genomic instability during S phase [44, 45], suggesting that the ATR-CHK1 pathway is important for the maintenance of genomic stability during DNA replication. In budding yeast and human cells, several Mec1 and ATR substrates are transiently phosphorylated during S phase even in the absence of extrinsic DNA damage [46]. In fission yeast, histone H2A, a substrate of the ATR homologue Rad3, is phosphorylated during S phase at natural replication fork barriers, retrotransposons, ribosomal DNA (rDNA) repeats, and heterochromatin in centromeres and telomeres [47]. These findings suggest that ATR plays an important role in protecting replication forks at specific loci in the genome during unperturbed S phase.
Compared to the function of ATR in unperturbed cells, the function of ATR in response to extrinsic DNA damage is more extensively characterized. Studies using yeast, Xenopus, and mammalian systems have shed light on several functions of ATR in DNA damage response, including (i) arresting the cell cycle, (ii) preventing firing of late replication origins, (iii) stabilizing stressed replication forks, and (iv) promoting DNA repair and restart of DNA replication. In response to ionizing radiation (IR) or UV-induced DNA damage, a large number of proteins are phosphorylated in a manner dependent upon ATM and/or ATR [48–50]. The potential substrates of ATR are involved in a wide range of cellular processes, including many that have not yet been linked to the DNA damage response.
Cell division control 25 (CDC25) phosphatases are key regulators of the cell cycle and critical targets of the ATR pathway in cell cycle arrest. In response to DNA damage, ATR activates its effector kinase CHK1, which in turn phosphorylates CDC25A. The phosphorylation of CDC25A by CHK1 and another kinase NEK11 promotes the ubiquitylation and subsequent degradation of CDC25A, leading to reduced cyclin-dependent kinase 2 (CDK2) activity in S phase [51–53]. In addition to CDC25A, CHK1 also phosphorylates CDC25C. This phosphorylation event generates a binding site for 14-3-3 proteins [54, 55]; the binding of 14-3-3 proteins inhibits CDC25C function and prevents activation of the CDK1–cyclinB kinase and mitotic entry.
In response to DNA damage, the yeast ATR homologue Mec1 has an important role in preventing firing of late replication origins, which limits the number of replication forks that could run into DNA lesions [56]. Rad53, the Mec1 effector kinase, phosphorylates Dbf4 and inhibits activity of the Cdc7–Dbf4 kinase [57]. The Mec1- and Rad53-mediated inhibition of Cdc7–Dbf4 prevents the initiation factor Cdc45 from binding to late replication origins [58]. Furthermore, Rad53 phosphorylates another initiation factor Sld3 and inhibits its function [59]. In Xenopus extracts, DNA damage-mediated Xatr activation also leads to inhibition of Xcdc7–Xdbf4 and reduced binding of Xcdc45 to chromatin [60]. In human cells, although there is evidence that the CDC45–chromatin binding is repressed by the ATR–CHK1 pathway, the CDC7–DBF4 kinase remains active after DNA damage [61, 62]. The inhibition of CDK2 by the ATR-CHK1 pathway and the phosphorylation of the histone methyltransferase MLL by ATR might contribute to the repression of late origin firing in human cells [63].
In budding yeast, both Mec1 and its effector Rad53 are important for the stability of replication forks in the presence of DNA damage or replication stress [64, 65]. In cells expressing a mutant allele of MEC1, DNA polymerases are destabilized at stalled forks [66]. Furthermore, loss of Rad53 leads to dissociation of the MCM helicase from stalled forks [66]. Interestingly, deletion of the exonuclease Exo1 suppresses the fork instability caused by RAD53 mutation, suggesting that the key function of Rad53 in stabilizing replication forks is to antagonize Exo1 [67]. Nevertheless, deletion of EXO1 does not suppress the fork instability caused by MEC1 mutation, suggesting that the function of Mec1 in fork stabilization is not completely dependent on Rad53. Recent studies have suggested that the phosphorylation of Mrc1, a replication fork component, by Mec1 is important for fork stability [68, 69]. In human cells, several proteins involved in the signaling of the ATR-CHK1 pathway, including TOPBP1, BRCA1, CLASPIN, and CHK1, are required for preventing fork collapse at common fragile sites [70]. A number of replication fork proteins are known or potential substrates of ATR [48, 50]; however, the mechanisms by which ATR stabilizes stressed replication forks in human cells remain to be elucidated.
