Abstract
Gluconeogenesis makes a major contribution to hepatic glucose production, a process critical for survival in mammals. In this study, we identify the p160 family member, SRC-1, as a key coordinator of the hepatic gluconeogenic program in vivo. SRC-1 null mice displayed hypoglycemia secondary to a deficit in hepatic glucose production. Selective re-expression of SRC-1 in the liver restored blood glucose levels to a normal range. SRC-1 was found induced upon fasting to coordinate in a cell-autonomous manner, the gene expression of rate-limiting enzymes of the gluconeogenic pathway. At the molecular level, the main role of SRC-1 was to modulate the expression and the activity of C/EBPα through a feed-forward loop in which SRC-1 used C/EBPα to transactivate pyruvate carboxylase, a crucial gene for initiation of the gluconeogenic program. We propose that SRC-1, acts as a novel and critical mediator of glucose homeostasis in the liver by adjusting the transcriptional activity of key genes involved in the hepatic glucose production machinery.
INTRODUCTION
Throughout evolution, organisms have devised metabolic strategies to survive during long periods of starvation. Some organisms are able to place their own metabolism on “pause” (hypometabolism or even ametabolism) to sustain them during extreme environmental conditions (Storey and Storey, 2007). Humans and most other higher mammals have abandoned this type of extreme adaptation by developing alternative regulatory circuits that allow for maintenance of cellular energy (ATP) in vital tissues. While metabolic adaptation during fasting involves a variety of tissues, the liver plays the key role in the orchestration of this complex phenomenon (Cahill, 2006). During fasting, hepatic glucose production is required to maintain blood sugar levels in a normal range, ensuring a sufficient supply of energy for the central nervous system (Roden et al., 2001). Shortly after food withdrawal, liver glycogen stores are depleted and gluconeogenesis becomes the main contributor to hepatic glucose production and survival (Newgard, 2004). The rate of gluconeogenic flux is controlled by the activities of key enzymes such as pyruvate carboxylase (PC), phosphoenolpyruvate carboxykinase (PEPCK), fructose-1, 6-bisphosphatase (FBP1) and glucose-6-phosphatase (G6Pase) (Granner and Pilkis, 1990). Due to the epidemic expansion of type 2 diabetes (Doria et al., 2008), and because dysregulation of gluconeogenesis is one of the major contributing factors of this metabolic disorder, there is currently a need for a better understanding of mechanisms and the transcriptional players that drive the gluconeogenic program in vivo. Recent studies using knock-out animals of transcriptional factors and coregulators including PGC-1α, CRTC2 and SIRT1 strongly suggest that additional factors are likely to be involved in the control of hepatic glucose production (Chen et al., 2008; Le Lay et al., 2009; Leone et al., 2005; Lin et al., 2004).
The p160 family of coactivators includes SRC-1 (NCOA1), SRC-2 (TIF2; GRIP1, NCOA2) and SRC-3 (AIB1; ACTR; TRAM1; RAC3; p/CIP; NCOA3). These coregulators have been studied extensively for their transcriptional control mechanisms in cell culture but their physiological roles in vivo are less understood. Nevertheless, previous studies have revealed regulatory roles for this family in certain aspects of metabolism (Louet et al., 2006; Louet and O'Malley, 2007; Picard et al., 2002). SRC-3 has been implicated in adaptive thermogenesis regulation via modulation of PGC-1α activity in skeletal muscle and brown adipose tissue (Coste et al., 2008). In the liver, SRC-2 interacts with RORα to control the expression of the glucose-6-phosphatase gene, and its ablation results in a glycogen storage-1a-like (Von Gierke's) syndrome (Chopra et al., 2008). The specific metabolic roles of SRC-1 remain unknown.
In the present study, we found that the hepatic expression of SRC-1 gene was increased during the fed-to-fasting metabolic switch. A critical role for SRC-1 in the control of hepatic glucose production was uncovered by studying SRC-1 null mice which were hypoglycemic in fed and in fasting states despite an absence of increased insulin secretion or peripheral insulin sensitivity. Screening for alterations of the gene expression of key metabolic enzymes revealed impairment in the gluconeogenic program in SRC-1 null mice. Dissection of the underlying molecular mechanisms identified SRC-1 as a critical mediator of glucose homeostasis in the liver in the fed-to-fasting transition.
RESULTS
SRC-1 knock-out mice are hypoglycemic due to a liver metabolic defect
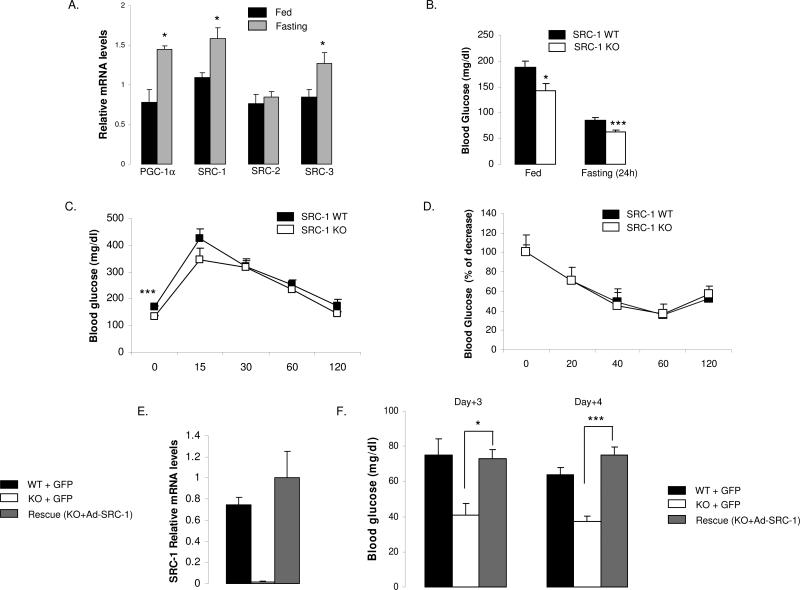
In an attempt to uncover new metabolic functions for the p160 family of coactivators, we monitored SRC-family gene expression in the liver by qPCR during the transition between the fed-to-fasting states and found that the hepatic expression of SRC-1 and SRC-3 were significantly increased upon fasting (Fig.1A). As previously described, PGC-1α mRNA was increased (Yoon et al., 2001) whereas SRC-2 expression was not changed (Fig.1A). Since one of the major functions of the liver during the fed-to-fasting transition is to maintain blood sugar in a normal range, we further characterized the importance of SRC-1 and SRC-3 by determining the blood glucose levels in animals with global KOs of these two coactivators. We observed a significant decrease in blood glucose levels in fasted (and also in randomly fed) SRC-1 null animals compared to wild type animals (Fig.1B); no significant differences were found in the SRC-3 KO mice (Fig.S1A). Based on this observation, we performed detailed phenotypic analyses of the SRC-1 null mice.
Figure 1. Impact of SRC-1 on fasting glycemia is liver dependent.
A) SRC-1 and SRC-3 gene expression are increased in the liver during fed-to-fasting transition. The gene expression of the p160 family of coactivators and PGC-1α was measured by qPCR in the liver of WT animals in the fed state (n=5 mice per group) and upon 24 hours of fasting (n= 5 mice per group).
B) Ablation of SRC-1 results in fed and fasting hypoglycemia. Blood glucose levels were determined in SRC-1 knockout (KO) and WT mice during ad-libitum feeding (n = 5 mice per group) and after 24 hours of fasting (n = 12-15 mice per group), using a hand-held glucometer.
