Abstract
Stuttering is a disorder which affects the fluency of speech. It has been shown to have high heritability, and has recently been linked to mutations in the GNPTAB gene. One such mutation, Glu1200Lys, has been repeatedly observed in unrelated families and individual cases. Eight unrelated individuals carrying this mutation were analyzed in an effort to distinguish whether these arise from repeated mutation at the same site, or whether they represent a founder mutation with a single origin. Results show that all 12 chromosomes carrying this mutation share a common haplotype in this region, indicating it is a founder mutation. Further analysis estimated the age of this allele to be ~572 generations. Construction of a cladogram tracing the mutation through our study sample also supports the founder mutation hypothesis.
INTRODUCTION
Stuttering is a common disorder of speech fluency characterized by repetitive fragmentation of the beginnings of words, prolongation of initial sounds, and large gaps between words or syllables, which are known as silent blocks.1
Stuttering has been shown to be highly heritable. It has been reported to aggregate in families 5–6, approximately half of stutterers have a family history of the disorder7–8 , and significant genetic linkage to stuttering has been observed on chromosome 12.9 We have shown that mutations in the GNPTAB (GlcNAc-phosphotransferase; EC 2.7.8.17) gene within this region are associated with this disorder.10 One mutation in GNPTAB, Glu1200Lys, was found in a number of Pakistani stuttering families, and several unrelated affected individuals of South Asian descent.10
The aim of the present study was to further characterize the Glu1200Lys mutation in GNPTAB. We first sought to determine whether this mutation represents a founder mutation descended from a common ancestor, or a recurrent mutation at the same position. We also sought to estimate the age of the mutation and to construct a cladogram tracing the mutation through our study population, to further understand the history of this mutation.
MATERIALS AND METHODS
Eight unrelated individuals previously shown to carry at least one copy of the Glu1200Lys mutation in the GNPTAB gene were examined in this study. Four of the eight subjects were heterozygous for the mutation, providing a total of 12 affected and 4 unaffected chromosomes from these 8 individuals. We genotyped 33 SNPs across the 650 kb region surrounding the Glu1200Lys mutation by sequencing genomic DNA. We also genotyped the 4 nearest microsatellite markers listed on the Marshfield Map Marker Database. Forty eight random Pakistani individuals from a geographically matched location in Pakistan (= 96 chromosomes) were also genotyped with all markers to generate control allele frequencies.
To identify additional informative markers in this region, the 20kb region immediately surrounding the Glu1200Lys mutation was sequenced to completion. This sequencing resulted in the discovery of 5 novel SNPs. The resulting genotypes were then analyzed with PHASE (v2.1.1) to determine the most likely haplotypes for each individual.12–13
The resulting haplotypes (12 mutation-containing chromosomes and 96 Pakistani control chromosomes) were analyzed with DMLE+ (version 2.2) to estimate the approximate age of the Glu1200Lys mutation.14 Trials were run using differing numbers of markers, ranging from all 33 informative SNP markers in the region to 7 markers immediately surrounding the Glu1200Lys mutation. The location of the Glu1200Lys mutation within the haplotype was defined as 0.022 cM from the first marker in the analysis using all 33 markers, and as 0.0039 cM when the 7 markers directly surrounding the mutation were analyzed.
TreeFinder 15 was used to generate a phylogenetic tree illustrating the relationships between the 16 chromosomes from the 8 individuals. The J3+I nucleotide substitution model was used, and a 1000× bootstrap analysis was run to generate possible phylogenic trees. A consensus program was run to choose the statistically most likely tree from those generated. The full sequence of the 650 kb region we had previously evaluated with SNP and microsatellite analysis was used. These sequences were derived from multiple sequencing traces aligned with the reference sequence using SeqMan, and included the relevant allele at each SNP or microsatellite position.