The ATR pathway needs to be deactivated for the restart of DNA replication after DNA damage. Paradoxically, ATR might have a positive role in replication restart by triggering a negative feedback loop. In Xenopus extracts, Xatr phosphorylates Xclaspin and Xmcm2, two components of replication forks, and promotes the recruitment of the Polo-like kinase Xplk1 [71–73]. The phosphorylation of Xclaspin by Xplk1 is needed for XChk1 inactivation and restart of DNA replication.
The role of ATR in DNA repair at replication forks
Although proteins in several DNA repair pathways, such as nucleotide excision repair (NER) and mismatch repair (MMR), contribute to ATR activation by processing damaged DNA or by direct protein interactions, how ATR regulates these DNA repair pathways is still poorly understood. Recent studies on the Fanconi anemia (FA) pathway have shed new light on the functions of ATR in DNA interstrand crosslink (ICL) repair at replication forks. FA is a recessive disorder associated with hypersensitivity to DNA-crosslinking agents. Thirteen FA complementation groups have been identified and their corresponding genes have been cloned. The FA proteins can be divided into three functional groups: the FA core complex and associated proteins (FANCA, -B, -C, -E, -F, -G, -L, and -M), the FANCD2–FANCI complex, and the BRCA-related FA proteins (FANCD1 (also called BRCA2), FANCN (also called PALB2), and FANCJ (also called BACH1)). Interestingly, several FA proteins have been implicated in the activation of ATR, and ATR has an important role in regulating ICL repair. Thus, the interplay between ATR and the FA pathway provides an excellent example of how ATR functions as an integral part of DNA repair at replication forks.
The roles of FA proteins in activation of the ATR pathway
FANCM and FAAP24 form a heterodimer that associates with the FA core complex. In vitro, FANCM preferentially binds Holliday junctions and fork-like DNA structures, and displays ATP-dependent translocase activity [74, 75]. In vivo, loss of FANCM increases the incidence of replication fork pausing and/or collapse [76, 77]. FANCM–FAAP24 was first implicated in ATR regulation by its interaction with TEL2 [78]. Depletion of FANCM and FAAP24 results in a modest reduction in CHK1 phosphorylation in response to replication stress [76–78]. The translocase activity of FANCM is required for its function in the checkpoint response. In cells lacking FANCM, the chromatin association of TOPBP1 is reduced [77]. FANCM–FAAP24 was recently shown to play an especially important role in ICL-induced ATR activation [79]. Unlike other types of DNA lesions, ICLs stall replication forks without inducing substantial amounts of ssDNA. Nonetheless, the replication forks stalled by ICLs are able to recruit RPA through a mechanism dependent on FANCM–FAAP24 (Fig. 3). Notably, RPA recruitment requires the DNA-binding activity of FAAP24, but not the translocase activity of FANCM. In vitro, FAAP24 preferentially binds dsDNA with ICLs and promotes the recruitment of RPA.
Figure 3. A model for the activation and function of ATR at replication forks stalled by ICLs.
FANCM and FAAP24 are structure-specific DNA-binding proteins. FAAP24 directly binds ICLs in vitro, and associates with ICLs in cells in the absence of DNA replication. When DNA replication forks are stalled by ICLs (red box), the FANCM–FAAP24 complex (blue ovals labeled as M and 24) facilitates the recruitment of RPA (yellow ovals) to the forks and the activation of ATR–ATRIP (red ovals). Subsequently, ATR phosphorylates the FANCD2–FANCI complex (blue ovals labeled as D2 and I), which promotes the monoubiquitylation of FANCD2 by the FA core complex (large blue oval) and the accumulation of monoubiquitylated FANCD2 at the stalled forks. Monoubiquitylated FANCD2 recruits the FAN1 nuclease (green oval) to the stalled forks, allowing FAN1 to carry out the incisions near ICLs. The incisions near ICLs enable a translesion DNA polymerase to pass the lesions. The DNA lesion can be subsequently removed by nuclear excision repair (NER), and the DNA ends resulting from ICL incisions rejoined by homologous recombination (HR).