C-D) SRC-1 KO mice exhibit normal insulin sensitivity. Glucose tolerance test and insulin tolerance tests were performed after 4 hours of fasting (n=6 mice per group)
E-F) Correction of hypoglycemia in SRC-1 KO mice by adenovirus-mediated re-expression of SRC-1. E: SRC-1 expression levels measured by qPCR in the liver (n=4 mice per group) of the WT and the KO groups treated with a control (empty) adenovirus (WT+GFP or KO+GFP) and the KO group treated with an adenovirus expressing SRC-1 (KO+SRC-1). F: Blood glucose levels were determined in mice (n=4 mice per group) fasted 16h on two consecutive days (Day+3=left panel; Day+4=right panel) after adenovirus treatment. Data are shown for the WT and KO groups treated with control adenovirus (WT+GFP or KO+GFP) and for the KO group treated with an adenovirus expressing SRC-1 (KO+SRC-1).
Data are represented as mean + SEM. Unpaired student's t-test was used for evaluation of statistical significance. One asterisk indicates p < 0.05, two asterisks indicate p < 0.01 and three asterisks indicate p < 0.005.
“See also Fig.S1”
Decreased blood glucose levels in SRC-1 null mice were not a consequence of increased secretion of pancreatic insulin in fasting conditions (Fig.S1B). Levels of glucagon, corticosteroids, and IGF-1, as well as circulating free fatty acids or triglycerides, were unchanged in plasma upon fasting (Fig.S1B+Fig.S1C). Global in vivo lipolysis was unimpaired in SRC-1 KO mice, as evidenced by equal increases in fatty acids and glycerol in blood of SRC-1 KO mice and WT mice following 4 hours of fasting, or after fasting and injection of CL316243, a beta-3 adrenergic receptor agonist; these results indicate no fundamental defects in regulation of lipolysis in white adipose tissue (Fig.S1D). Insulin sensitivity of SRC-1 KO mice was similar to wild-type (WT) animals based on glucose and insulin tolerance tests (Fig.1C-D). Finally, no differences were found in physical activity, body weight, food consumption, percentage of fat mass and energy expenditure between the KO and WT animals (Figs.S1E+S1F). Therefore, the hypoglycemia observed in SRC-1 null mice suggested a hepatic defect.
SRC-1 depletion impairs hepatic glucose production
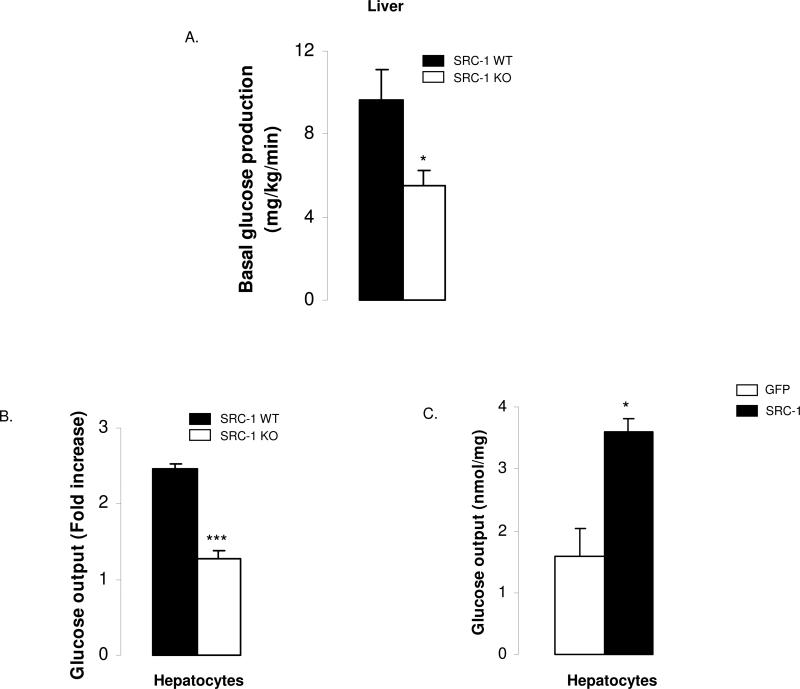
To demonstrate that the liver was the primary cause of the hypoglycemia in SRC-1 null animals, we re-expressed the SRC-1 coactivator selectively in the liver through the injections of an adenovirus encoding SRC-1. This approach restored hepatic expression of SRC-1 to levels similar to WT animals (Fig.1E) and resulted in complete normalization of blood glucose levels after 16h of fasting (Fig.1F). To substantiate this finding, we determined in vivo glucose production and found a clear defect in hepatic glucose production in the SRC-1 KO mice upon fasting (Fig.2A). In primary hepatocytes from SRC-1 KO mice, hormonal induction of glucose production by glucocorticoids and cAMP was significantly decreased compared to WT cells (Fig.2B). Conversely, adenovirus-mediated overexpression of SRC-1 in primary hepatocytes increased glucose output (Fig.2C). Thus, SRC-1 appears to function as an important regulator of hepatic glucose production in response to fasting.
Figure 2. Hepatic glucose production is impaired in SRC-1 KO mice.
A) In vivo glucose production is decreased in SRC-1 KO mice. Glucose production was performed on SRC-1 null and WT mice after an overnight fast (16h) in order to assess basal hepatic glucose production (n = 5 mice per group).
B-C) SRC-1 affects glucose production in primary hepatocytes. B: Measurement of glucose production in primary hepatocytes from WT or SRC-1 null animals after induction of gluconeogenesis with dexametasone and forskolin. C: Measurement of basal glucose production 48 h after overexpression of SRC-1, or as a control, GFP.
Data are represented as mean ± SEM. Unpaired student's t-test was used for evaluation of statistical significance. One asterisk indicates p < 0.05, two asterisks indicate p < 0.01 and three asterisks indicate p < 0.005.
SRC-1 controls the gene expression of key gluconeogenic enzymes in vivo
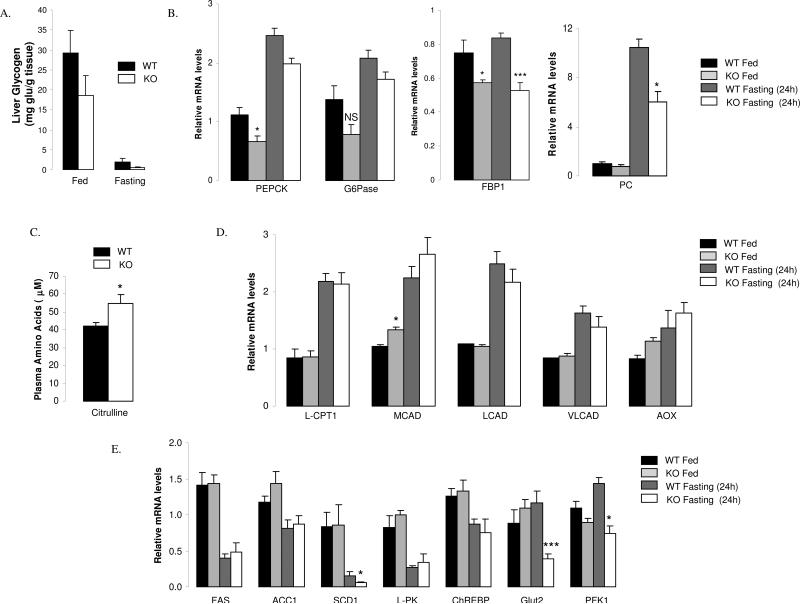
In order to understand the molecular mechanisms underlying the impact of SRC-1 on the control of hepatic glucose production, we next focused on glycogenolysis and gluconeogenesis, two major pathways involved in glucose production by the liver. No significant difference was found between the hepatic glycogen content of knock-out vs. wild type animals regardless of the nutritional status (Fig.3A). We also found that key glycogenolysis genes were not impaired upon fasting (Fig.S2A), suggesting that glycogenolysis was not affected by the absence of SRC-1. We therefore shifted our attention to key genes that control critical steps of gluconeogenesis. Interestingly, PEPCK mRNA levels were found to be significantly decreased in fed status in SRC-1 KO mice compared to WT without reaching significance upon fasting, whereas G6Pase gene expression was globally not significantly impaired (Fig.3B). A significant decrease was also noted for FBP1 gene expression in fed and in fasting, suggesting a control of this major gluconeogenic enzyme by SRC1 (Fig.3B). Additionally, mRNA levels of pyruvate carboxylase (PC) were decreased in SRC-1 KO mice compared to WT littermates upon fasting (Fig.3B). This is of importance since one of the critical checkpoints for the adequate metabolic adaptation of the liver to fasting is the increase of the activity of PC, which allows the liver to switch from an organ oxidizing glucose and esterifying fatty acids into triglycerides during the fed condition, to an organ mainly producing glucose and oxidizing fat upon fasting (Desvergne et al., 2006). Of note, accumulation of citrulline was found in the liver and the plasma of SRC-1 KO animals compared to WT (Fig.S2B+Fig.3C), similarly to that found in human patients presenting with PC deficiency (Garcia-Cazorla et al., 2006). Re-expression of SRC-1 in the liver of SRC1 KO mice restored plasma citrulline levels to a normal range (Fig.S2C), thus reinforcing the finding that SRC-1 is a major regulator of PC in vivo.
