Abstract
Estrogen Receptor-α (ER) is the key feature in the majority of breast cancers and ER binding to the genome correlates with the Forkhead protein FOXA1 (HNF3α). We now show that FOXA1 is a critical determinant that can influence differential ER-chromatin interactions. We show that almost all ER-chromatin interactions and gene expression changes are dependent on the presence of FOXA1 and that FOXA1 influences genome-wide chromatin accessibility. Furthermore, we show that CTCF is an upstream negative regulator of FOXA1-chromatin interactions. In ER responsive breast cancer cells, the dependency on FOXA1 for tamoxifen-ER activity is absolute and in tamoxifen resistant cells, ER binding occurs independently of ligand, but in a FOXA1 dependent manner. Importantly, expression of FOXA1 in non-breast cancer cells can alter ER binding and function. As such, FOXA1 is a major determinant of estrogen-ER activity and endocrine response in breast cancer cells.
Introduction
Estrogen Receptor is the defining feature of luminal breast cancers, where it functions as a transcription factor to regulate cell division. Luminal breast cancers compose the majority of all breast cancers and are generally treated with endocrine therapies, including the antiestrogen tamoxifen1, although resistance does occur in a significant fraction of women2. Defining the basis of drug resistance requires understanding the molecular mechanisms of ER activity. Our knowledge of ER activity has evolved significantly in recent years and we now appreciate the multitude of factors that augment or inhibit ER transcriptional activity3. Coupled with this, a better understanding of the cis-regulatory elements and enhancers of ER target genes has evolved4,5.
A number of studies have mapped Estrogen Receptor binding events genome-wide in MCF-7 breast cancer cells, by combining Chromatin Immunoprecipitation (ChIP) with microarrays or high throughput sequencing6-8. The general conclusions were that ER rarely binds to promoter proximal regions and in the majority of studies, Forkhead motifs were enriched in the ER binding events. The Forkhead protein FOXA1/HNF3α has been shown to be present at many ER binding regions9-11 and at a few tested ER binding events, FOXA1 was shown to be required for ER binding9,10, where it likely functions as a pioneer factor12,13. The global importance of FOXA1 in mediating ER function and the underlying factors that determine FOXA1 specificity are not completely clear, although specific histone marks (H3K4me1 and H3K4me2) appear to demarcate FOXA1 regulatory regions11. Clinically, FOXA1 was shown to predict outcome in ER positive breast cancer patients14 and it is one of the minimal gene markers of ER positive luminal tumors (also called HNF3-α)15,16.
The interaction between ER and FOXA1 has been limited to correlation of binding events in one cancer cell line11 and mechanistic analysis on a genomic level is lacking. Whole genome analysis of ER-FOXA1 interactions is required to understand molecular mechanisms of ER activity. We now explore the dependency of ER on FOXA1 on a global scale and in multiple cellular contexts. We show that FOXA1 dictates global chromatin structure and is necessary for ER-chromatin interaction in breast cancer cells under different ligand conditions, including the breast cancer treatment tamoxifen and that FOXA1 is sufficient to permit ER-chromatin interactions and transcriptional activity in diverse target tissues.
Results
Differential ER binding correlates with FOXA1 binding
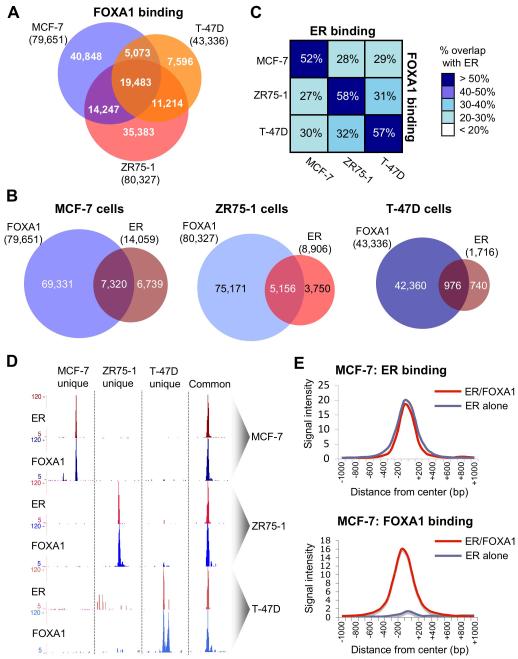
We mapped FOXA1 binding events by ChIP-seq in asynchronous MCF-7, ZR75-1 and T-47D breast cancer cells, the three most commonly studied estrogen responsive breast cancer cell lines17. Binding events were called using MACS18, resulting in 79,651 FOXA1 binding events in MCF-7 cells, 80,327 in ZR75-1 cells and 43,336 in T-47D cells (Figure 1A). The overlap among the three cell lines would suggest that FOXA1 binding events can be dynamic and cell line specific. ER ChIP-seq experiments were also conducted in the three breast cancer cell lines and the ER and FOXA1 binding information was integrated (Figure 1B and Supplementary figure 1). We considered an ER-FOXA1 overlapping region if the binding events shared at least one base pair. The overlap between ER and FOXA1 binding within the same cell line ranged from 52-58% (Figure 1B), but there was significantly lower overlap when comparing between different cellular contexts (Figure 1C). Importantly, the cell line-unique ER binding events were also significantly more likely to be cell line-unique FOXA1 binding regions (data not shown). Examples of cell line specific ER and FOXA1 binding is shown in Figure 1D. By selecting similar numbers of random genomic regions and overlapping with the FOXA1 binding events, we found ER overlap was significantly more enriched (p < 0.00001) than expected by chance. By chance, the expected overlap (derived from hundreds of random comparisons), is ~%2, as compared to the >50% overlap experimentally observed. All together, these data showed that differential ER binding correlated with FOXA1 binding, suggesting genome-wide co-operativity between these factors in a context specific manner.
Figure 1.
