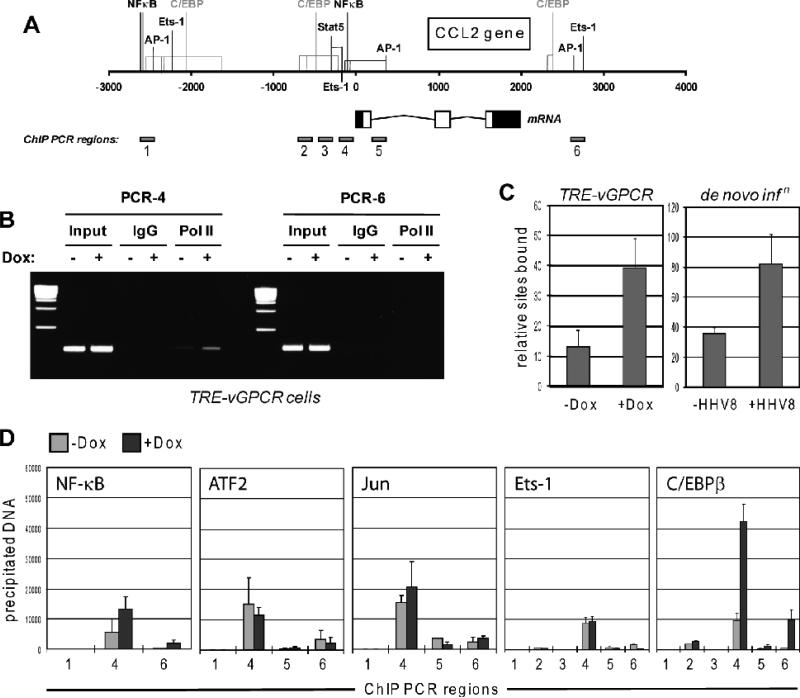
Figure 2.
Transcriptional analysis of vGPCR-mediated CCL2 induction. (A) Diagrammatic representation of the CCL2 gene, indicating regions and associated putative or demonstrated transcription factor binding sites implicated by previous studies in transcriptional regulation. Predicted C/EBP binding sites, identified by application of TFSEARCH (www.cbrc.jp/research/db/TFSEARCH.html) are shown in grey. The CCL2 mRNA structure is indicated, along with the map positions of the products of PCR primer pairs used for ChIP assays. (B) Ethidium bromide agarose gel analysis of PCR products derived from chromatin immunoprecipitation (ChIP) assays using antibodies to RNA polymerase II for precipitation of PolII-bound DNA. Two sets of primer pairs, one directed to proximal promoter sequences and the other to potential regulatory sequences 3' of the CCL2 gene, were used for PCR-mediated amplification of precipitated DNA. The former, specifically, was able to give rise to PCR products, which were increased in amplitude as a function of vGPCR expression (+Dox), indicative of receptor-mediated induction of CCL2 gene transcription. Normal IgG was used as a negative control to ensure specificity of DNA precipitation by PolII antibody. (C) Quantified ChIP data using primer pair 4 for qPCR analysis of PolII-associated and -precipitated DNA derived from Dox-treated or mock-treated TIME-vGPCR cells (left, TRE-vGPCR), confirming the data in panel B. Equivalent analysis of CCL2 promoter-associated PolII in response to HHV-8 de novo infection of TIME cells provided similar evidence of transcriptional induction of CCL2, 24 h post-inoculation (right, de novo infn). Error bars indicate deviation from mean values obtained from duplicate PCR reactions. (D) Analogous ChIP assays to investigate the potential relevance of a panel of transcription factors of possible relevance to vGPCR-mediated regulation of CCL2. Several PCR primer pairs (1-6) directed to different regions of the CCL2 locus were used for PCR amplification of precipitated DNA. Each was pre-validated to ensure ability to amplify “input” cellular DNA (data not shown). vGPCR-induced promoter-association of C/EBPβ, exclusively, was detected, specifically with regions amplified by PCR primer pairs 4 and 6 (right chart). Error bars indicate deviation from mean values obtained from duplicate PCR reactions.