RESULTS
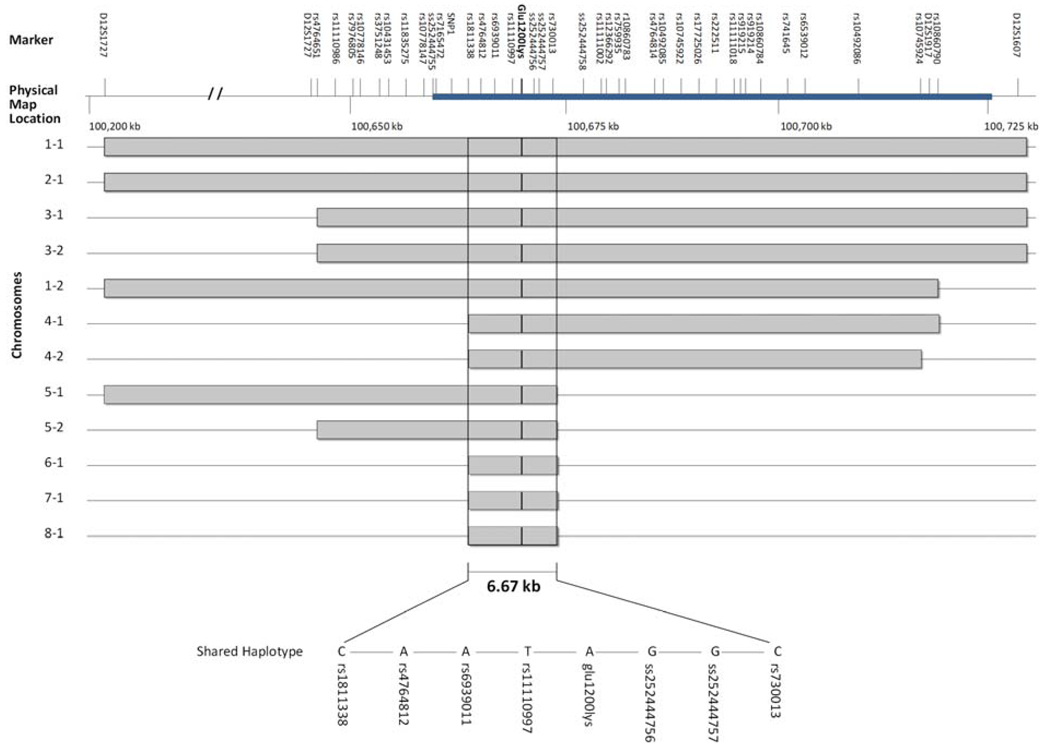
Eight unrelated individuals carrying one or two copies of the 0Glu1200Lys mutation were analyzed by a combination of sequencing and genotyping in a 650 kb region surrounding the mutation. The resulting genotypes were then analyzed using PHASE to determine the most likely haplotypes carried by each of these 8 individuals and 48 control individuals. All 12 chromosomes containing the Glu1200Lys mutation were found to share a common haplotype immediately surrounding this position (Figure 1 and Supplementary Table 1). At its minimum, this haplotype consists of a unique combination of alleles at the 7 SNPs nearest to the mutation, and is approximately 6.67 kb in length. This haplotype was not found in any of the 96 chromosomes in the control sample.
Figure 1.
The Glu1200Lys mutation is surrounded by a shared haplotype. The marker identifiers are listed above the topmost line. SNPs newly discovered in this study are ss252444755, ss252444756, ss252444757, ss252444758, and SNP 1, which was not assigned an rs/ss designation because it was monomorphic in our normal control sample. The horizontal blue bar illustrates the location of the GNPTAB gene, and the lysine mutation is shown by the vertical black line in the center. The grey horizontal bars indicate regions of shared haplotype in each chromosome.
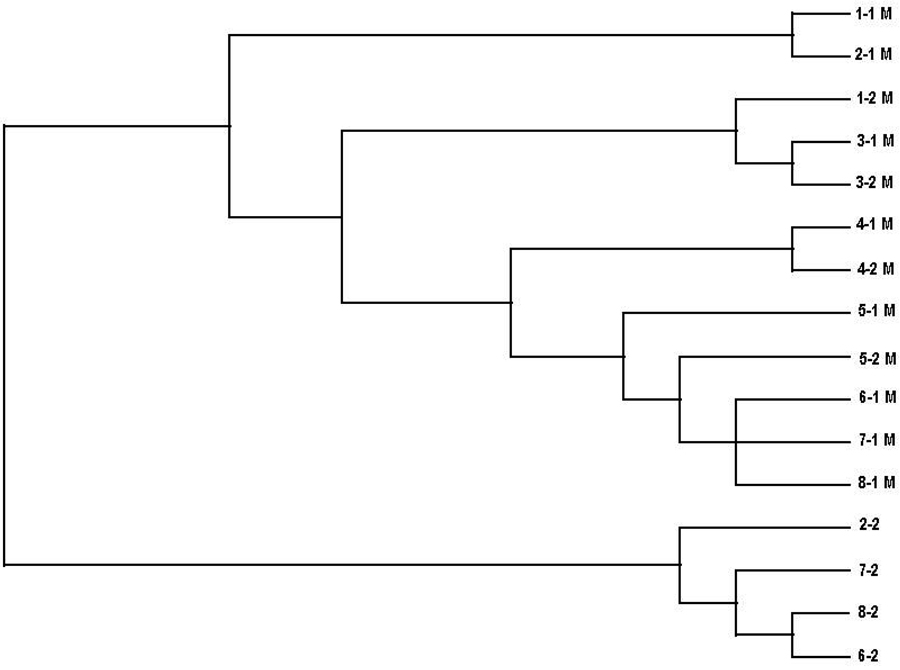
Because these results suggest a single origin of the Glu1200Lys mutation, we sought to estimate the age of this allele. In our analysis using DMLE+, the estimated age of Glu1200Lys mutation was 572 generations (95% credible set: 467–697), or 14,300 years based on a 25 year generation time16. We also constructed a phylogenetic tree of the 16 chromosomes in the 8 unrelated individuals carrying the Glu1200Lys mutation using genotypes and DNA sequence spanning the 650 kb region shown in Figure 1. The consensus tree is illustrated in Figure 2. Chromosomes segregate into two distinct branches, one that contains the 12 chromosomes carrying the mutation, and another branch containing the four that do not. Additionally, the chromosomes that carry the mutation tend to segregate with other chromosomes that contain a similar amount of the shared haplotype. This can be seen in a comparison between Figure 1 and Figure 2.