Differential FOXA1 and ER binding overlaps in a cell context manner. A. Overlap in FOXA1 binding events between MCF-7, ZR75-1 and T-47D cells. B. Overlap between ER and FOXA1 in the three ER positive breast cancer cell lines. C. Relative overlap in ER and FOXA1 binding events within and between the three cell lines. The percentages represent the fraction of ER binding events in that cell line. An overlap was considered if the peaks shared at least one base pair. D. Examples of regions showing cell line specific ER and FOXA1 binding. Also shown in an example of a common region bound by ER and FOXA1 in all three cell lines. E. Average signal intensity of ER binding sites that are shared with FOXA1 binding regions or ER binding events that are not shared with FOXA1. The signal intensity of ER binding events that are not shared with FOXA1 are similar to those that overlap with FOXA1. Also included is average signal intensity for FOXA1 binding at these two separate ER binding categories.
When comparing the signal intensity of ER binding events in MCF-7 cells that either overlapped with FOXA1 or did not overlap with FOXA1, we found equivalent binding intensity for both categories of ER binding events (Figure 1E), implying that both categories are direct ER binding events. Similarly, when we overlap the ER only or ER/FOXA1 shared binding regions with genomic regions involved in active chromatin loops (ChIP-PET data)5, we find comparable overlaps. Approximately 25% of the ER/FOXA1 shared regions correlate with active chromatin loops versus 26% of the ER alone binding events, suggesting that there is no difference in functionality between ER binding events that are shared with FOXA1 versus ER binding events that do not occur at FOXA1 binding regions.
FOXA1 is essential for ER binding to chromatin and for transcriptional activity
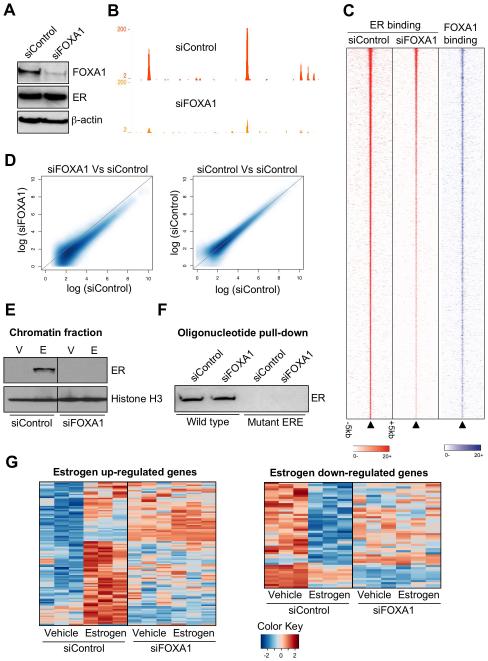
We aimed to determine what impact FOXA1 played on regulating ER-chromatin interactions on a genome-wide scale. We transfected hormone depleted MCF-7 cells with siRNA to control or to FOXA1 (Figure 2A) resulting in no appreciable change in ER protein (Figure 2A) or mRNA levels (Supplementary figure 2). After silencing of FOXA1 (or control), we mapped ER binding events by ChIP-seq. In control transfected MCF-7 cells, we found a total of 13,631 high confidence estrogen-induced ER binding events, in line with our previous genome-wide ER map19. When we mapped ER binding events after silencing FOXA1, ER binding decreased significantly (an example is shown in Figure 2B). The signal intensity of more than 90% of all ER binding events decreased by at least 50% (Figure 2C and 2D), despite the fact that ER protein levels did not change (Figure 2A). This was validated with an independent siRNA and re-expression of FOXA1 rescued the decreased binding (Supplementary figure 3). Only 595 of all the ER binding events did not decrease by more than 50% after silencing FOXA1 (Supplementary figure 2). However, these 595 regions were also the strongest ER binding regions and since silencing was not complete, these likely represent the strongest ER binding regions that decrease, but not by 50%. As such, there is no specificity or distinct feature to these 595 remaining regions. We validated these findings by silencing FOXA1 and Western blotting for ER on the chromatin fraction (Figure 2E). Interestingly, ER binding to unchromatinized, naked DNA was not influenced by silencing FOXA1, since an oligonucleotide pulldown using a 40bp double stranded ER binding region (from an experimentally identified ER binding event)19 was still bound by ER even in the absence of FOXA1 (Figure 2F). These data suggest that ER binding capacity to non-chromatinized DNA does not require FOXA1 (Figure 2F), but that ER association with chromatin is dependent on FOXA1. It is unclear why most ER binding events require FOXA1, yet only ~50% of the ER binding events overlap exactly with a FOXA1 binding event. Two possibilities exist to explain this: the first is that ER binding is stabilised by a FOXA1 binding event that does not occur at the exact same region or that FOXA1 stabilises a secondary factor that subsequently regulates ER binding.
Figure 2.
ER binding to chromatin and transcriptional activity requires FOXA1. A. Western blot of cells transfected with siControl or siFOXA1. B. An example of ER binding in siControl or siFOXA1 transfected cells. C. Heatmap showing the signal intensity of ER binding in siControl or siFOXA1 transfected cells in a window of −/+ 5kb. Also shown in the signal intensity for FOXA1 at the equivalent genomic region. The heatmap represents binding events ranked from the strongest to weakest ER binding (in the siControl condition) and the adjacent columns represent the signal from the corresponding genomic region, but under the different experimental conditions. D. Smoothened scatterplot comparing ER binding intensity in siControl versus siFOXA1 transfected cells. As a control, a scatterplot representing two different siControl experiments is shown. E. Cells were transfected with siControl or siFOXA1, treated with vehicle (V) or estrogen (E) and cells were fractionated to enrich for the chromatin fraction, which was Western blotted. Histone H3 functioned as a loading control. The uncropped Western blot is in Supplementary figure 2. F. Oligonucleotide pulldown using total protein from siControl or siFOXA1 transfected cells. A double stranded biotin labelled oligonucleotide containing a perfect ERE or a mutant sequence was used and protein enriched by the oligonucleotide was Western blotted. G. Gene expression microarray analysis following transfection of siControl or siFOXA1 and treatment with vehicle or estrogen for 6hr.
