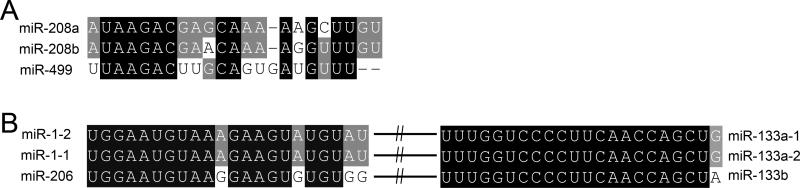Abstract
MicroRNAs (miRs) regulate protein expression by inhibiting translation of expressed mRNAs. Targeting by one or more miRs of multiple mRNA transcripts encoding proteins with common functions confers nodal control over cardiac development and stress-response. Dynamic co-regulation of miRs and their mRNA targets has complicated understanding their biology, but also provides opportunities for clinical diagnostics and therapeutics. Here, the biology of miRs is reviewed as it relates to the cardiac system, recent findings are described that illuminate miR control of cardiac development and myofiber identity, and the clinical ramifications of miR expression profiling are illustrated.
Introduction: miRs and their mRNA targets
MicroRNAs (miRs) are small (18-25 nucleotide) non-coding RNAs that fine-tune protein expression by recruiting target mRNAs into macromolecular extra-nuclear complexes termed RISCs (RNA-induced silencing complexes), and suppressing protein translation or destabilizing the mRNA (reviewed in 1). Sequence analysis suggests that there are over 1,000 miRs in the human genome, many of which have been cloned, expression profiled in different tissues, and interrogated for functional significance. miRs bind via Watson-Crick pairing to complementary sequences in target mRNA transcripts; bioinformatics algorithms predict that approximately one third of all gene transcripts are miR targets. miR-mRNA pairing is determined by nucleotide sequences within the mRNA 3’ untranslated regions and complementation to one or more miR sequences. The prevailing view is that critical miR “seed sequences” (bases 2-8) determine miR-mRNA interactions, although evidence is beginning to accumulate that extra-seed sequence pairing may play a larger role in fine discrimination of miR targets than previously recognized. A single mRNA may have one or more binding sites for a given miR, and binding sites for multiple miRs 2. Intriguingly, binding sequences for a miR or for closely related miR family members can be present on multiple mRNA transcripts that are involved in common cellular responses. Thus, miRs can exert “nodal” control over multiple aspects of such functions as apoptosis, cell proliferation, and organ development. Given the multiplicity of miRs that can target mRNAs within a particular functional pathway, and the likelihood that specific mRNAs with overlapping or complementary functions are controlled by multiple different miRs, the exact mechanisms of biological control by miRs are anticipated to be complex and involve numerous direct and indirect regulatory events. Even if it were possible to precisely categorize all of the direct mRNA targets of all miRs expressed in a given biological context (which it currently is not), indirect effects will determine some of the observed changes in protein expression, and therefore of biological effect. This is a rapidly evolving area of investigation, and it is certain that our current understanding of specific miR-mRNA pairings and their biological effects will continue to change.
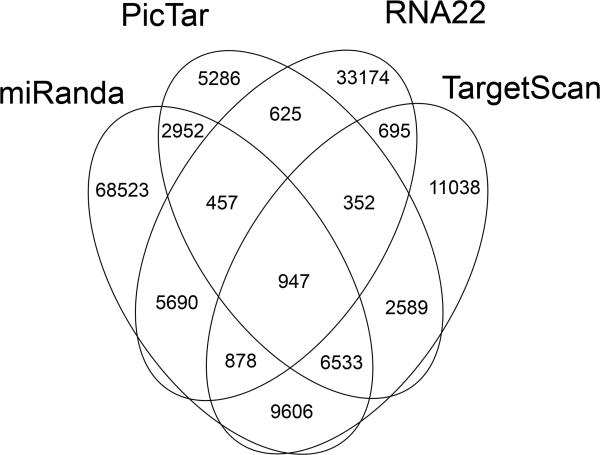
The two mechanisms by which miRs prevent mRNAs translation are mRNA destabilization by de-adenylation, and translational suppression. The molecular and biochemical details of both pathways have been reviewed recently 1. Current analytical methods do not distinguish between these two mechanisms for translational inhibition induced by a given miR-mRNA pairing event. However, the anticipated reciprocal relationship between a miR and its destabilized mRNA target suggested that comparative assays of miR and mRNA levels would help delineate specific miR-mRNA targeting events. Unfortunately, this approach has met with limited success because of degenerate and extra-seed sequence miR-mRNA binding, indirect mRNA regulation by other miR targets, and translational inhibition by miRs without mRNA destabilization 3. Bioinformatics analytical platforms are also