In addition to FANCM–FAAP24, several other FA proteins are also implicated in activation of the ATR pathway. In human cells, FANCJ facilitates the recruitment of RPA to chromatin following replication stress [30]. Although loss of FANCD2 does not affect ICL-induced CHK1 phosphorylation in human cells [80], depletion of either Xfancd2 or Xfancl from Xenopus extracts prevents the ICL-induced Xchk1 phosphorylation [81]. Moreover, Xfancl is specifically required for the binding of Xrpa to ICLs but not UV-induced DNA damage.
Together, the studies suggest that the FA proteins have both general and ICL-specific functions in ATR activation. Furthermore, they might contribute to several distinct steps during ATR activation, including generation of ssDNA, recruitment of RPA and ATR–ATRIP, and recruitment of ATR regulators such as TOPBP1.
The role of ATR in ICL repair
The FANCD2–FANCI complex is a critical player in ICL repair. In response to DNA damage and replication stress, both FANCD2 and FANCI are phosphorylated and monoubiquitylated [82–84]. FANCD2 phosphorylation is ATR-dependent in cells treated with the DNA-crosslinking agent mitomycin C (MMC) [85]. Both ATR and the MMC-induced phosphorylation of FANCI are required for FANCD2 monoubiquitylation [86, 87]. When a FANCI mutant carrying phosphomimetic mutations is expressed in cells, it induces constitutive FANCD2 monoubiquitylation and protects cells from MMC-dependent killing [87], suggesting that ATR promotes FANCD2 monoubiquitylation by phosphorylating FANCI (Fig. 3). In Xenopus extracts depleted of Xatr, the ICL-induced chromatin associations of both Xfancl and Xfancd2 are reduced [81], suggesting that Xatr might stabilize both the FA core and the Xfancd2-Xfanci complexes at ICL sites.
In a cell-free assay for replication-coupled ICL repair using Xenopus extracts, Xfancd2 depletion inhibits the nucleolytic incisions near the ICL and the subsequent translesion DNA synthesis (TLS) past the lesion [88]. This function of Xfancd2 in incision is dependent upon its monoubiquitylation site. Recent studies showed that monoubiquitylated FANCD2 recruits FAN1, a protein with a ubiquitin-binding UBZ domain and a structure-specific endonuclease activity [89–92]. Thus, the ATR-dependent localization of FANCD2 to ICLs promotes incisions near the ICLs, which enables subsequent TLS (Fig. 3). Furthermore, both FAN1 and FANCD2 possess exonuclease activities [89–93] that might contribute to the later steps during ICL repair to rejoin DNA ends.
Additional functions of ATR in DNA repair at replication forks
In addition to the FANCD2–FANCI complex, a number of other DNA repair proteins are known or potential substrates of ATR. In budding yeast, the repair scaffold complex consisted of Slx4 and Rtt107 interacts with Dpb11 in response to the DNA damage induced by the DNA-alkylating agent methyl methanesulfonate (MMS) [94]. The interaction between Slx4–Rtt107 and Dpb11 is dependent upon Mec1 and the damage-induced phosphorylation of Slx4. Given that MMS is known to interfere with DNA replication and to activate Mec1, these results suggest that Mec1 activation promotes the recruitment of Slx4–Rtt107 to stressed replication forks by Dpb11. The Slx4–Rtt107 scaffold interacts with several protein complexes involved in DNA repair, including Slx1, Smc5–Smc6, and Rtt101–Mms22–Mms1. Human SLX4 forms a multi-protein complex with the XRF–ERCC1, MUS81–EME1, and SLX1 endonucleases [95, 96]. In vitro, the Slx1-Slx4 complex functions a Holliday junction resolvase. Cells lacking SLX4 are not only sensitive to MMS, but also DNA crosslinking agents, suggesting that the SLX4 complex might function in multiple DNA repair pathways. Interestingly, SLX4 is phosphorylated at multiple sites by ATM and/or ATR after DNA damage [50], raising the possibility that it might mediate some of the DNA repair functions of ATR at stressed replication forks.