Figure 3. SRC-1 controls the hepatic gluconeogenic program.
A) SRC-1 does not influence the glycogenolysis pathway in the liver. Liver glycogen content was measured in liver of WT and SRC-1 KO animals under fed (n=5 mice per group) or 24 hour fasted (n= 5 mice per group) conditions.
B) SRC-1 regulation of key genes in the gluconeogenic pathway in liver during the fasting adaptation. mRNA levels for the indicated genes were measured by qPCR in the liver of WT animals under fed (n=5 mice per group) and fasted (n= 5 mice per group) conditions. PEPCK: Phosphoenolpyruvate carboxykinase, cytosolic isoform; G6Pase: Glucose-6-phosphatase; FBP1: Fructose-1,6-bisphosphatase; PC: pyruvate carboxylase.
C) Hypercitrullinemia in SRC-1 KO mice. Citrulline levels in plasma of fasted (24 hours) WT and SRC-1 KO mice (n= 5 mice per group).
D) Impact of SRC-1 on important genes of the β-oxidation pathway in the liver. mRNA levels for the indicated genes were measured qPCR in the liver of WT and SRC-1 KO animals in fed (n=5 mice per group) and 24 hour-fasted mice (n= 5 mice per group). LCPT1: Carnitine palmitoyltransferase 1a, liver; MCAD: Acyl-Coenzyme A dehydrogenase, medium chain; LCAD: Acyl-Coenzyme A dehydrogenase, long chain, VLCAD: Acyl-Coenzyme A dehydrogenase, very long chain; AOX: Acyl-Coenzyme A oxidase 1.
E) mRNA levels for several important genes involved in glycolysis and lipogenesis pathways. FAS, ACC1, SCD1 (lipogenesis) and L-PK, ChREBP, Glut2, PFK1 (glycolysis) mRNA levels were measured by qPCR in the liver of SRC-1 KO and WT animals following 24 hours of fasting (n= 5 mice per group). FAS: fatty acid synthase, ACC1: acetyl-CoA carboxylase, SCD1: stearoyl-CoA desatuase 1, L-PK: pyruvate kinase, liver isoform, ChREBP: carbohydrate responsive element binding protein, Glut2: Glucose transporter 2, PFK1: phosphofructose kinase 1.
Data are represented as mean ± SEM. Unpaired student's t-test was used for evaluation of statistical significance. One asterisk indicates p < 0.05, two asterisks indicate p < 0.01 and three asterisks indicate p < 0.005.
“See also Fig.S2”
Since impairment in the hepatic β-oxidation pathway was found associated with hypoglycemia in humans (Eaton, 2002; Foster, 2004), the impact of SRC-1 on this pathway was investigated by testing the gene expression profile of key genes of this pathway. Only minor differences in mRNA levels of fatty oxidation genes compared to WT were detected (Fig.3D) suggesting that SRC-1 was not a key regulator of β-oxidation in the liver.
Finally, we also assessed the expression levels of hepatic genes involved in glycolysis and lipogenesis, two pathways that are normally decreased during fasting, in order to spare glucose. An abnormally high activity of these pathways can indeed result in hypoglycemia under food restriction (Magnuson et al., 2003). In SRC-1 null mice, none of the tested glycolytic or lipogenic genes showed an increase of their gene expression (Fig.3E). Instead, two glycolytic genes, the glucose transporter 2 (Glut2) and phosphofructokinase1 (PFK1), were found decreased in fasted SRC-1 KO mice compared to WT mice (Fig.3E).
Together, numerous important metabolic genes were quantified by qPCR for their in vivo expression in SRC-1 depleted livers during the transition from fed to fasting states. From this analysis, we concluded that SRC-1 is a crucial coordinator of the expression of specific key gluconeogenic regulators in the liver, but that it does not exert appreciable control of expression of genes dedicated to glycogenolysis, β-oxidation, glycolysis and lipogenesis.
Cell-autonomous action of SRC-1 on the control of critical gluconeogenic genes
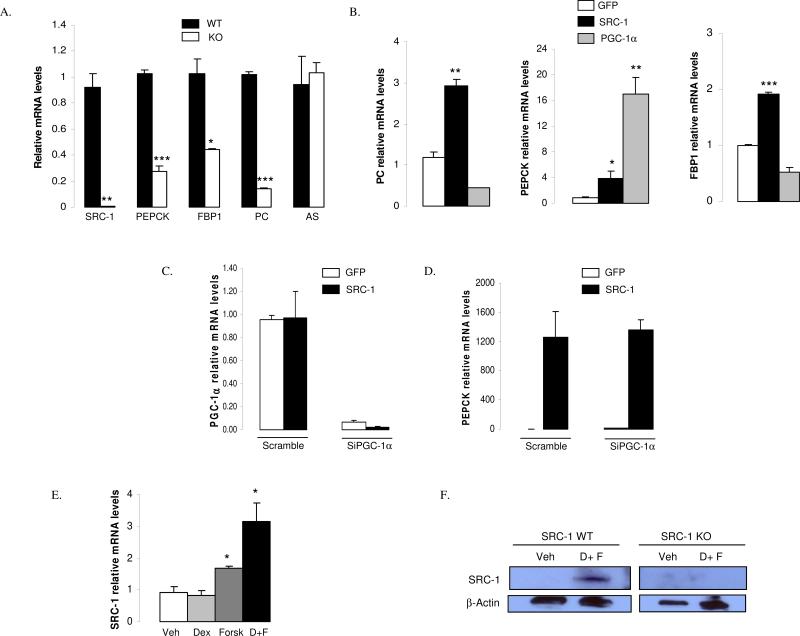
To test for a cell autonomous effect of SRC-1 on the gluconeogenic program, we compared the mRNA levels of key gluconeogenic genes in primary hepatocytes isolated from SRC-1 KO mice and their WT littermates. As expected, we found that ablation of SRC-1 significantly affected the transcription of FBP1 and PC (Fig.4A), confirming our in vivo results. No defect was found on the argininosuccinate synthetase (AS) gene expression, an important hepatic gene involved in the urea cycle (Fig.4A). Interestingly, whereas the impact of the abrogation of SRC-1 on PEPCK in intact animals was observed only in a randomly fed status and not in response to a long-term fasting, isolated hepatocytes from SRC-1 knockout mice did show a lower expression of the endogenous PEPCK compared to WT (Fig.4A). This observation suggested that (i) PEPCK could be a potential target gene of SRC-1 and the role of which would be to maintain the basal activity of this enzyme in between meals and (ii) that in vivo complex inter-tissular and hormonal interactions occurring upon fasting might “mask” this transcriptional regulation.