limited in their applicability because they tend to rely heavily upon one aspect of miR-mRNA pairing to rank interactions, such as miR seed sequence complementarity to mRNA 3’ untranslated regions, evolutionary conservation, and the calculated binding energy of miR-mRNA duplexes. Indeed, when we asked four different bioinformatic algorithms to predict which of 10,790 cardiac-expressed mRNAs are likely targets of 139 validated cardiac-expressed miRs 4, the results showed little agreement (Figure 1). Thus, the best information about miR-mRNA targeting has been derived from luciferase reporter studies performed in transfected cultured cells 5. Results of reporter studies indicate that a miR-mRNA interaction can have functional consequences, but do not tell you if that interaction actually takes place in a particular in vivo pathophysiological context, when levels of miR and mRNA are subject to regulatory influences. I believe that identifying relevant miR-mRNA targeting events requires direct measurements of the interaction and its functional consequence in the specific cell/organ of interest, and in the proper in vivo context. Recent techniques that assay mRNA content of miR-programmed RISC complexes 6, that delineate miR-mRNA interaction sites by RNA footprinting 7, and that assess the impact of miRs on the proteome 8, are advancing our ability to define relevant miR-mRNA interactions.
Figure 1. Bioinformatic predictions of cardiac-expressed mRNA targets for cardiac-expressed miRs.
~10,000 cardiac-expressed mRNAs were examined for targeting of 136 cardiac miRs. Note, multiple mRNAs can be targeted by any given miR, and one mRNA can be targeted by many miRs, accounting for the large numbers. Note the absence of consensus results from the different prediction algorithms.
Genetic models interrogating the roles of miRs in cardiac disease
Primary miR transcripts (“pri-miRs”) undergo intra-nuclear splicing by the Drosha/Dgcr8 enzyme complex, generating a characteristically hair-pin folded “pre-miR” that is exported from the nucleus. Cytoplasmic pre-miRs are cleaved by a Dicer-containing complex to generate miR-miR* duplexes containing a mature miR on one strand and a non-functional miR* on the other, or sometimes two complementary functional miRs. Because Drosha and Dicer uniquely process miRs, it has been possible to assess the overall impact of miRs on heart development and function by ablating each of these essential miR-processing enzymes in mice.
Initial studies ablated the Dicer gene, which interrupts miR processing at its penultimate step. Not surprisingly, the germ-line Dicer knockout (i.e. no Dicer in the whole mouse) produced an embryonic lethal phenotype 9, consistent with a general requirement for miR processing during development. A series of cardiac-specific mice were therefore engineered in which Dicer was ablated in the embryonic heart (directed by Nkx2.5-Cre), after the heart was fully developed shortly after birth (directed by MYH6-Cre), or in juvenile and adult mice (directed by MYH6-driven tamoxifen activated MER-Cre-MER) 10 11, 12. In each study, interrupting miR biogenesis in cardiac myocytes induced cardiomyopathic phenotypes: embryonic cardiac Dicer deletion caused lethality early in cardiac development associated with cardiac hypoplasia whereas postnatal and adult cardiac Dicer ablation produced lethal cardiomyopathies having pathological cardiac gene expression, abnormal sarcomeric structure, and cardiomyocyte hypertrophy and/or apoptosis. Likewise, interrupting miR formation by striated muscle-specific ablation of DiGeorge syndrome critical region 8 (Dgcr8), which participates with Drosha in intra-nuclear miR processing, produced left ventricular remodeling that progressed to lethal heart failure 13. Taken together, these studies uncover a central role for miRs in post-developmental cardiac health. Other studies described below have implicated specific miR members or families in different aspects of cardiac development and function.