In addition to proteins directly involved in DNA repair, ATR and Mec1 also phosphorylate histone H2AX (H2A in yeast) in response to DNA replication stress. Phosphorylated H2AX recruits a number of DNA repair proteins and chromatin-remodeling complexes to the chromatin adjacent to stressed replication forks. For example, in yeast the Ino80 chromatin-remodeling complex is known to interact with phosphorylated H2A. Recent studies suggest that that the Ino80 complex might regulate Rad51-mediated recombination at stressed forks and determine the stability of stressed forks [97–99]. Thus, ATR and Mec1 might contribute to DNA repair at stressed replication forks at multiple levels, including through the modulation of adjacent chromatin, recruitment of DNA repair proteins, and regulation of the functions of DNA repair proteins.
Concluding remarks
Although our knowledge of the ATR checkpoint pathway has been significantly expanded in the past few years, there are still many important questions waiting to be addressed. How ATR is regulated by DNA damage remains a major question. Although a number of DNA damage sensors and ATR regulators have been identified, how these proteins function in concert on damaged DNA to transform ATR into a fully active kinase remains poorly understood. Studies on how ATR responds to different types of DNA damage, particularly those on the response to ICLs, suggest that the activation and function of ATR is highly context-specific. Furthermore, how ATR responds to intrinsic replication stress in unperturbed cells and how it responds to oncogenic stress in tumor cells remains largely unknown [100]. Understanding the regulation and function of ATR in the context of different types of cellular stresses will require extensive studies in the future. Last but not least, the putative substrates of ATR are involved in a broad spectrum of cellular processes that go well beyond DNA replication and repair. It is conceivable that many novel functions of ATR await discovery.
Acknowledgments
L. Z. is supported by a NIH grant (GM076388) and is an Ellison New Scholar on Aging. R. L. F. is supported by an ACS fellowship 0902501. We thank Dr. Bunsyo Shiotani for helpful comments, and apologize to our colleagues whose work we could not cover owning to length constrains.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 2.Cortez D, et al. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 3.Takai H, et al. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131:1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 4.Takai H, et al. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev. doi: 10.1101/gad.1956410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurov KE, et al. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 24:1939–1950. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 7.Majka J, et al. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacDougall CA, et al. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou L, et al. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci U S A. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou L, et al. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonilla CY, et al. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell. 2008;30:267–276. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, et al. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 14.Delacroix S, et al. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumagai A, et al. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 16.Shiotani B, Zou L. ATR signaling at a glance. J Cell Sci. 2009;122:301–304. doi: 10.1242/jcs.035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou L. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev. 2007;21:879–885. doi: 10.1101/gad.1550307. [DOI] [PubMed] [Google Scholar]
- 19.Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jazayeri A, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 21.Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byun TS, et al. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 24.Gong Z, et al. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell. 37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Postow L, et al. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem. 2009;284:35951–35961. doi: 10.1074/jbc.M109.048330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan J, et al. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009;23:2394–2399. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yusufzai T, et al. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 2009;23:2400–2404. doi: 10.1101/gad.1831509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciccia A, et al. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009;23:2415–2425. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bansbach CE, et al. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23:2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michael WM, et al. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- 31.Lupardus PJ, et al. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes Dev. 2002;16:2327–2332. doi: 10.1101/gad.1013502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van C, et al. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J Cell Biol. 189:233–246. doi: 10.1083/jcb.200909105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopes M, et al. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Callegari AJ, et al. Postreplication gaps at UV lesions are signals for checkpoint activation. Proc Natl Acad Sci U S A. 107:8219–8224. doi: 10.1073/pnas.1003449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navadgi-Patil VM, Burgers PM. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell. 2009;36:743–753. doi: 10.1016/j.molcel.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem. 2008;283:35853–35859. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mordes DA, et al. Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci U S A. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toledo LI, et al. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev. 2008;22:297–302. doi: 10.1101/gad.452308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mordes DA, et al. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J, Dunphy WG. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell. 21:926–935. doi: 10.1091/mbc.E09-11-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi JH, et al. Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proc Natl Acad Sci U S A. 107:13660–13665. doi: 10.1073/pnas.1007856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 44.Syljuasen RG, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casper AM, et al. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 46.Sorensen CS, et al. ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle. 2004;3:941–945. [PubMed] [Google Scholar]