Figure 4. SRC-1 controls gluconeogenic genes in a cell-autonomous manner.
A) mRNA levels for several important gluconeogenic genes are decreased in primary hepatocytes from SRC-1 KO mice. AS: argininosuccinate synthetase
B) SRC-1 controls gluconeogenic genes that are not responsive to PGC-1α. Primary hepatocytes were treated with SRC-1 or PGC-1α adenoviruses and studied 48 hours after viral treatment. The gene expression level of specific target genes was evaluated by qPCR.
C-D) Specific knock-down of PGC-1α does not influence the activation of PEPCK by SRC-1. The mRNA levels of PGC-1α were decreased via SiRNA in Hepa1.6 cells and then over-expressed by SRC-1 using adenoviruses during 48h. The gene expression level of PEPCK was evaluated by qPCR
E) SRC-1 mRNA levels (measured by qPCR) are increased in WT hepatocytes by 24 hours of exposure to dexamethasone or forskolin or dexamethasone + forskolin.
F) SRC-1 protein levels increase in hepatocytes treated with dexamethasone + forskolin. SRC-1 protein levels were measured by immunoblot (western) analysis in hepatocytes from SRC-1 KO and WT littermates following 24 hours of treatment with dexamethasone and forskolin. D: dexamethasone; F: forskolin
Data are represented as mean ± SEM of three independent experiments. Unpaired student's t-test was used for evaluation of statistical significance. One asterisk indicates p < 0.05; two asterisks indicate p < 0.01 and three asterisks indicate p < 0.005.
“See also Fig.S3”
We next compared the action of SRC-1 and PGC-1α, another important metabolic coactivator known to control gluconeogenesis. By using an adenoviral strategy in primary hepatocytes, we found that SRC-1 controlled PC and FBP1 while overexpressed PGC-1α did not upregulated them (Fig.4B). PEPCK was found regulated by both SRC-1 and PGC-1α (Fig.4B).
To investigate the possibility that SRC-1 might regulate transcription via molecular mechanisms distinct from those used by PGC-1α, we first checked the impact of SRC-1 on the PEPCK gene expression in hepatocytes where PGC-1α gene expression was specifically decreased by siRNA. We found that a strong decreased of PGC-1α mRNA (Fig.4C) levels had no effect on the SRC-1 dependent activation of PEPCK (Fig.4D). This result strongly suggest that SRC-1 and PGC-1α control PEPCK via independent mechanisms.
Finally, we reproduced in the context of isolated hepatocytes, the fasting induction of the expression of endogenous SRC-1 gene by using glucocorticoids and cAMP (Fig.4E) whereas insulin was without significant effect (Fig.S3A). These observations confirmed that SRC-1 is regulated at the level of transcription by hormonal and cellular signals specific to the fasting liver. Induction of the SRC-1 protein also occurred in a cellular context as well as in vivo (Fig.4F+Fig.S3B).
SRC-1 controls the gene expression and the transcriptional activity of C/EBPα upon fasting
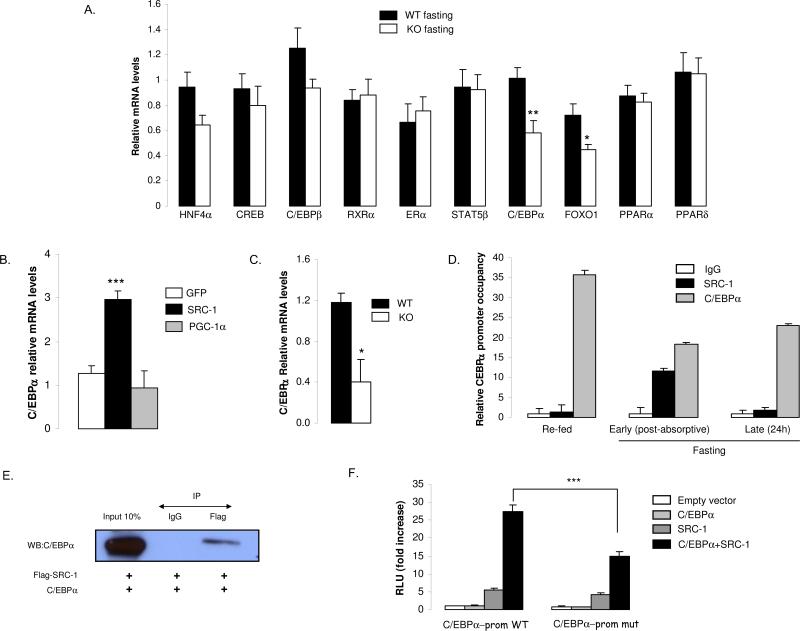
To determine the molecular mechanisms underlying the impact of SRC-1 on gluconeogenesis, we next investigated hepatic mRNA levels of several important transcription factors known to be involved in the fasting metabolic transition (Desvergne et al., 2006). Out of 10 tested transcriptional factors, only C/EBPα and FOXO1 were found to be significantly decreased in fasted SRC-1 null animals compared to WT mice (Fig.5A). These transcription factors are key players in the control of glucose production by the liver in vivo, as their respective KO mice present with profound hypoglycemia (Darlington et al., 1995; Matsumoto et al., 2007). While FOXO1 is known to act via PGC-1α to control hepatic glucose production (Puigserver et al., 2003), the molecular players with C/EBPα are still unknown. In our hands, when C/EBPα mRNA levels were assessed in primary hepatocytes in which SRC-1 was overexpressed, we found that the SRC-1 coactivator induced endogenous C/EBPα gene expression (Fig.5B). Moreover, this effect was selective to SRC-1 as overexpressing PGC-1α had no effect on C/EBPα mRNA levels (Fig.5B). We then checked the expression level of C/EBPα in the isolated SRC-1 KO hepatocytes and found a significant decrease in KO cells compared to WT (Fig.5C). Then, specific binding of SRC-1 on the C/EBPα promoter was investigated in mouse liver using chromatin immunoprecipitation assays in different key metabolic conditions which are refed, post-absorptive (fed) and fasting (24h) animals. Interestingly, SRC-1 was not found in the C/EBPα promoter when animals were re-fed (Fig.5D). In contrast, a strong binding of SRC-1 was demonstrated on this promoter in post-absorptive status and declined after 24h of fasting (Fig.5D). We also confirmed the constitutive presence of C/EBPα on its own promoter (Fig.5D), as demonstrated by others (Timchenko et al., 1995).
Figure 5. The coactivator SRC-1 controls the expression and the transcriptional activity of C/EBPα gene.
A) C/EBPα and FOXO1 mRNA levels are decreased in the liver of SRC-1 KO mice upon fasting. Gene expression levels of several transcription factors were determined by qPCR in the liver (n=5 mice per group). HNF4α: Hepatic nuclear factor 4 alpha; CREB: cAMP response element (CRE)-binding protein; C/EBPα: CAAT-enhancer binding protein alpha; C/EBPβ: CAAT-enhancer binding protein beta; ERα: Estrogen receptor alpha, STAT5beta: Signal transducer and activator of transcription 5 beta; RXRα: retinoid X receptor alpha; FOXO1: Forkhead box O1, PPARα: Peroxysome proliferator activated receptor alpha, PPARδ: Peroxysome proliferator activated receptor delta.
B) Overexpression of SRC-1 activates C/EBPα gene expression in primary hepatocytes. Mouse hepatocytes were treated with SRC-1 or PGC-1α adenoviruses and harvested 48 h later for measurement of C/EBPα mRNA by qPCR.
C) mRNA levels for C/EBPα are decreased in primary hepatocytes from SRC-1 KO mice.