MyomiR regulation of cardiac myofiber identity
Olson and colleagues have elucidated a pathway by which expression of myosin heavy chain isoforms is directed by miRs. A detailed description of these fascinating studies can be found in their recent review 14. The miR regulatory scheme for myosin heavy chain protein is best understood in the context of the evolution of the myosins. All 11 human MYH genes appear to be derived from a single ancient ancestor. The two cardiac-expressed MYH genes are MYH6, encoding αMHC, and MYH7, encoding βMHC. In humans, βMHC predominates, with αMHC normally comprising less than 8% of total ventricular MHC 15. In mice, βMHC is the fetal cardiac isoform, but is replaced in the ventricles by αMHC shortly after birth. In both mice and humans αMHC and βMHC are reciprocally regulated in health and disease via the influence of thyroid hormone on the two MYH gene promoters 16: Under euthyroid conditions or when thyroid hormone (T3) is added to cultured cardiomyocytes, the balance shifts in favor of αMHC expression, whereas hypothyroidism induced by propyl-thiouracil (PTU) or pressure overload hypertrophy favors βMHC, the so-called “α to β myosin isoform shift” 17
The human MYH6 and MYH7 genes occur in tandem on chromosome 14q12, as the consequence of a relatively recent gene duplication event 18. The genetic and protein structures of these two genes are highly homologous in mouse and man. Phylogenetic analysis has suggested that a third MYH gene, MYH7b located on human chromosome 20q11.22, is closely related to the two cardiac myosin heavy chain genes, even though its expression is low in the heart 19. Importantly, within each of these recently duplicated MYH genes are intronic miRs that were also duplicated, miR-208a in αMHC/MYH6, miR-208b in βMHC/MYH7, and miR-499 in MYH7b. Collectively, these three miRs share sequence homology (Figure 2a) and are called the “MyomiRs” 20.
Figure 2.
A. Sequence alignment of the MyomiRs. B. Sequence alignment of the miR-1 – miR-133 cluster as duplicated twice in the genome.
Because the MyomiR genes are in the introns of their parent MYH genes, they are transcribed into the same precursor mRNAs and subsequently spliced out and independently processed. It might therefore be expected that MyomiR expression will mirror that of its parent gene transcript. This has not turned out to be the case because of elegant feedback regulation between MYH genes 20, 21 and differences in stability between miR-499 and an alternately spliced form of the parent MYH7b mRNA 22. These events are of special importance because the above-described counter-regulation of αMHC and βMHC by thyroid hormone appears to be driven by the MyomiRs embedded with their genes, and to also require miR-499.
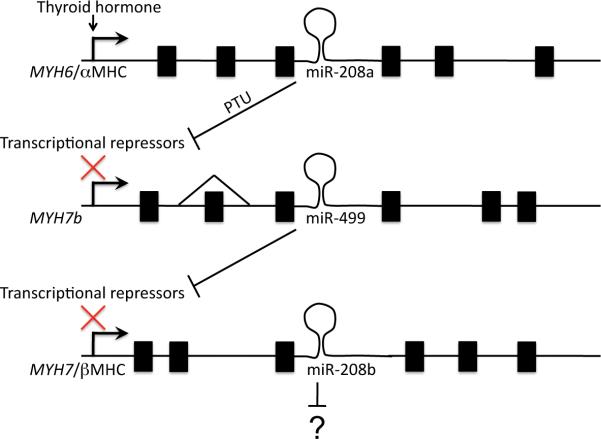
Like its parent transcript (MYH6/αMHC), the cardiac-specific MyomiR miR-208a is potently induced by T3 and suppressed by PTU. However, because miRs are more stable than mRNAs, αMHC mRNA declines in response to PTU, whereas miR208a is not rapidly degraded and its levels are maintained for an extended period of time 21. Residual miR-208a therefore suppresses the translation of several repressors of MYH7 and MYH7b transcription (Figure 3). Thus, the net effect of retained miR-208a transcribed with αMHC is to de-repress expression of βMHC (and miR-208 encoded within). Accordingly, mice lacking miR-208a do not exhibit normal βMHC upregulation with PTU-induced hypothyroidism 21. Expression of MYH7b (and its MyomiR, miR-499) parallels that of MYH7 in this system. Thus, MYH7b and miR-499 are also not detected in miR-208a knockout mice 20. Intriguingly, directed expression of miR-499 in the miR-208a knockout model restores MYH7/miR-208b expression, although the miR-499 knockout does not mimic the miR-208 knockout phenotype. Collectively, these findings indicate that miR-499 is one essential downstream effector of miR-208a regulated βMHC expression, and a critical transducer of the α/βMHC “switch” (Figure 3). Importantly, cardiac development and myosin heavy chain identity in normal adult hearts were not affected by miR-208a/b or miR-499 ablation, demonstrating that the determinants of myosin heavy chain expression differ in response to developmental cues and to external stress.
Figure 3. Schematic depiction of MyomiR regulation of myosin heavy chain isoform expression by thyroid hormone.
PTU = propyl-thiouracil, which induces hypothyroidism. miR-499 within the MYH7b gene is essential to expression of βMHC, although MYH7b mRNA and protein are rare in the heart due to exon 7 skipping. All exons are not shown and distances are not to scale.