- 47.Rozenzhak S, et al. Rad3 decorates critical chromosomal domains with gammaH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet. 6:e1001032. doi: 10.1371/journal.pgen.1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stokes MP, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mu JJ, et al. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem. 2007;282:17330–17334. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- 50.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 51.Melixetian M, et al. NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat Cell Biol. 2009;11:1247–1253. doi: 10.1038/ncb1969. [DOI] [PubMed] [Google Scholar]
- 52.Busino L, et al. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- 53.Jin J, et al. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–3074. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng CY, et al. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 55.Sanchez Y, et al. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 56.Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 57.Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. Embo J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aparicio OM, et al. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci U S A. 1999;96:9130–9135. doi: 10.1073/pnas.96.16.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zegerman P, Diffley JF. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature. doi: 10.1038/nature09373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costanzo V, et al. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 61.Tsuji T, et al. The role of Dbf4/Drf1-dependent kinase Cdc7 in DNA-damage checkpoint control. Mol Cell. 2008;32:862–869. doi: 10.1016/j.molcel.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu P, et al. The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25A/Cdk2-independent mechanism. J Biol Chem. 2006;281:30631–30644. doi: 10.1074/jbc.M602982200. [DOI] [PubMed] [Google Scholar]
- 63.Liu H, et al. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lopes M, et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 65.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 66.Cobb JA, et al. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 2005;19:3055–3069. doi: 10.1101/gad.361805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Segurado M, Diffley JF. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev. 2008;22:1816–1827. doi: 10.1101/gad.477208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lou H, et al. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol Cell. 2008;32:106–117. doi: 10.1016/j.molcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Naylor ML, et al. Mrc1 phosphorylation in response to DNA replication stress is required for Mec1 accumulation at the stalled fork. Proc Natl Acad Sci U S A. 2009;106:12765–12770. doi: 10.1073/pnas.0904623106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arlt MF, et al. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst) 2006;5:1126–1135. doi: 10.1016/j.dnarep.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 71.Yoo HY, et al. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J Biol Chem. 2004;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- 72.Yoo HY, et al. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell. 2004;117:575–588. doi: 10.1016/s0092-8674(04)00417-9. [DOI] [PubMed] [Google Scholar]
- 73.Trenz K, et al. Plx1 is required for chromosomal DNA replication under stressful conditions. Embo J. 2008;27:876–885. doi: 10.1038/emboj.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gari K, et al. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 75.Gari K, et al. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci U S A. 2008;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwab RA, et al. ATR activation and replication fork restart are defective in FANCM-deficient cells. Embo J. 29:806–818. doi: 10.1038/emboj.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luke-Glaser S, et al. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. Embo J. 29:795–805. doi: 10.1038/emboj.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Collis SJ, et al. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 79.Huang M, et al. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol Cell. 39:259–268. doi: 10.1016/j.molcel.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pichierri P, Rosselli F. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. Embo J. 2004;23:1178–1187. doi: 10.1038/sj.emboj.7600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ben-Yehoyada M, et al. Checkpoint signaling from a single DNA interstrand crosslink. Mol Cell. 2009;35:704–715. doi: 10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taniguchi T, et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 83.Sims AE, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 84.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ho GP, et al. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol. 2006;26:7005–7015. doi: 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andreassen PR, et al. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ishiai M, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Knipscheer P, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu T, et al. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 90.Smogorzewska A, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kratz K, et al. /FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 1018;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 92.MacKay C, et al. /FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 1018;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pace P, et al. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 94.Ohouo PY, et al. DNA damage signaling recruits the Rtt107-Slx4 scaffolds via Dpb11 to mediate replication stress response. Mol Cell. 39:300–306. doi: 10.1016/j.molcel.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 95.Fekairi S, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Svendsen JM, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Falbo KB, et al. Involvement of a chromatin remodeling complex in damage tolerance during DNA replication. Nat Struct Mol Biol. 2009;16:1167–1172. doi: 10.1038/nsmb.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shimada K, et al. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- 99.Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- 100.Bartek J, et al. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]