Figure 2.
Cladogram of 16 chromosomes from 8 unrelated individuals carrying the Glu1200Lys mutation. A total of 16 chromosomes from 8 individuals are included. Chromosomes are labeled by subject number, followed by the chromosome number within that subject. Chromosomes carrying the mutation are designated by an M.
DISCUSSION
Our results reveal that all chromosomes carrying the Glu1200Lys mutation in the GNPTAB gene share a single, apparently unique haplotype surrounding this mutation. This indicates that the Glu1200Lys mutation is a founder mutation that occurred once and has been inherited by all of the affected individuals in our sample. The shared haplotype was found to be as short as 6.67kb in length, suggesting that this mutation may be relatively old. Our estimation of the age of this mutation supports this hypothesis. Using a variety of parameters, we obtained an age estimate of 572 generations, or 14,300 years.
The phylogenetic tree generated from the data also supports the founder mutation hypothesis. The chromosomes with a larger portion of the mutation-carrying haplotype are clustered closer together than chromosomes sharing a lesser amount. All chromosomes carrying the mutation are separated on the cladogram from those shown not to carry the mutation, further supporting the conclusion that they derive from a common ancestor.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Hashim Raza for helpful discussion and T. Friedman and K. Noben-Trauth for valuable comments on the manuscript. This work was supported by NIDCD intramural research grant Z01-000046-10.
REFERENCES
- 1.Bloodstein O, Ratner N. A handbook on stuttering. 6th Edn. New York: Thomson Delmar Learning; 2008. [Google Scholar]
- 2.Andrews G, Morris-Yates A, Howie P, Martin NG. Genetic factors in stuttering confirmed. Arch. Gen. Psychiatry. 1991;48:1034–1035. doi: 10.1001/archpsyc.1991.01810350074012. [DOI] [PubMed] [Google Scholar]
- 3.Felsenfeld S, Kirk KM, Zhu G, Statham DJ, Neale MC, Martin NG. A study of the genetic and environmental etiology of stuttering in a selected twin sample. Behavior. Genetics. 2000;30:359–366. doi: 10.1023/a:1002765620208. [DOI] [PubMed] [Google Scholar]
- 4.Howie PM. Concordance for stuttering in monozygotic and dizygotic twin pairs. J. Speech. Hear. Res. 1981;24:317–321. doi: 10.1044/jshr.2403.317. [DOI] [PubMed] [Google Scholar]
- 5.Kidd KK, Heimbuch RC, Records MA. Vertical transmission of susceptibility to stuttering with sex-modified expression. Proc. Natl. Acad. Sci. USA. 1981;78:606–610. doi: 10.1073/pnas.78.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yairi E, Ambrose N, Cox N. Genetics of stuttering: a critical review. J. Speech. Hear. Res. 1996;39:771–784. doi: 10.1044/jshr.3904.771. [DOI] [PubMed] [Google Scholar]
- 7.Drayna D, Kilshaw J, Kelly J. The sex ratio in familial persistent stuttering. Am. J. Hum. Genet. 1999;65:1473–1475. doi: 10.1086/302625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viswanath N, Lee HS, Chakraborty R. Evidence for a major gene influence on persistent developmental stuttering. Hum. Biol. 2004;76:401–412. doi: 10.1353/hub.2004.0050. [DOI] [PubMed] [Google Scholar]
- 9.Riaz N, Steinberg S, Ahmad J, Pluzhnikov A, Riazuddin S, Cox NJ, et al. Genomewide significant linkage to stuttering on chromosome 12. Am. J. Hum. Genet. 2005;76:647–651. doi: 10.1086/429226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang C, Riazuddin S, Mundorff J, Krasnewich D, Friedman P, Mullikin JC, et al. Mutations in the lysosomal enzyme-targeting pathway and persistent stuttering. N. Engl. J. Med. 2010;362:677–685. doi: 10.1056/NEJMoa0902630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome. Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeve JP, Rannala B. DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics. 2002;18:894–895. doi: 10.1093/bioinformatics/18.6.894. [DOI] [PubMed] [Google Scholar]
- 15.Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC. Evol. Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.World Marriage Data. 2008. 2008. http://www.un.org/esa/population/publications/WMD2008/WP_WMD_2008/Data.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.