When we perform the reciprocal experiment, inhibiting ER and mapping FOXA1 binding, we do not find any appreciable differences in FOXA1 binding in the presence or absence of ER (Supplementary figure 4), confirming that FOXA1 is upstream of ER-chromatin interactions.
We assessed the global role that FOXA1 plays on gene expression. Hormone-deprived MCF-7 cells were transfected with siControl or siFOXA1 and treated with vehicle or estrogen for 6hr. Gene expression microarray analysis was performed and differentially regulated genes were identified. Specific silencing of FOXA1 globally affected the estrogen-mediated transcriptome (Figure 2G), with more than 95% of all estrogen regulated genes requiring FOXA1 for estrogen regulation. This suggests that the primary role for FOXA1 in breast cancer cells is to facilitate ER-mediated transcription. These findings were validated with an independent siRNA and with the FOXA1 rescue experiment (Supplementary Figure 3). Furthermore, silencing of FOXA1 resulted in significant (p = 2.6 × 10−6) growth arrest of MCF-7 cells (Supplementary figure 2), confirming an absolute requirement of FOXA1 for the estrogen response in breast cancer cells. This was validated using an independent measure of cell proliferation (% S-phase) (data not shown). This finding was recapitulated in the ZR75-1 ER positive breast cancer cell line (Supplementary figure 3). The growth arrest mimics the results observed by inhibiting ER in MCF-7 cells (data not shown).
FOXA1 is required for tamoxifen action in drug sensitive and resistant contexts
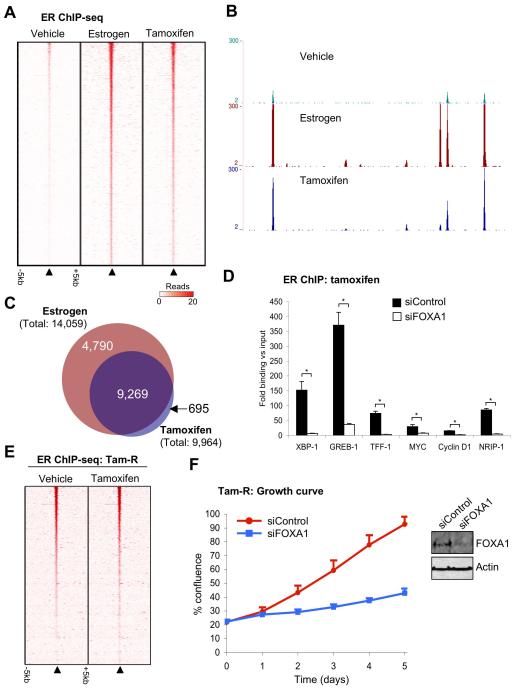
The effectiveness of tamoxifen, one of the most successful cancer drugs20, is primarily due to its ability to inhibit estrogen-ER activity in breast cancer cells, where tamoxifen-ER is recruited to chromatin21 to repress transcription. Previous data implied that in MCF-7 cells, estrogen and tamoxifen could induce significant differences in ER binding profiles8, suggesting that mechanisms must exist for regulating ligand specific ER binding events. We replicated the ER ChIP-seq data in MCF-7 cells following estrogen or tamoxifen treatment and the consensus of the triplicate experiments were considered. We also included vehicle treated ER ChIP-seq. We found that ~93% of the tamoxifen-ER binding events were also estrogen-ER binding events (Figure 3A), a figure that represents natural variations between identical experiments. As such, we did not find significant tamoxifen-ER unique binding events (Figure 3A) as previously described8, possibility due to differences in replicate numbers (Supplementary figure 5). Examples of binding is shown in Figure 3B. However we did find that tamoxifen-ER binding intensity was substantially weaker than estrogen-ER binding (Supplementary figure 5), resulting in less binding events in total (Figure 3C). We also performed gene expression microarray analysis in hormone deprived MCF-7 cells treated with vehicle, estrogen, tamoxifen or estrogen plus tamoxifen for 6hr. This analysis confirmed that tamoxifen mostly antagonises estrogen-mediated gene expression (Supplementary figure 6), with only 27 genes regulated by tamoxifen, but not estrogen. Of these 27 genes, almost all were shown in previous studies to be estrogen-regulated6,22, suggesting these are not true tamoxifen unique gene targets. At later time points tamoxifen may be able to regulate unique genes22, but at the early time point, tamoxifen almost exclusively antagonises estrogen function. Therefore, tamoxifen induces similar ER binding events when compared to estrogen and this results in common genes being regulated by estrogen and tamoxifen.
Figure 3.
Tamoxifen induces similar ER binding events to estrogen, in a FOXA1 dependent manner. A. Heatmap representing ER binding signal intensity in hormone deprived MCF-7 cells treated with vehicle, estrogen or tamoxifen for 45 minutes. The window represents −/+ 5kb. The heatmap represents binding events ranked from the strongest to weakest ER binding (in the estrogen condition). B. Example of ER binding under the different ligand conditions. C. Venn diagram representing the overlap in ER binding events between estrogen and tamoxifen treatment. Comparisons with published data are provided in Supplementary figure 5. D. Hormone deprived MCF-7 cells were transfected with siControl or siFOXA1 and treated with tamoxifen. ER ChIP was performed followed by real time PCR of known ER binding regions. The data are the fold enrichment over Input. The data are the average of independent replicates −/+ Std Dev. E. ER ChIP-seq binding data in hormone deprived tamoxifen resistant MCF-7 cells (Tam-R), treated with vehicle or tamoxifen. F. Tam-R cells were hormone deprived, transfected with siControl or siFOXA1 and treated with tamoxifen. Total cell growth was assessed. The data are the average of independent replicates −/+ Std Dev.