Another wrinkle to the MyomiR story involves alternate splicing of MYH7b transcripts22. MYH7b protein is present in skeletal muscle, but as noted above is not measurable in hearts. Nevertheless, myocardial levels of the MyomiR encoded within MYH7b, miR-499, are quite high. The Leinwand laboratory discovered that exon 7 is spliced out of the vast majority (~95%) of mouse cardiac MYH7b transcripts, but of only 60% in soleus muscle. Fully 99% of human heart and skeletal muscle MYH7B transcripts lack exon 7. It was discovered that the alternately spliced MYH7B transcripts lacking exon 7, in which a premature translation termination signal is introduced, are degraded through the mechanism of nonsense-mediated mRNA decay 23. Importantly, since nonsense mRNA decay occurs in the cytosol at the ribosome, nuclear processing of the primary miR-499 transcript is not affected by exon 7 splicing, and production of mature miR-499 is functionally uncoupled from transcript expression of the parent MYH7b gene mRNA (Figure 3). In human hearts therefore, the MYH7b gene is almost entirely a nonfunctioning carrier for its incorporated miR (miR-499), which is essential for expression of the major cardiac myosin heavy chain isoform, MYH7/βMHC. These fascinating findings add even more complexity to regulation of cardiac muscle identity by miRs.
miR1/133 regulation of cardiac development and remodeling
Development of the embryonic heart and cardiac remodeling in response to injury or hemodynamic stress are mechanistically linked, as evidenced by recapitulation of an embryonic gene expression profile in hearts reacting to pressure or volume overload 24. Cardiac development and stress response are also connected by actions of the miR1/133 cluster. miR-1 (or related miR-206) and miR-133 occur as bi-cistronic transcripts that have been twice duplicated, i.e. there are three copies of both miRs, each linked with the other. As with the MyomiR family, the striated muscle-specific miR-1/miR-133 cluster is characterized by close sequence homology between duplicated miRs (Figure 2b).
miR-1 is most recognized for its essential role in cardiac development. Reduction of cardiac miR-1 levels by ~50% through miR-1-2 ablation (leaving miR-1-1 intact) predisposes to ventricular septal defects that can be prematurely lethal, and dilated cardiomyopathy in mice that survive to adulthood 10. The Srivastava laboratory has identified a number of critical miR-1 targets, including HAND2, HDAC4, and MEF2a, each of which is central to cardiac development and/or cardiomyocyte growth. For this reason, forced expression of miR-1 in the embryonic heart (directed by the MYH7 promoter) has a profound inhibitory effect on normal cardiomyocyte proliferation and formation of the fetal myocardium, resulting in embryonic lethality at approximately the developmental stage where cardiac pump function becomes necessary for viability 25.
Although they do not share sequence homology (see Figure 2b), the expression partner of miR-1, miR-133a, has overlapping effects: While individual ablation of either of the two cardiac-expressed miR-133a loci produced no detectable phenotype, combined ablation of miR-133a-1 and miR-133a-2 resulted in a high incidence of ventricular septal defects and dilated cardiomyopathy in mice that survive to adulthood (like the miR-1-2 knockout), and molecular abnormalities consistent with increased proliferative stimuli 26. In adult hearts, inhibition of miR-133a using an antagomiR produced a hypertrophic response 27. In contrast, and as with miR-1, forced expression of miR-133a in the embryonic heart suppressed cardiomyocyte proliferation, producing embryonic lethality associated with hypoplastic ventricular myocardium 26. However, expression of miR-133a in the postnatal heart (directed by the MYH6 promoter that turns on shortly after birth) did not alter normal cardiac growth or the hypertrophic response to sustained pressure overload 28. Rather, the miR-133a transgenic heart exhibited diminished myocardial fibrosis and improved diastolic function after surgical aortic coarctation, consistent with a more important regulatory function for myocardial fibrosis in adult hearts 29.
Other aspects of cardiac pathophysiology regulated by miRs
Two additional areas in which miRs have been pathologically implicated (and may therefore be potential therapeutics targets) are regulation of angiogenesis and myocardial fibrosis. Roles for miRs in arrhythmogenesis and the response to ischemic injury are also likely, but direct data are less certain.