D) In the mouse liver, SRC-1 binds the C/EBPα promoter in the region containing C/EBPα responsive elements in fasting condition. ChIP assays were performed in re-fed, post-absorptive and fasting (24h) conditions with 221 bp amplicons flanking the region containing the C/EBPα motifs. qPCR (normalized to input) was used to assess SRC-1 and C/EBPα occupancy of the C/EBPα promoter upon chromatin immunoprecipitation, using SRC-1 and C/EBPα specific antibodies.
E) SRC-1 and C/EBPα are found in the same complexes. Co-immunoprecipitation was performed after over-expression of SRC-1/flag and C/EBPα in the 293/T cell line.
F) SRC-1 controls the C/EBPα promoter by using C/EBPα as its transcriptional factor partner. Hela cells were transfected with the C/EBPα promoter in the native context (WT) or mutated for major C/EBP-RE elements (mut) in the presence of SRC-1, C/EBPα, or both expression vectors. Luciferase activity was measured after an additional 48 hours of cell culture and is presented as fold increase compared to empty vector.
Data are represented as mean ± SEM of two independent experiments performed in triplicate. Unpaired student's t-test was used for evaluation of statistical significance. One asterisk indicates p < 0.05; two asterisks indicate p < 0.01 and three asterisks indicate p < 0.005.
“See also Fig.S4”
We next investigated the potential physical interaction between C/EBPα and SRC-1 by co-immunoprecipitation and found them in the same protein complexes in a cellular context and in the liver (Fig.5E+Fig.S4A). Furthermore, we demonstrated a functional effect of SRC-1 on the promoter of C/EBPα by using C/EBPα itself as the transcription factor, as we observed a strong synergistic effect when these two proteins were co-expressed (Fig.5F). The effect of SRC-1 on the C/EBPα promoter was dependent upon the C/EBP response element (C/EBP-RE), since (i) the specific mutation of C/EBP-RE in the context of the C/EBPα natural promoter significantly reduced the synergistic effect observed when SRC-1 and C/EBPα were co-expressed (Fig.5F), (ii) an artificial promoter containing only multi-copies of this C/EBP-RE motif was also strongly activated after co-expression of SRC-1 and C/EBPα (Fig.S4B). The functional cooperation between C/EBPα and SRC-1 was not due to the histone acetylase domain of SRC-1 (HAT) but in part due to one of its activation domains (AD2) (Fig.S4C). Taken together, we found strong evidence supporting a direct control of SRC-1 on both the gene expression and the transcriptional activity of C/EBPα.
A central role for the C/EBPα-PC axis in the control of hepatic glucose production by SRC-1
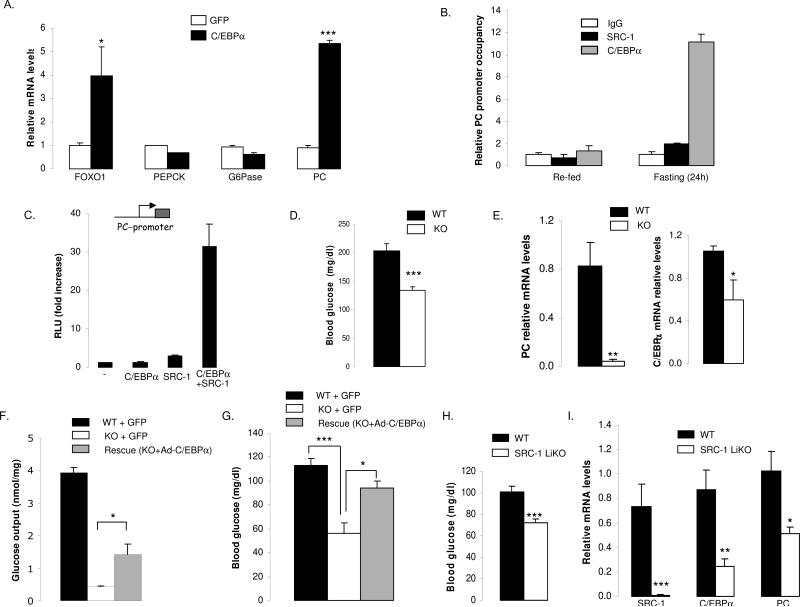
To identify the molecular targets of C/EBPα in the hepatic gluconeogenic program, we used recombinant adenoviruses to over-express it in primary hepatocytes. We first observed that increasing the mRNA levels of C/EBPα (Fig.S5A) affected the gene expression of FOXO1 (Fig.6A), confirming a tight functional cooperation between these two transcription factors in the liver as suggested in a previous study (Sekine et al., 2007). This result could explain why the FOXO1 mRNA levels dropped in SRC-1 KO consequent to a reduction of C/EBPα gene expression (Fig.5A). No effect was observed on the classical key enzymes of the gluconeogenic program such as PEPCK and G6Pase which seems surprising considering in vitro promoter studies previously performed on these genes, especially PEPCK (Park et al., 1993; Park et al., 1990; Roesler et al., 1998). However, this result mirrored the absence of altered PEPCK and G6Pase expression observed in liver specific C/EBPα KO mice (Inoue et al., 2004). Interestingly, we found a robust increase on the gene expression levels of PC (Fig.6A). Based on this result, we decided to study in more depth the transcriptional effect of C/EBPα on the PC gene. By using an in silico approach, we identified a potential C/EBP-response element in close proximity to the start site of the PC promoter. Chromatin-immunoprecipitation analysis revealed direct binding of both C/EBPα and SRC-1 to this region of the PC promoter and this specifically in fasting condition as no recruitment of these two proteins was found in the liver of re-fed animals (Fig.6B). Moreover, a transactivation assay using the PC promoter revealed extensive synergism between C/EBPα and SRC-1 (Fig.6C).
Figure 6. Glucose production impairment in SRC-1 KO hepatocytes is rescued by C/EBPα re-expression.
A) C/EBPα controls PC and FOXO1 gene expression but not other important known genes of the gluconeogenic pathway. Primary hepatocytes were treated with a C/EBPα adenovirus or empty virus (GFP) and used 48 hours later for measurement of gluconeogenic gene mRNA by qPCR.
B) In the mouse liver, C/EBPα and SRC-1 bind to the PC promoter in the region containing C/EBPα responsive elements specifically upon fasting. ChIP assays were performed with 100 bp amplicons flanking the region containing the C/EBPα motifs of the PC promoter. qPCR (normalized to input) was used to assess SRC-1 and C/EBPα occupancy of the PC promoter upon chromatin immunoprecipitation, using SRC-1 and C/EBPα specific antibodies.
C) The combination of C/EBPα and SRC-1 controls the PC promoter in a synergistic manner. Hela cells were transfected in the presence of the proximal promoter of the PC gene in presence of C/EBPα, SRC-1 or both expression vectors.
D) SRC-1 KO mice placed on a high protein diet are hypoglycemic. Blood glucose levels were determined in mice in the fed state (n = 5 mice), using a hand-held glucometer.
E) In SRC-1 KO mice placed on a high protein diet, PC and C/EBPα gene expression are decreased. PC and C/EBPα mRNA levels were measured by qPCR in the liver of KO and WT animals in the fed state (n=5 mice per group).
F) Acute overexpression of C/EBPα reactivates glucose production in primary hepatocytes from SRC-1 KO mice. Primary hepatocytes from SRC-1 KO mice and WT littermates were treated with C/EBPα or GFP adenoviruses and cultured for an additional 48 hours prior to measurement of glucose production over a period of 6 hours in the presence of forskolin and dexamethasone.
G) Re-expression of C/EBPα in liver rescued the glucose production in SRC-1 KO mice. Adenoviruses (GFP and C/EBPα) were injected via the tail vein of each mouse (n=5-6 for each group). Blood glucose was checked after 7 days of infection using a hand-held glucometer.