Since tamoxifen induced similar ER binding profiles to estrogen, one could hypothesise that the same mechanisms are used for chromatin interactions. Indeed our data showed that silencing of FOXA1 inhibited tamoxifen-mediated ER binding to chromatin (Figure 3D). Our experimental conditions preclude the possibility for determining if FOXA1 is required for the tamoxifen-mediated growth arrest of breast cancer cells (i.e. cells transfected with siRNA to FOXA1 could not circumvent tamoxifen-ER growth arrest an acquire increased proliferation, since estrogen activity is also inhibited). To assess the requirement for FOXA1 in the tamoxifen-mediated growth of breast cancer cells, we focused on a tamoxifen resistant (Tam-R) MCF-7 breast cancer cell line23, since this would allow us to determine if endocrine resistant cells still maintain a requirement for FOXA1. Initially, we mapped ER binding events in hormone deprived Tam-R cells that were treated with vehicle or tamoxifen. Surprisingly, in the Tam-R cells, the ER binding profile was substantially different from the wild type MCF-7 cells and ER binding occurred independently of tamoxifen treatment (Figure 3E and Supplementary figure 7). The ER binding in Tam-R cells was similar when comparing vehicle or tamoxifen treatment (Figure 3E). The differences in ER binding were not due to mutations in ER, since ER was sequenced in the Tam-R cells and was not mutated. We mapped FOXA1 binding events by ChIP-seq in tamoxifen treated Tam-R cells and found that the FOXA1 binding events were significantly different in Tam-R cells when compared to wild type MCF-7 cells (Supplementary figure 7), but the novel Tam-R FOXA1 binding events correlated with the Tam-R specific ER binding regions (~55% of the Tam-R ER binding events overlapped with a FOXA1 region) (Supplementary figure 7). Furthermore, silencing of FOXA1 in the Tam-R cells resulted in decreased ER binding (Supplementary figure 7) and significant growth inhibition (p-value < 0.001) (Figure 3F), confirming that the altered ER binding and ligand independent growth of the Tam-R cells was still dependent on the presence of FOXA1.
FOXA1 is sufficient to render ER functional in non-breast cancer cells
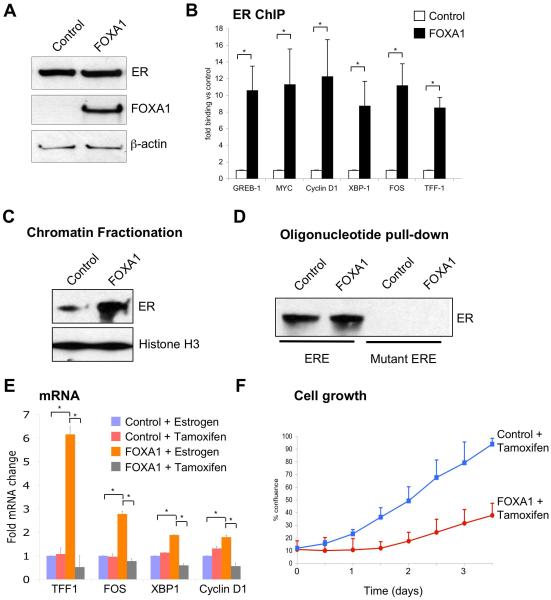
Although FOXA1 is necessary for ER binding to the genome in a breast cancer cell line, it is unknown if FOXA1 is sufficient for ER binding to occur. To address this, we utilised the U20S oesteosarcoma cell line that stably expresses exogenous ER24, but express low levels of FOXA1 (Figure 4A). We transfected U20S-ER cells with control plasmid, or a FOXA1 expression construct (Figure 4A) and assessed ER binding at known breast cancer cis-regulatory elements by ChIP. In U20S-ER cells transfected with FOXA1, increased ER binding was observed at every tested region (Figure 4B).
Figure 4.
FOXA1 expression in U20S-ER osteosarcoma cancer cells renders ER functional. A. FOXA1 or control plasmids were transfected in U20S-ER cells and Western blotting was used to confirm expression. B. In control or FOXA1 expressing U20S-ER cells, ER ChIP was performed followed by real-time PCR of known ER binding events derived from breast cancer cells. * denotes p < 0.01. The data are the average of independent replicates −/+ Std Dev. C. U20S-ER cells were transfected with control or FOXA1 expressing vector and total chromatin fraction was collected and Western blotted for ER. Histone H3 functioned as a control. D. Oligonucleotide pulldown using total protein from control or FOXA1 transfected U20S-ER cells. A double stranded biotin labelled oligonucleotide containing a perfect ERE or a mutant sequence was used and protein enriched by the oligonucleotide was Western blotted. E. Control or FOXA1 expressing U20S-ER cells were treated with estrogen or tamoxifen and mRNA levels of known breast cancer associated target genes were assessed. The data are the average of independent replicates −/+ Std Dev. * denotes p < 0.05. Specifically for TFF1, the fold change is x10 of the y-axis values. F. Cell proliferation was performed in which cell confluence was assessed in U20S-ER cells transfected with control or FOXA1 and treated with tamoxifen. The data are the average of independent replicates −/+ Std Dev.
A global FOXA1-mediated enrichment of chromatin associated ER was assessed in transfected U20S-ER cells. Western blot analysis for ER on the chromatin fraction showed a FOXA1-mediated increase in chromatin bound ER (Figure 4C). Interestingly, ER binding to unchromatinized DNA occurred in U20S-ER cells independently of FOXA1 and FOXA1 expression did not increase ER binding to naked DNA (Figure 4D). Therefore, in osteosarcoma cells ER binding is dictated by other, currently unknown factors25, but the expression of FOXA1 can alter ER binding events.