Vascularization
The roles played by angiogenesis-regulating miRs in the heart (“angiomiRs”) in cardiac disease, and the potential for their therapeutic applications, were recently reviewed in depth 30. The most compelling data describe a critical role for endothelial cell-specific miR-126 in developmental formation of the vascular system. In companion papers published in Developmental Cell in 2008, the Olson laboratory used gene ablation in mice and the Srivastava laboratory used morpholino knockdown in zebrafish to first demonstrate that vascular endothelial formation and angiogenesis required miR-126 31, 32. In the murine system, a consequence of miR-126 deficiency was defective vessel formation after experimental myocardial infarction 31. Recently, an elegant two-photon study of zebrafish more fully described the mechanisms linking miR-126 and angiogenesis: blood flow activates the mechano-sensitive transcription factor, klf2a, which induces expression of miR-126 that enhances angiogenic Vegf signaling 33. It is interesting to speculate that mimicking miR-126 with an agomiR could bypass the requirement for blood flow in this angiogenic pathway and promote revascularization of tissues (such as myocardial infarcts) lacking significant blood flow.
Anti-angiogenic effects of endothelial cell-expressed miR-92a have been found to play an important role in the response to myocardial ischemia. Bonauer, et al 34 described increased miR-92a expression after experimental myocardial infarction, and found that miR-92a inhibited vessel formation in vitro and in vivo. Accordingly, they treated myocardial infarcted mice with a miR-92a antagomir, and found that inhibition o miR-29a increased vascularization of the infarct border zone, diminished infarct size, and improved post-infarction ventricular function. These studies in the heart and related studies in the hind limb ischemia model strongly support potential therapeutic efficacy for miR inhibition to enhance revascularization after tissue ischemia.
Myocardial fibrosis
The roles of miRs in cardiac fibrosis, and whether observed effects relate to cardiomyocyte- or fibroblast-specific expression, are somewhat controversial. The preponderance of evidence supports a role for miR-21 in fibroblasts or cardiomyocytes or both: miR-21 is one of the “oncomiRs”, i.e. it functions under certain conditions as an oncogene by suppressing pro-apoptotic factors 35. miR-21 expression is increased in the border zone of myocardial infarcts 36, 37 and in the fibroblasts of failing human hearts, where it enhances fibroblast survival/myocardial fibrosis by activating the ERK MAP kinases 38. Consequently, miR-21 antagomiR treatment reduces myocardial fibrosis and reverses pathological remodeling in the mouse aortic banding pressure overload model 38. Interestingly, cardiomyocyte (not fibroblast) miR-21 expression is downregulated by hypoxia, perhaps as part of the Akt survival pathway 39.
Like miR-21, miR-29 expression is greater in cardiac fibroblasts than in cardiac-myocytes. However, miR-29 is downregulated in the border zone of infarcted hearts. Yet, this antithetic regulation is still linked to myocardial fibrosis by de-repression of collagen and elastin translation 36.
As noted above, miR-133a expression was associated with suppression of myocardial fibrosis after surgical pressure overloading 28. Duisters, et al have suggested that this may relate to miR-133a regulation of connective tissue growth factor 29, although the mRNA of CTGF was not significantly regulated in the miR-133a transgenic mouse. In the same study, Duisters, et al also proposed a role in myocardial fibrosis for miR-30, which like miR-133a is downregulated in pressure overload hypertrophy and (in vitro) can target the same connective tissue genes.
microRNA expression profiling in cardiac disease
As noted above, regulated expression of cardiac miRs is observed throughout cardiac development and in response to various forms of cardiac stress. Accordingly, there is tremendous interest in determining whether regulation of cardiac-expressed miRs may provide a clinically useful molecular signature for specific cardiac syndromes. Of the ~1,000 human miRs identified to date, approximately 200 are expressed in myocardium, 70% of which are reportedly up- or down-regulated in either clinical heart disease or experimental models of cardiac hypertrophy or heart failure 4. ~60% of the genes encoding human miRs are distinct genetic entities, located in the spaces between conventional genes. Expression of these miRs is individually regulated by conventional transcriptional machinery. In contrast, transcriptional control for the ~40% of miRs that are located within other, “parent”, genes is determined by the parent gene, although mRNA splicing and differences in stability between mRNA and miR can affect steady-state expression levels (see below).