H) SRC-1 LiKO mice are hypoglycemic upon fasting. Blood glucose levels were determined in mice (n = 5 mice) in the fasted state (24h), using a hand-held glucometer. I) In SRC-1 liKO mice placed on fasting conditions, PC and C/EBPα gene expression are decreased. PC and C/EBPα mRNA levels were measured by qPCR in the liver of KO and WT animals (n=3-4 mice per group) upon fasting (24h).
Data are represented as mean ± SEM. Unpaired student's t-test was used for evaluation of statistical significance. One asterisk indicates p < 0.05; two asterisks indicate p < 0.01 and three asterisks indicate p < 0.005.
“See also Fig.S5”
To further investigate SRC-1 as a key regulator of the C/EBPα and the PC genes in vivo, we put KO and WT mice on a high protein diet, a diet known to activate the gluconeogenic pathway in vivo (Jungas et al., 1992). After three weeks on this diet, we found that SRC-1 KO mice were clearly hypoglycemic compared to WT animals (Fig.6D). In this context, PC mRNA and protein levels were both significantly reduced (Fig.6E+Fig.S5C), as well as the C/EBPα, PEPCK and FOXO1 mRNA levels (Fig.6E+Fig.S5B).
To validate the importance of C/EBPα in the control of the glucose production by SRC-1, we decided to acutely re-expressed C/EBPα in the context of primary hepatocytes and also specifically in the liver of SRC-1 KO mice using adenovirus approach. Importantly, we observed that the re-expression of C/EBPα alone was able to partially rescue the impaired glucose production in primary hepatocytes (Fig.6F), and in the liver (Fig.6G), therefore validating our model placing C/EBPα-PC axis in the center of the control of glucose production by SRC-1. It is noteworthy however to mention that re-expression of C/EBPα in vitro and in vivo did not restore glucose production to the level observed in WT cells most likely because (i) SRC-1 is necessary to the maximal action of C/EBPα as we showed previously in this study, and (ii) FBP1 gene, another key target of SRC-1 in the liver, is not under the control of C/EBPα (Fig.S5D) while still contributing to the impairment of glucose output observed in SRC-1 null hepatocytes.
Finally, to further substantiate the importance of the C/EBPα-PC axis in the control of hepatic gluconeogenesis by SRC-1, we recently were able to obtain a newly generated mouse model in which the SRC-1 coactivator was deleted specifically in the liver (SRC-1 LiKO). As a strong confirmation of our conclusions, these SRC-1 liver specific KO mice, fasted for 24h, revealed significant hypoglycemia compared to WT (Fig.6H); they also presented with a clear decrease in expression of the C/EBPα and PC genes (Fig.6I). Taken together, the totality of our data confirms the central role that the C/EBPα-PC axis plays in the control of glucose production by SRC-1 in the liver.
Gene expression of SRC-1 correlates with the expression levels of C/EBPα and PC during the fed-to-fasting transition in vivo
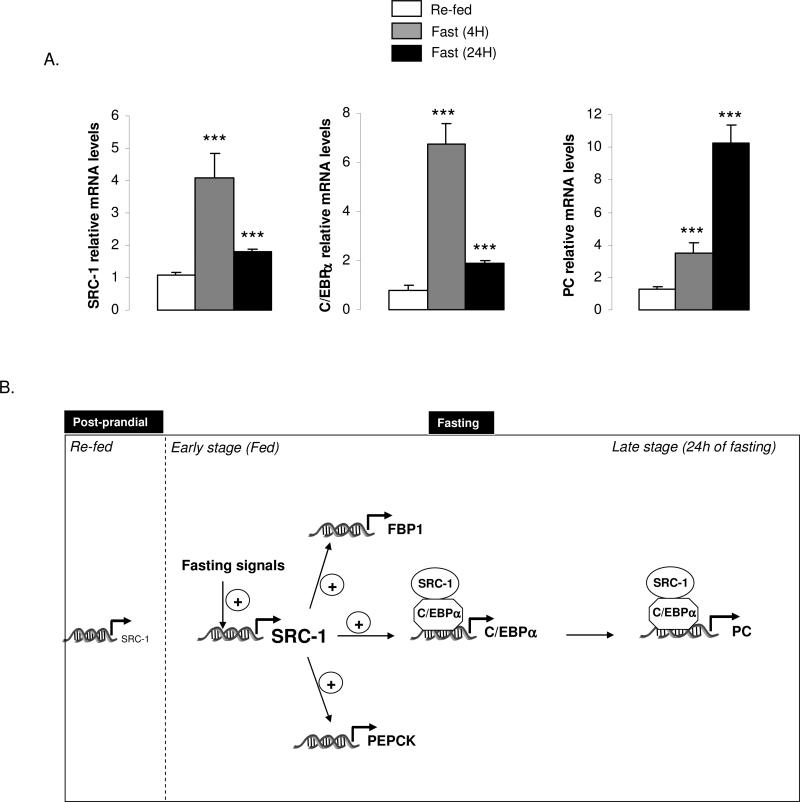
To further characterize the importance of SRC-1 on the hepatic glucose production during the fed-to-fasting condition in vivo, we investigated its kinetic of induction during key metabolic checkpoints which are re-fed, short (4h) and long (24h) fasting animals. Importantly, we found that SRC-1 gene expression is significantly activated after a short period of fasting (4h) and was maintained up-regulated during 24h (Fig.7A). This result which suggests an early role of this coactivator on the gluconeogenic pathway upon fasting, correlates well with our observation showing cAMP as a major fasting signal activating SRC-1 gene expression in primary hepatocytes with only a cooperative effect for glucocorticoids (Fig.4E). Moreover, by testing the gene expression of C/EBPα and PC in those same conditions, we found that C/EBPα was mainly activated during early fasting (4h) whereas PC reached its highest expression level only in late fasting (24h) (Fig7A). These observations combined with the totality of our data validated our proposed model.
Figure 7. Kinetics of induction of SRC-1, C/EBPα and PC during fed-to-fasting transition in liver.
A) SRC-1, C/EBPα and PC mRNA levels are increasing during early and late fasting in vivo. The gene expression of SRC-1, C/EBPα and PC was measured by qPCR in the liver of WT animals in the re-fed state (n=5 mice per group), and upon 4 hours (n= 5 mice per group) and 24 hours of fasting (n= 5 mice per group).
B) Schematic representation of the SRC-1 role on the gluconeogenic pathway in vivo. SRC-1 controls the gene expression of key enzymes involved in gluconeogenesis such as PEPCK, FBP1 and PC. During a short-term fasting (randomly fed status), SRC-1 controls the gene expression of PEPCK and FBP1. Upon a long-term fasting, after its own accumulation in the liver, SRC-1 functions to modulate the expression of C/EBPα through a feed-forward loop in which SRC-1 uses C/EBPα to transactivate PC. These sequential molecular events explain how SRC-1 could coordinate hepatic glucose production in fed and in fasting conditions.
“See also Fig.S6”
Finally, to analyze whether SRC-1 may have an impact in pathological situations, we checked the level of SRC-1 gene expression in db/db and ob/ob mice, two commonly studied animal models for insulin resistance and type 2 diabetes. These mice are strongly hyperglycemic compared to WT during fasting (Fig.S6A). Interestingly, we observed a significant reduction of the SRC-1 gene expression in these prediabetic models (Fig.S6B). These results suggests a key role of SRC-1 as an original sensor of glucose homeostasis as the gene expression of this coactivator is increased in fasting when the glucose and the insulin levels drop and is decreased in situations where production of glucose and insulin is ‘abnormally’ high. To validate this hypothesis, we checked whether the glucose and the insulin could together modulate the gene expression of SRC-1. Accordingly, we found that a high level of insulin combined with a high level of glucose significantly repressed the gene expression of SRC-1 (Fig.S6C).