U20S-ER cells expressing ER possess a gene expression programme and binding profile that is different from the breast cancer context25. We transfected U20S-ER cells with vector or FOXA1, after which we assessed for gene expression of known ER breast cancer target genes. In the presence of FOXA1, estrogen-rich complete media was able to induce expression of a number of breast cancer genes and tamoxifen was able to down-regulate all the tested genes (Figure 4E). U20S-ER cells do not growth arrest in the presence of tamoxifen, but instead increase growth26. We hypothesised that the expression of FOXA1 may alter the tamoxifen response of U20S-ER cells. In control transfected U20S-ER cells, no anti-proliferative response was observed following tamoxifen treatment26, yet expression of FOXA1 was sufficient to impart tamoxifen sensitivity (p-value < 0.05) (Figure 4F). This tamoxifen-mediated growth repression of U20S-ER cells did not occur in control transfected cells (Figure 4F) and was not due to FOXA1 expression alone. We could reproduce these findings in ovarian cancer cells, where expression of FOXA1, could alter ER binding and target gene expression (Supplementary figure 8).
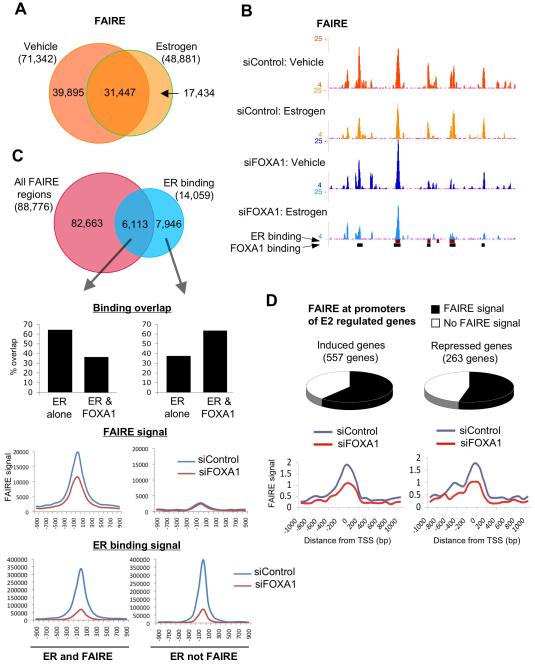
FOXA1 is required for maintaining ligand dependent and independent euchromatic conditions
FOXA1 had previously been shown to be required for maintaining euchromatic conditions at specific cis-regulatory elements11,27. In order to glean insight into the global impact that FOXA1 plays on regulating chromatin structure, we performed FAIRE (formaldehyde-assisted isolation of regulatory elements), a method for identifying nucleosome-free, euchromatic regions of the genome28,29. We coupled FAIRE with high-throughput sequencing. Hormone-deprived cells were transfected with siControl or siFOXA1 and treated with vehicle or estrogen for 1hr. FAIRE was performed and processed for high-throughput sequencing. In control transfected cells we found 71,342 FAIRE region in vehicle treatment and 48,881 in estrogen treatment. We found significant overlap between vehicle and estrogen treatment, with 31,447 regions occurring in a ligand independent manner (Figure 5A). We performed an independent FAIRE replicate and observed similar overlap (data not shown). Examples of FAIRE signal, representing nucleosome depleted chromatin regions, are shown in Figure 5B. When we overlap the FAIRE data (FAIRE signal from vehicle and estrogen treatment combined) with ER binding data, we find substantial, but not complete overlap (Figure 5C). The ER binding and FAIRE shared regions are less likely to be a FOXA1 binding region (Figure 5C). Similarly, the ER binding that occurs in nucleosome-rich chromatin regions (no FAIRE signal), are more likely to be a shared ER and FOXA1 binding region. This suggests that ER binding to nucleosome-free, euchromatic regions of the genome are less likely to occur at regions pre-bound by FOXA1, whereas ER binding to more condensed, nucleosome-rich chromatin requires the pioneering function of FOXA1. When we compare the FAIRE signal in the presence or absence of FOXA1, we find substantial decreases in global FAIRE signal in siFOXA1 transfected cells (Figure 5C). As such, at specific regions, FOXA1 is required for effective maintenance of nucleosome-depleted ‘euchromatic’ conditions within the genome, in many cases in a ligand-independent manner.
Figure 5.
FOXA1 is required for maintaining chromatin structure. A. Genome-wide FAIRE (Formaldehyde-assisted isolation of regulatory elements) was performed in MCF-7 cells transfected with siControl or siFOXA1 and treated with vehicle or estrogen for 1hr. A. Overlap in FAIRE regions between vehicle and estrogen treatment in control cells. B. An example of FAIRE regions, some that are dependent on FOXA1 and adjacent regions that are independent of FOXA1. C. Overlap between FAIRE (both vehicle and estrogen combined) and ER binding. The different categories (ER and FAIRE positive regions versus ER but not FAIRE signal) were assessed for the fraction that represent either ER but not FOXA1 binding or shared ER and FOXA1 binding regions. Also included are the changes in FAIRE and ER binding signal within the two categories. D. Fraction of promoter proximal regions of 6hr estrogen induced or estrogen repressed genes that possess FAIRE signal. The relative difference in FAIRE signal in cells transfected with siControl or siFOXA1 is shown.
Given the observation that promoter regions are traditionally represented by nucleosome-free chromatin, we integrated the FAIRE data with estrogen-mediated gene expression data30. We could show that the majority of the estrogen induced or repressed genes contained FAIRE signal at their promoter regions (Figure 5D). Importantly, the global FAIRE signal was decreased ~50% at the transcription start site (TSS) of genes in cells transfected with siFOXA1 (Figure 5D), confirming a requirement for FOXA1 in maintaining optimal chromatin conditions at promoters of estrogen target genes.