Van Rooij, et al were the first to describe regulated miRs in human heart disease, using microarrays to characterize miR expression in mouse models. Changes in the levels of five miRs were also observed by Northern analysis of a handful of human heart failure specimens 40. Results of comprehensive miR expression profiling in human heart disease were first described by Thum, et al 41, who compared mRNA and miR expression profiles from four non-failing and six failing hearts using Ambion miR microarrays that assayed 384 different miRs. The pattern of 67 upregulated and 43 downregulated miRs in failing hearts was similar to the miR profile in six fetal hearts. Although this clinical study is very small by today's standards, the results provided evidence for regulated miR expression in heart failure recapitulating miR expression in the embryonic heart (as is the case for mRNAs 42). Ikeda et al subsequently reported expression profiling of 428 different miRs in twenty-five human dilated cardiomyopathy hearts, nineteen human ischemic cardiomyopathy hearts, thirteen human aortic stenosis hearts, and ten normal human hearts 43. They detected 87 cardiac-expressed miRs, 43 of which were regulated in at least one of the disease groups, providing a diagnostic accuracy rate of approximately 70%. The notion that miR signatures might have diagnostically utility received additional support by Sucharov, et al 44, who profiled expression of 470 miRs in five ischemic cardiomyopathy, five non-ischemic cardiomyopathy, and six non-failing hearts. Naga Prasad et al also used a microarray to identify eight miRs that were upregulated in fifty heart failure cardiac samples, compared to twenty nonfailing specimens 45, and validated the clinical associations in an independent case-control cohort (twenty dilated cardiomyopathy and ten nonfailing). Seven of the eight regulated miRs had previously been identified as regulated in either human or mouse heart failure, increasing confidence in these results. Taken together, these studies demonstrate that miR expression signatures can be sufficiently distinct to be useful as a clinical diagnostic tool in cardiac disease.
Our laboratory assessed the utility of diagnostic miR profiling in end-stage human heart failure by comparing mRNA and miR profiles in heart failure from myocardial specimens of subjects with and without mechanical left ventricular support (LVAD) 46. We were examining the notion that LVAD-supported hearts manifest a “recovery” molecular phenotype, and that the miR expression signature would better track this phenotype than mRNA profiles, which are relatively invariants with this form of functional “recovery” 47. Microarrays were used to interrogate expression of 467 human miRs and of the complete human transcriptome; expression signatures were generated from seventeen cardiomyopathic hearts not treated with LVAD (“heart failure”), ten hearts treated with LVADs (“failure recovery”), and eleven non-failing control hearts. Of twenty-eight miRs that were significantly upregulated in heart failure, twenty were fully normalized in the LVAD failure recovery group, with a strong trend toward normalization of the other eight regulated miRs. Consistent with previous findings however 47, the mRNA expression signature did not distinguish between heart failure and failure recovery groups. These findings show the exquisite sensitivity of miR expression profiling to clinical status in end-stage heart failure. The notion that miR expression reflects acute changes in cardiac function was further validated by a recent report assaying MyomiR expression levels in myocardial biopsy specimens. MyomiR expression was increased in dilated cardiomyopathy. Importantly, levels of miR-208a, but not of the other two MyomiRs (miR-208b and miR-499) not only correlated with myocardial βMHC transcript levels and collagen content, but was also with adverse clinical outcomes 48.
miRs as circulating biomarkers of cardiac disease
A major obstacle preventing miR expression profiling from being routinely used as a clinical test for heart disease is the requirement for myocardial biopsy to obtain myocardium as a source for this biomarker. However, recent studies have shown that circulating miRs may be useful for this purpose: Mitchell et al were the first to carefully validate the concept of assaying circulating miRs to detect cancer in experimental models and human subjects 49. Their seminal findings are noteworthy because they found that endogenous circulating miRs are unexpectedly stable, persisting in serum samples for days, whereas exogenous miRs “spiked” into serum were rapidly degraded. Subsequent studies demonstrated that miRs are secreted into the blood within small cell-derived microvesicles, or “exosomes” 50. The tissue-specificity of circulating miRs (for prostate cancer) in Mitchell paper raised the possibility that circulating levels of the more cardiac-specific miRs might prove useful to monitor cardiac status. Indeed, the Mitchell study found that regulated expression of miRs in cancer was accurately reflected in their circulating serum levels.
In the wake of this initial report a flurry of relatively small clinical serum miR profiling studies have been published relating to heart disease. Tijsen et al found that circulating miR-423-5p was increased in heart failure 51, although like most studies of this type, there was no comparison with a conventional heart failure biomarkers such as BNP 52. Other studies have since described up- and down-regulation of a host of other miRs in chronic heart failure and/or coronary artery disease 53-55. At this point, all of these findings require independent validation in separate cohorts and prospective evaluation to see if miR levels add value to currently available tests, and under what clinical scenarios they will be most useful (early rejection after cardiac transplantation deserves careful evaluation).