Taken together, our data indicate that SRC-1 acts as a new coordinator of the gluconeogenic program in vivo. These functions are summarized in the model in Fig.7B. SRC-1 controls key rate limiting genes involved in the gluconeogenic program such as PEPCK, FBP1 and PC. At the molecular level, the SRC-1 gene is activated by fasting signals (glucagon and glucocorticoids). The main role of SRC-1 then is to modulate the expression and the activity of C/EBPα through a feed-forward loop in which the coactivator SRC-1 uses C/EBPα as the transcription factor to transactivate PC, the rate limiting gene for initiation of the gluconeogenic program. These sequential molecular events explain how PC is induced upon fasting in the liver and also introduce a new role for C/EBPα as an important transcription factor for initiation of expression of the PC gene.
DISCUSSION
The transcriptional coactivator SRC-1 was the first cloned nuclear receptor coactivator (Onate et al., 1995), but its physiologic roles in vivo remained unclear until recently (O'Malley et al., 2008). Our interest in this coregulator as a potential coordinator of metabolic processes emerged from a series of recent observations. SRC-1 appears to be an important player in global energy expenditure, as SRC-1 KO animals fed on a high fat diet are prone to develop obesity due to a net decrease in adaptive thermogenesis (Picard et al., 2002). Gene expression microarray analysis of liver target genes of SRC-1 revealed that this coactivator could control as many as 2% of the genes expressed in this tissue, placing SRC-1 as an important putative regulator of common hepatic functions (Jeong et al., 2006). A report of a genome-wide scan for genes with very strong positive selection during evolution of populations revealed that the SRC-1 locus presented the highest rate of genetic variation and selection in the ancestral African population (Voight et al., 2006). The fact that evolution mandated retention of a greater number of genetic modifications in the SRC-1 gene suggests that a major biological activity of this gene could be to mediate adaptations to environmental changes in nutrient supplies during population migrations. In support of this hypothesis, this same study found evidence for a positive genetic selection in the leptin receptor (Voight et al., 2006).
To unmask the in vivo impact of the SRC-1 coactivator in the fed-to-fasting transition, we used KO mice and analyzed the consequences of SRC-1 deletion on the expression of key metabolic regulatory genes. Our data suggested a primary role for SRC-1 in the regulation of the hepatic glucose production. We were able to exclude an active role for SRC-1 in the control of several pathways involved in the fed-to-fasting adaptation such as glycogenolysis, lipolysis and glycolysis. This result is of particular interest when juxtaposed with our recent parallel study showing that SRC-2 is a crucial modulator of the glycogenolysis pathway; SRC-2 KO animals mimic many of the clinical features of the human glycogen storage disease (type 1a) called Von Gierke's disease (Chopra et al., 2008). Taken together, these observations provide strong evidence highlighting the importance of this family of coregulators as crucial integrators of metabolic pathways, and underline the selective and non-redundant modes of action of the p160 family members in the control of specific metabolic programs.
Our present study revealed that one of the major roles of SRC-1 during fasting is to control the gene expression and the activity of C/EBPα. This transcription factor is known to be a key player in the hepatic glucose production as its genetic invalidation in mice resulted in a lethal hypoglycemia occurring a few hours after birth (Wang et al., 1995). Nevertheless, the exact mechanisms underlying the action of C/EBPα on the gluconeogenic pathway remained unclear. Indeed, in C/EBPα KO mice, G6Pase and PEPCK mRNA levels were found to be low just after birth and then increased to normal and remained high until death (Wang et al., 1995). The fact that blood glucose concentrations were dropping despite normal expression levels of PEPCK and G6Pase strongly suggested that C/EBPα is able to control other key players of the liver glucose production. Based on our finding, PC is likely one of these critical missing regulators of gluconeogenesis. Indeed, we suggest that C/EBPα via SRC-1 influences the gene expression of PC which acts as (i) a critical first step in the gluconeogenic program by providing oxaloacetate for subsequent conversion into phosphoenolpyruvate by PEPCK and acts as (ii) a carrier of reducing equivalents (NADH) from mitochondria to the cytoplasm. It is noteworthy that a recent study revealed that gluconeogenic flux was still observed despite the abrogation of 80 % of the PEPCK activity, suggesting that PEPCK could not be considered anymore as the primary control point of gluconeogenesis (Burgess et al., 2007). Clearly, this unexpected observation shed light on the potential role of other rate-limiting enzymes of glucose production such as PC, as suggested by the authors (Burgess et al., 2007). In fact the concept that PC could have greater control strength than ‘traditional’ rate-limiting enzyme such as PEPCK and G6Pase has been previously suggested (Groen et al., 1986). However, one of the main conclusions of this study is that the metabolic flux is under the control of the entire metabolic network rather than each individual enzyme. Consequently, modifications in the expression or activity of individual enzyme do not predictably impact flux through the intact pathway. Clearly, a coregulator such as SRC-1 which acts as a “master regulator” by controlling the expression of multiple enzymes of the same pathway is a strong candidate to fulfill this complex coordination of metabolic flux. Interestingly, important target genes of SRC-1 such as PC, FBP1 and C/EBPα are not under the control of PGC-1α suggesting that SRC-1 employs unique mechanisms to coordinate this set of genes in the liver and reinforcing the hypothesis that the mechanism of action of SRC-1 is distinct from that of PGC-1α.
Because of the clear defect observed in glucose production in SRC-1 KO mice, metabolic compensations were expected to occur in order to counter-balance this impairment as it has been reported in numerous other studies done in vivo (Lin et al., 2004; Rhee et al., 2003). Effectively, a net compensatory increase in β-oxidation flux (Fig.S2DE) and a decrease in glucose oxidation were seen in our mouse models (Fig.S2F). The fact that β-oxidation is known to play an important role in gluconeogenesis by providing NADH and ATP as energy sources, as well as acetyl-CoA which serves as an allosteric activator of PC enzyme activity (Newgard, 2004) support our conclusion. The decrease in glycolytic flux and glucose oxidation in SRC-1 -/- mice is suggests that these animals are striving to prevent lethal hypoglycemia during fasting.
In conclusion, we propose an original mode of action of SRC-1 as a central transcriptional player orchestrating the gluconeogenic program with only a few overlapping roles with the already known and important regulatory molecules in this pathway - PGC-1α, CRTC2 and SIRT-1. Clarification of the specific role for SRC-1 presents a more complete picture of the important metabolic coordination that exists among this group of coactivators and extends the emerging concept that select coregulators indeed are “master genes” with pleiotropic effects on several crucial physiologic programs. Importantly, a greater understanding of gluconeogenesis enhances our effort toward new therapeutic avenues for metabolic diseases such as type 2 diabetes.
MATERIALS AND METHODS
Mice
SRC-1 mice were maintained on a pure C57BL/6J background. SRC-3 knockout mice were maintained on a mixed background (C57BL/6J and SV129). SRC-1 F/F mice (Yamada et al., 2004) were crossed with Albumin-Cre (Postic and Magnuson, 2000) mice to generate liver-specific SRC-1 knockout mice (SRC-1 LiKO). We used 8-16 week old male mice and sex-matched WT for all in vivo studies. For the liver selective SRC-1 re-expression, 16 weeks old male mice were exposed to adenoviral mediated transgenesis (1011 virus particles per mouse), via tail-vein injections as previously described (Chopra et al., 2008). Mice were sacrificed 4 days after viral infusion and blood glucose and MS experiments were performed on fasted mice (16 hours). For the liver selective C/EBPα re-expression, 8 weeks old male mice were exposed to adenoviral mediated transgenesis (0.5×1010 virus particles per mouse), and sacrificed 7 days after viral infusion and blood glucose were performed on fasted (24h) mice. High protein diet (Harlan TD 90018) was provided to the mice during three weeks. The 8 weeks db/db and ob/ob mice and WT littermates were purchased from Harlan. The Baylor College of Medicine Institutional Animal Care and Utilization Committee approved all experiments.