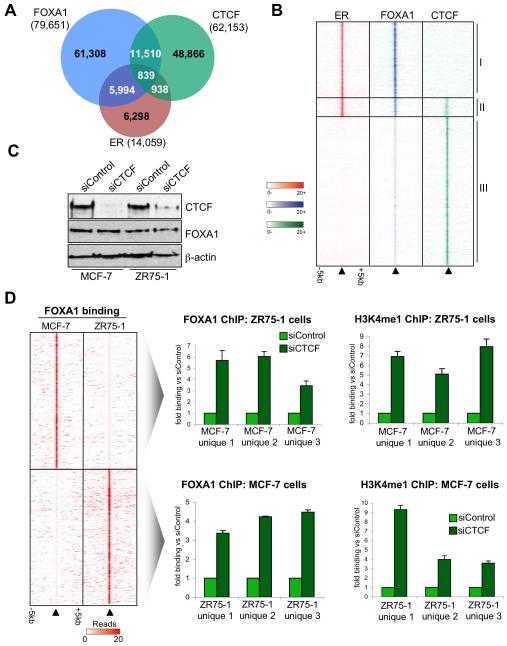
Differential FOXA1 binding and function is influenced by CTCF
Given the context dependent differential FOXA1 binding (Figure 1A), we investigated possible factors that may influence FOXA1 binding: one of which is the insulator protein CTCF, a factor shown to demarcate and influence functional ER regions of the genome31,32. CTCF binding events were mapped in MCF-7 cells by ChIP-seq, resulting in 62,153 binding regions that occurred in multiple replicates (Figure 6A). Interestingly, although a significant number of FOXA1 sites overlapped with CTCF (~15.5%), only ~1% of FOXA1 regions overlapped with both CTCF and ER (Figure 6A). In fact the ER and FOXA1 shared binding events were almost exclusively not bound by CTCF (~88% of the ER and FOXA1 co-bound regions were independent of CTCF binding). These ER and FOXA1 (but not CTCF) binding domains were enriched near most of the estrogen regulated genes (data not shown), suggesting that the presence of CTCF may negatively influence transcriptional activity. It is unclear if this is a direct effect or whether CTCF can alter chromatin state and indirectly influence ER and FOXA1 binding. Although FOXA1 binding did correlate with CTCF binding, these tended to be weak (non-stabilized) FOXA1 binding events (Figure 6B) and were are almost always devoid of ER binding and transcriptional potential.
Figure 6.
ER and FOXA1 shared binding events are exclusively independent of CTCF and CTCF can repress FOXA1 binding and activity. A. Overlap between FOXA1, ER and CTCF binding events in MCF-7 cells. B. Heatmap representing binding signal from regions where ER, FOXA1 and/or CTCF overlap. The categories are: I. ER and FOXA1 shared (but not CTCF) binding events, II. Regions bound by ER, FOXA1 and CTCF, III. FOXA1 and CTCF shared (but not ER) binding regions. The window represents −/+ 5kb. The FOXA1 binding events that overlap with CTCF tend to be the weakest FOXA1 binding events. C. Western blot of MCF-7 and ZR75-1 cells transfected with siControl or siCTCF. D. Heatmap showing the MCF-7 and ZR75-1 unique FOXA1 binding regions. ZR75-1 and MCF-7 cells were transfected with siControl or siCTCF. FOXA1 and H3K4me1 ChIP was performed, followed by real-time PCR of three regions that were FOXA1 binding regions exclusively in the other cell line. The data are the average of independent replicates −/+ Std Dev.
We hypothesized that the CTCF binding may be negatively influencing FOXA1 binding or ability to associate with additional transcription factors such as ER. We specifically silenced CTCF in MCF-7 or ZR75-1 cells (Figure 6C) and performed ChIP of FOXA1 at regions that were unique FOXA1 binding regions in the other cell line. Surprisingly, we found that specific silencing of CTCF in ZR75-1 cells allowed FOXA1 to bind to regions in ZR75-1 cells that were previously unique FOXA1 binding regions in MCF-7 cells (Figure 6D). The silencing of CTCF also resulted in increased H3K4me1 at these regions, a histone mark previously shown to occur at distal ER cis-regulatory elements in a FOXA1 independent manner11. Similarly, we could show that silencing of CTCF altered the chromatin in MCF-7 cells such that FOXA1 and H3K4me1 could be detected in regions that were previously exclusive FOXA1 binding regions in ZR75-1 cells (Figure 6D). These data confirmed that CTCF imposes pressures that inhibit stability of FOXA1 binding.
Discussion
Previous data had shown that FOXA1 (HNF3-α) is one of the minimal signature genes defining ER positive luminal breast cancers15 and that it can predict outcome in breast cancers14. Our current data show that mechanistically, FOXA1 is required for almost all ER binding events in breast cancer cells. This was a surprising finding given the fact that only ~half of the ER binding events directly overlap with a FOXA1 binding event11 (Figure 1). However, FOXA1 may indirectly stabilise the additional ER binding events (potentially by regulating an additional factor that influences ER-chromatin interaction) or may stabilise ER from a distance, possibly mediated via chromatin loops that form between distinct ER binding regions.
Our data also showed that tamoxifen recruits ER to a subset of the regions bound by estrogen-ER, contrary to previous findings8 (Supplementary figure 5). The tamoxifen ‘unique’ regions in the Welboren study were mostly estrogen-ER binding events, precluding the possibility that differences in reagents were contributing factors to the discrepancy. The most likely explanation is the lack of replicates in the Welboren study. Furthermore, the Welboren study suggested that Forkhead motifs are not enriched in ER binding events, clearly a conclusion contradicted by the mechanistic data in this investigation and the discovery of Forkhead motifs in every other ER mapping investigation6,7,10,11,33. Our experimental data would suggest that FOXA1 is required for estrogen-ER function, but also for tamoxifen-ER function. This potentially explains why high FOXA1 levels predict a positive response to tamoxifen in ER positive patients14; namely that functional tamoxifen-ER also requires the same pioneering machinery as estrogen-ER and the absence of FOXA1 negatively impacts on the chromatin binding capacity of tamoxifen-ER and the efficacy of tamoxifen. Another possibility is that tumors with low FOXA1 also possess low ER transcriptional activity and have acquired growth that is less dependent on ER. It is currently unclear if additional breast cancer endocrine therapies utilise similar mechanisms for growth inhibition or if resistance to these treatments are also dependent on FOXA1. Since Aromatase Inhibitors (AIs) deplete tumors of estrogen, one could speculate that any resistance to AIs may be due to acquisition of hypersensitivity to low levels of estrogen, as previously suggested34. If this is the case, ER may simply switch on the endogenous transcriptional pathways, either in low levels of estrogen or in a ligand independent manner, both of which may still require FOXA1 for ER binding to chromatin and for effective transcriptional potential.