On the other hand, the results of assaying circulating miR levels after acute myocardial infarction have been more consistent and are more easily understood. Four independent studies to date indicate that one or more MyomiRs are increased in the serum after experimental or clinical myocardial infarction, presumably reflecting their release from injured myocardium in the same manner as myocardial-specific enzymes 56-59. Again however, comparison with standard clinical biomarkers is needed to determine if miR assays will increase sensitivity or specificity of myocardial infarction diagnoses.
miRs as therapeutic targets
Nodal control by a miR or miR cluster over multiple effectors of complex intracellular pathways makes miRs tremendously attractive as therapeutic targets, distinguishing them from the single protein targets (i.e. receptor or enzyme) of most conventional drugs. miRs are themselves “small molecules” and various biochemical means of stabilizing and delivering them have proven successful for miR antagonists (less so for miR mimetics). Care et al were the first to use an antagomiR for in vivo modulation of cardiac disease 27. After observing downregulation of miR-133a during cardiac hypertrophy, they further observed that forced expression of miR-133a in cultured cardiac myocytes prevented hypertrophy, whereas chronic infusion of a miR-133a antagomiR into mice produced hypertrophy. Therefore, they used transcoronary gene delivery of a synthetic miR-133a to inhibit development of spontaneous hypertrophy in a well characterized genetic mouse model. The utility of miR mimicry or antagonism in rescuing experimental cardiac phenotypes has since been repeatedly demonstrated.
The main uncertainty standing in the way of further developing miR-directed therapeutics for heart disease may be defining the targets of endogenous or synthetic miRs in the in vivo heart. Tissue culture experimentation does not provide the complexity of the in vivo myocardial milieu, in which miR and their mRNA targets are dynamically regulated. We have to develop techniques to delineate the strong and weak targets of miRs under varying conditions introduced into the experimental design. These methods can then be used to clearly define the consequences of specific engineered changes in custom miRs undergoing evaluation as possible therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 2.Standart N, Jackson RJ. MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes & Development. 2007;21:1975–1982. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Condorelli G, Latronico MV, Dorn GW., II microRNAs in heart disease: putative novel therapeutic targets? Eur Heart J. 2010;31:649–658. doi: 10.1093/eurheartj/ehp573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, Holland EC. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Ransom JF, Li A, Vedantham V, von DM, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 11.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, Meissner G, Patterson C, Hannon GJ, Wang DZ. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci USA. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, Molkentin JD, De Windt LJ. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567–1576. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- 13.Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, Reinhardt F, Liao R, Krieger M, Jaenisch R, Lodish HF, Blelloch R. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res. 2009;105:585–594. doi: 10.1161/CIRCRESAHA.109.200451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyata S, Minobe W, Bristow MR, Leinwand LA. Myosin heavy chain isoform expression in the failing and nonfailing human heart. Circ Res. 2000;86:386–390. doi: 10.1161/01.res.86.4.386. [DOI] [PubMed] [Google Scholar]
- 16.Morkin E. Control of cardiac myosin heavy chain gene expression. Microsc Res Tech. 2000;50:522–531. doi: 10.1002/1097-0029(20000915)50:6<522::AID-JEMT9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Nakao K, Minobe W, Roden R, Bristow MR, Leinwand LA. Myosin heavy chain gene expression in human heart failure. J Clin Invest. 1997;100:2362–2370. doi: 10.1172/JCI119776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desjardins PR, Burkman JM, Shrager JB, Allmond LA, Stedman HH. Evolutionary implications of three novel members of the human sarcomeric myosin heavy chain gene family. Mol Biol Evol. 2002;19:375–393. doi: 10.1093/oxfordjournals.molbev.a004093. [DOI] [PubMed] [Google Scholar]
- 19.McGuigan K, Phillips PC, Postlethwait JH. Evolution of sarcomeric myosin heavy chain genes: evidence from fish. Mol Biol Evol. 2004;21:1042–1056. doi: 10.1093/molbev/msh103. [DOI] [PubMed] [Google Scholar]