Our data would support a role for FOXA1 in mediating ER function in a non-breast cancer context. We observed a FOXA1 dependent response to estrogen and tamoxifen in both ovarian and osteosarcoma cell lines, suggesting that the estrogen-mediated effects in these cellular contexts may also be influenced by FOXA1. Since ovarian and bone tissue are typically ER positive and genetic ablation of ER can impact the physiology in these tissues35, it is possible that ER binding and activity may be dictated by FOXA1 in these target tissues. It is interesting to speculate that ER transcriptional activity in non-breast cancer target tissues, including osteosarcoma and ovarian, may be influenced by the presence or absence of FOXA1.
FOXA1 can mimic linker histone and can bind directly to compacted chromatin12,13 thereby providing the opportunity for other transcription factors to subsequently associate with the chromatin. We found that ER binding events that co-occurred with FOXA1 were more likely to be condensed chromatin (chromatin lacking in FAIRE signal), whereas ER binding events that were not shared with FOXA1 binding were more likely to be nucleosome-depleted and accessible (positive FAIRE signal). As such, ER cis-regulatory elements occurring at condensed chromatin are more dependent on FOXA1 to maintain chromatin interactions. However, FOXA1 binding is not a stable event and appears to possess some context specificity. Our data would suggest that one factor that can influence FOXA1 activity is the insulator protein CTCF, although it is currently unclear if CTCF can directly influence FOXA1 binding or simply alters chromatin state, resulting in an indirect impact on FOXA1. A recent study suggested that CTCF forms demarcated domains within which ER functionality is promoted and that loss of CTCF binding in a different cell line may contribute to decreased ER transcriptional activity32. One clear role for CTCF is its involvement in mediating chromatin loops36, presumably different chromatin loops than those that involve ER and FOXA1 during estrogen-mediated transcription of breast cancer cells5. Our data would suggest that a number of FOXA1 binding events do occur adjacent to CTCF sites, but these tend to be the weakest FOXA1 binding events and rarely recruit ER. As such, the presence of CTCF at these exact genomic regions may destabilise FOXA1 binding, rendering them non-functional or may change chromatin conditions that indirectly influence FOXA1 accessibility.
These findings suggest that FOXA1 is the primary determinant of ER binding and transcriptional activity in breast cancer cells, under both estrogenic and tamoxifen conditions. Furthermore, FOXA1 expression in non-breast cancer cells can enhance ER-chromatin interactions and may be the defining feature that renders ER functional in other cellular contexts.
Supplementary Material
Acknowledgements
We thank Gordon Brown, Ros Russell and Rory Stark for bioinformatics support, James Hadfield for genomic help and Sarah Vowler for statistical advice. We thank Dr. Spelsberg and Dr. Hutcheson for cell lines. We would like to acknowledge the support of The University of Cambridge, Cancer Research UK and Hutchison Whampoa Limited. C.R-I is supported by a Commonwealth fellowship, K.H is supported by a Breast Cancer Campaign grant, J.S.C and A.H are supported by an ERC Starting Grant.
Appendix
Experimental procedures
Cell lines
MCF-7 cells were grown as previously described37. U20S-ER cells (a kind gift from Dr. Thomas Spelsberg, Mayo clinic, Minnesota) were grown in DMEM supplemented with 10% FCS. Tam-R cells (a kind gift from Dr. Iain Hutcheson, Tenovus Centre, Cardiff University) were grown in phenol red free DMEM as previously described23. OVCAR3 cells were obtained from ATCC and grown in RPMI with 10% FCS. Estrogen was used at a final concentration of 100nM and tamoxifen at a final concentration of 1μM.
Chromatin immunoprecipitations (ChIP)
ChIP experiments were performed as described previously38. Antibodies used were anti-ER (sc-543) from Santa Cruz Biotechnologies (USA), FOXA1 (ab5089 and ab23738) and H3K4me1 (ab8895) from Abcam (UK) and CTCF from Millipore (07-729). Primers used for ChIP are given in Supplementary Table 1.
ChIP-sequencing (ChIP-seq)
ChIP DNA was amplified as previously described38. Sequences generated by the Illumina GAIIx genome analyzer (using 36bp reads) were aligned against NCBI Build 36.3 of the human genome using MAQ (http://maq.sourceforge.net/) with default parameters. For each biological replicate, a corresponding set of Input sequence reads of similar size was obtained by random sampling from the full set of Input sequence reads. Peaks were called using Model-based Analysis for ChIP-Seq (MACS)18, run using default parameters.
RT-PCR
Cells were deprived of hormones as previously described39. Total RNA was collected and RT-PCR was performed as previously described6. Primer sequences are given in Supplementary Table 1.
siRNA
siRNA experiments were as previously described6. The sequence of the siRNAs can be found in Supplementary Table 1. All siRNAs were from Dharmacon (Thermo Scientific Dharmacon RNAi Technologies).
Plasmids
FOXA1 expression was from pcDNA3.1-FOXA1 (a kind gift from Dr. Jerome Eeckhoute, University of Rennes, France). ER expression was from pcDNA3.1-ER. As a control, empty pcDNA3.1 was used.
Western blotting
Cells were deprived of hormones as previously described39. For chromatin fractionation experiments, the chromatin fraction was collected as previously described40. Antibodies used were: ERα (sc-543) from Santa Cruz Biotechnologies (USA), FOXA1 (ab23738), β-Actin (ab6276) and histone H3 (ab1791) from Abcam (UK) and CTCF from Millipore (07-729).