- 20.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr., Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 22.Bell ML, Buvoli M, Leinwand LA. Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping. Mol Cell Biol. 2010;30:1937–1945. doi: 10.1128/MCB.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- 24.Dorn GW., II The fuzzy logic of physiological cardiac hypertrophy. Hypertension. 2007;49:962–970. doi: 10.1161/HYPERTENSIONAHA.106.079426. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 26.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW, II, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 28.Matkovich SJ, Wang W, Tu Y, Eschenbacher WH, Dorn LE, Condorelli G, Diwan A, Nerbonne JM, Dorn GW., II MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ Res. 2010;106:166–175. doi: 10.1161/CIRCRESAHA.109.202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van dMI, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–178. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- 30.Wang S, Olson EN. AngiomiRs--key regulators of angiogenesis. Curr Opin Genet Dev. 2009;19:205–211. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 35.Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37:918–925. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 36.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, Wang D, Krall TJ, Delphin ES, Zhang C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009;284:29514–29525. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 39.Sayed D, He M, Hong C, Gao S, Rane S, Yang Z, Abdellatif M. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J Biol Chem. 2010;285:20281–20290. doi: 10.1074/jbc.M110.109207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, vanLaake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J. MicroRNAs in the human heart: A clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 42.Chien KR, Knowlton KU, Zhu H, Chien S. Regulation of cardiac gene expression during myocardial growth and hypertrophy: molecular studies of an adaptive physiologic response. FASEB J. 1991;5:3037–3046. doi: 10.1096/fasebj.5.15.1835945. [DOI] [PubMed] [Google Scholar]
- 43.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B, Pu WT. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 44.Sucharov C, Bristow MR, Port JD. miRNA expression in the failing human heart: functional correlates. J Mol Cell Cardiol. 2008;45:185–192. doi: 10.1016/j.yjmcc.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naga Prasad SV, Duan ZH, Gupta MK, Surampudi VS, Volinia S, Calin GA, Liu CG, Kotwal A, Moravec CS, Starling RC, Perez DM, Sen S, Wu Q, Plow EF, Croce CM, Karnik S. Unique microRNA profile in end-stage heart failure indicates alterations in specific cardiovascular signaling networks. J Biol Chem. 2009;284:27487–27499. doi: 10.1074/jbc.M109.036541. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW., II Reciprocal Regulation of Myocardial microRNAs and Messenger RNA in Human Cardiomyopathy and Reversal of the microRNA Signature by Biomechanical Support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Margulies KB, Matiwala S, Cornejo C, Olsen H, Craven WA, Bednarik D. Mixed messages: Transcription patterns in failing and recovering human myocardium. Circ Res. 2005;96:592–599. doi: 10.1161/01.RES.0000159390.03503.c3. [DOI] [PubMed] [Google Scholar]
- 48.Satoh M, Minami Y, Takahashi Y, Tabuchi T, Nakamura M. Expression of microRNA-208 is associated with adverse clinical outcomes in human dilated cardiomyopathy. J Card Fail. 2010;16:404–410. doi: 10.1016/j.cardfail.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. Detection of microRNA expression in human peripheral blood microvesicles. Plos One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, Pinto YM. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–1039. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 52.Kumarswamy R, Anker SD, Thum T. MicroRNAs as circulating biomarkers for heart failure: questions about MiR-423-5p. Circ Res. 2010;106:e8. doi: 10.1161/CIRCRESAHA.110.220616. author reply e9. [DOI] [PubMed] [Google Scholar]
- 53.Taurino C, Miller WH, McBride MW, McClure JD, Khanin R, Moreno MU, Dymott JA, Delles C, Dominiczak AF. Gene expression profiling in whole blood of patients with coronary artery disease. Clin Sci (Lond) 2010;119:335–343. doi: 10.1042/CS20100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voellenkle C, van Rooij J, Cappuzzello C, Greco S, Arcelli D, Di Vito L, Melillo G, Rigolini R, Costa E, Crea F, Capogrossi MC, Napolitano M, Martelli F. microRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genomics. 2010 doi: 10.1152/physiolgenomics.00211.2009. [DOI] [PubMed] [Google Scholar]
- 55.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Roxe T, Muller-Ardogan M, Bonauer A, Zeiher AM, Dimmeler S. Circulating MicroRNAs in Patients With Coronary Artery Disease. Circ Res. 2010 doi: 10.1161/CIRCRESAHA.109.215566. in press. [DOI] [PubMed] [Google Scholar]
- 56.Cheng Y, Tan N, Yang J, Liu X, Cao X, He P, Dong X, Qin S, Zhang C. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119:87–95. doi: 10.1042/CS20090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, Qin YW, Jing Q. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 58.Adachi T, Nakanishi M, Otsuka Y, Nishimura K, Hirokawa G, Goto Y, Nonogi H, Iwai N. Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin Chem. 2010;56:1183–1185. doi: 10.1373/clinchem.2010.144121. [DOI] [PubMed] [Google Scholar]
- 59.D'Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, Rubino M, Carena MC, Spazzafumo L, De Simone M, Micheli B, Biglioli P, Achilli F, Martelli F, Maggiolini S, Marenzi G, Pompilio G, Capogrossi MC. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010 doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]