Oligonucleotide Pull-down
The oligonucleotide pull-down was as previously described19. The sequences used were as previously described19.
FAIRE (Formaldehyde assisted isolation of regulatory elements)
MCF-7 cells were transfected with siControl or siFOXA1 and treated with vehicle or estrogen for 1hr. FAIRE was performed as previously described29. The enriched chromatin regions were processed for Illumina sequencing as previous described38.
Microarray analysis
MCF-7 cells were transfected with siControl or siFOXA1 and treated with vehicle or estrogen for 6hr. For tamoxifen and estrogen microarray experiments, hormone-deprived MCF-7 cells were treated with vehicle, estrogen (100nM), tamoxifen (1μM) or estrogen plus tamoxifen for 6hr. Gene expression analysis was conducted as previously described 19.
Cell growth assay
Cells were plated at equal confluence, grown in hormone-depleted DMEM media and treated with vehicle, estrogen (100nM) or 1μM tamoxifen. Confluence of cells was analysed using the live-cell imaging Incucyte™ Analyzer (Bucher Biotec AG, Switzerland). This approach has been validated as a robust method for assessing cell growth (www.essenbioscience.com).
Statistics
Statistical analysis was performed using two tailed paired T-tests. P-value cut-offs are indicated in the relevant figures.
Footnotes
Competing financial interests
None of the authors have any competing financial interests related to this work.
Accession numbers
All ChIP-seq and FAIRE data is deposited under ArrayExpress number E-MTAB-223. All microarray data is deposited under GEO number GSE25316.
References
- 1.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–12. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 2.Clarke R, Leonessa F, Welch JN, Skaar TC. Cellular and molecular pharmacology of antiestrogen action and resistance. Pharmacol Rev. 2001;53:25–71. [PubMed] [Google Scholar]
- 3.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- 4.Deblois G, Giguere V. Nuclear receptor location analyses in mammalian genomes: from gene regulation to regulatory networks. Mol Endocrinol. 2008;22:1999–2011. doi: 10.1210/me.2007-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–97. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 7.Lin CY, et al. Whole-Genome Cartography of Estrogen Receptor alpha Binding Sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welboren WJ, et al. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. Embo J. 2009;28:1418–28. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laganiere J, et al. Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A. 2005;102:11651–56. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cirillo LA, et al. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. Embo J. 1998;17:244–54. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–89. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 14.Badve S, et al. FOXA1 expression in breast cancer--correlation with luminal subtype A and survival. Clin Cancer Res. 2007;13:4415–21. doi: 10.1158/1078-0432.CCR-07-0122. [DOI] [PubMed] [Google Scholar]
- 15.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 16.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lacroix M, Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat. 2004;83:249–89. doi: 10.1023/B:BREA.0000014042.54925.cc. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross-Innes CS, et al. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 2010;24:171–82. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan VC. Tamoxifen: a most unlikely pioneering medicine. Nat Rev Drug Discov. 2003;2:205–13. doi: 10.1038/nrd1031. [DOI] [PubMed] [Google Scholar]
- 21.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 22.Frasor J, et al. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64:1522–33. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- 23.Knowlden JM, et al. Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology. 2003;144:1032–44. doi: 10.1210/en.2002-220620. [DOI] [PubMed] [Google Scholar]
- 24.Monroe DG, et al. Estrogen receptor isoform-specific regulation of endogenous gene expression in human osteoblastic cell lines expressing either ERalpha or ERbeta. J Cell Biochem. 2003;90:315–26. doi: 10.1002/jcb.10633. [DOI] [PubMed] [Google Scholar]
- 25.Krum SA, et al. Unique ERalpha cistromes control cell type-specific gene regulation. Mol Endocrinol. 2008;22:2393–406. doi: 10.1210/me.2008-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monroe DG, et al. Estrogen receptor alpha and beta heterodimers exert unique effects on estrogen- and tamoxifen-dependent gene expression in human U2OS osteosarcoma cells. Mol Endocrinol. 2005;19:1555–68. doi: 10.1210/me.2004-0381. [DOI] [PubMed] [Google Scholar]
- 27.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–26. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–85. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eeckhoute J, et al. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res. 2009;19:372–80. doi: 10.1101/gr.084582.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt D, et al. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–88. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan CS, Song JS. CCCTC-binding factor confines the distal action of estrogen receptor. Cancer Res. 2008;68:9041–9. doi: 10.1158/0008-5472.CAN-08-2632. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. CCCTC-binding factor acts upstream of FOXA1 and demarcates the genomic response to estrogen. J Biol Chem. 2010;285:28604–13. doi: 10.1074/jbc.M110.149658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charn TH, et al. Genome-wide dynamics of chromatin binding of estrogen receptors alpha and beta: mutual restriction and competitive site selection. Mol Endocrinol. 2010;24:47–59. doi: 10.1210/me.2009-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin LA, et al. Enhanced estrogen receptor (ER) alpha, ERBB2, and MAPK signal transduction pathways operate during the adaptation of MCF-7 cells to long term estrogen deprivation. J Biol Chem. 2003;278:30458–68. doi: 10.1074/jbc.M305226200. [DOI] [PubMed] [Google Scholar]
- 35.Couse JF, Korach KS. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev. 1999;20:358–417. doi: 10.1210/edrv.20.3.0370. [DOI] [PubMed] [Google Scholar]
- 36.Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–54. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt D, et al. ChIP-seq: Using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009 doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carroll JS, Prall OW, Musgrove EA, Sutherland RL. A pure estrogen antagonist inhibits cyclin E-Cdk2 activity in MCF-7 breast cancer cells and induces accumulation of p130-E2F4 complexes characteristic of quiescence. J Biol Chem. 2000;275:38221–9. doi: 10.1074/jbc.M004424200. [DOI] [PubMed] [Google Scholar]
- 40.Narita M, et al. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126:503–14. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.