Metabolic studies
We measured blood glucose using a hand-held glucometer (One Touch Ultra, Lifescan). Insulin levels were measured by ELISA (Mercodia), triglyceride by colorimetric assays (Roche), free fatty acids by colorimetric assays (Wako Pure Chemicals). Hepatic glycogen measurements have been described previously (Chopra et al., 2008). Glucose tolerance test (GTT), Insulin tolerance test (ITT) and indirect calorimetry were performed as described (Coste et al., 2008). Glucagon, corticosteroids, IGF-1 and GH were measured at the Hormone Assay & Analytical Services Core at the Vanderbilt University (http://hormone.mc.vanderbilt.edu). Glucose production using a tritiated glucose (Glucose, D-[3-3H]), lypolysis and physical activity were measured at the BCM Diabetes & Endocrinology Research Center (DERC) by the Mouse Metabolism Core (http://www.bcm.edu/diabetescenter/?PMID=9028) as described in (Chopra et al., 2008; Martinez-Botas et al., 2000). In vivo glucose production was performed as follows: a micro catheter was inserted into the jugular vein by survival surgery and we waited for 4-5 days for complete recovery. The study was then realized in conscious mice. Overnight-fasted conscious mice (16h) received a priming dose of HPLC-purified [3-3H] glucose (10μCi) and then a constant infusion (0.1μCi/min) of label glucose for ~90 minute. Blood samples were collected from the tail vein at 0, 50, 60, 75 and 90 min to measure the basal glucose production rate. Steady states were reached within one hour of infusion.
Metabolites profiling studies
Acylcarnitines and amino acids were analyzed in plasma of fed and fasted mice. The same metabolites and a panel of organic acids were also measured in liver extracts of fed and fasted mice. Methods of tissue handling and extraction have been described previously (Ferrara et al., 2008; Ronnebaum et al., 2006). Acylcarnitines and amino acids were analyzed by tandem mass spectrometry (MS/MS) as described (An et al., 2004; Ferrara et al., 2008; Newgard et al., 2009; Ronnebaum et al., 2006). All MS analyses employed stable-isotope-dilution with internal standards from Isotec (St. Louis, MO), Cambridge Isotope Laboratories (Andover, MA), and CDN Isotopes (Pointe-Claire, Quebec, CN). A list of all internal standards utilized in these studies has been published (Ferrara et al., 2008; Newgard et al., 2009).
Flux measurements
After isolation hepatocytes were incubated for 24 h in DMEM containing 5.5 mM glucose (Invitrogen), 1 X BSA/oleate-palmitate mixture (1%BSA, 0.5 mM oleate-palmitate (oleate:palmitate 2:1)), 10 nM Dexamethsone, and 1mM carnitine. Prior to the fuel oxidation assays, the cells were changed to a balanced oxidation medium containing 1 mM carnitine and 12.5 mM HEPES, and 1-3 μCi/ml 14C-labeled tracer. U-[14C]glucose, and 1-[14C]oleate were purchased from Amersham. 200 μl 1N NaOH was added into a center well that was carefully set into the culture well. The culture plate was then sealed with a microplate adhesive film and incubated at 37 °C for 3 h. 100-200 μl 70% perchloric acid was injected into the culture well and the plate was sealed with another layer of the microplate adhesive film and incubated at room temperature on a shaker for 1 h at 50 rpm. The NaOH in the center well was then taken into a scintillation vial and mixed with scintillation fluid for the analysis of 14C. The remaining acid-soluble metabolites (ASM) were collected into an eppendorf tube and kept at room temperature overnight. After centrifuged at 10000g for 5 min, approximately 200 μl of supernatant were used for scintillation counting. A parallel set of hepatocytes was analyzed for total protein content and used for normalization.
RNA analysis
We used standard RNA extraction procedures (RNeasy Mini Kit from Qiagen). Reverse transcription was carried out using the Superscript III kit (Invitrogen). For gene expression analysis, qPCR was performed using sequence-specific primers and probes from Roche (Universal Probe Library). GAPDH was used as an internal control for all gene-expression assays. All qPCR primers are available upon request.
Chromatin Immunoprecipitation
In vivo ChIP was performed by using liver tissue from mice placed at different key metabolic status (re-fed, post-absorptive (fed) and fasting (24h)) . Formaldehyde (1%) was added to produce cross linking during 10 minutes at room temperature. The rest of the ChIP procedure was performed using the EZ ChIP kit (Millipore) following the manufacturer's protocol. SRC-1 antibody is from Milipore and C/EBPα antibody is from SantaCruz. qPCR for ChIP was performed using the Sybr-Green technology (Applied Biosystems) using sequence specific primers. Results were normalized to input in each case. Primer sequences are available upon request.
Cell culture, overexpression and transient transfection experiments
Primary mouse hepatocytes were isolated from 8- to 12-week-old mice as described previously (Chopra et al., 2008). Cells were incubated overnight in Williams E media (Invitrogen) containing 10% FBS and dexamethasone 10-8M before each experiment for attachment. For adenoviral transduction experiments, SRC-1, PGC-1α and C/EBPα adenoviruses were used as described (Lerin et al., 2006; Martinez-Jimenez et al., 2006; Wang et al., 2004). RNA and protein isolation or glucose production assays were carried out 48 hours after treatment. For transfection experiments, HeLa cells were transfected with several reporter-gene plasmids containing either the native C/EBPα promoter (Wang et al., 2004), or a construction with specific mutants of C/EBP-RE sequences within the C/EBPα promoter realized with a site-directed mutagenesis kit (Agilent Technologies), or the -600/+300 bp PC proximal promoter cloned in the pGL-3 plasmid after PCR amplification from liver genomic DNA (Biochain). We also used pSG5-SRC-1-Flag and different mutants of SRC-1: ΔHAT (deletion of the histone acetylase domain); ΔAD2 (deletion of the activating domain 2) and ΔAD2+NR (deletion of the activating domain 2+ the nuclear receptor domain). These constructs were cotransfected as indicated in each experiment by different expression vectors encoding SRC-1 in PCR3.1 vector (Invitrogen) as well as C/EBPα that have been already described in (Louet et al., 2006) . Reporter-gene levels were determined 48 hours after transfection using Promega reagent.
Co-immunoprecipitation assays and western blot
For co-immunoprecipitation, 293/T cells were transiently transfected with Flag-SRC-1 and pSV-SPORT1-C/EBPα. Then, Hela cell lysates were incubated with anti- Flag (Invitrogen) antibody. The immune complexes were eluted and subjected to SDS-PAGE. The immunoblot detection was done using anti-C/EBPα antibody (Santa Cruz). For western blot, we used the SRC-1 antibody from Santa Cruz. Co-immunoprecipitation in the liver was performed by using tissue from fasting (16h) SRC-1 WT mice.
In-Vitro glucose production assay
Primary hepatocytes were cultured in Williams E media (Invitrogen) supplemented with 10% FBS and measured glucose production in glucose and phenol red-free DMEM (pH 7.4) supplemented with sodium lactate and pyruvate using a colorimetric assay (Biovision).
Statistical analysis
All results are presented as mean ± SEM. P value was calculated by unpaired Student's t test.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Pere Puigserver, Nikolai Timchenko and Ramiro Jover for respectively the PGC-1α, C/EBPα and the SRC-1 adenovirus constructs. Adenoviral vectors were produced by the Gene Vector Core in the Diabetes and Endocrinology Research Center at Baylor College of Medicine. We would like to thank also, Mao Jianqiang for the liver tissue specific SRC-1 KO and WT animals, Rainer Lanz for help with qPCR setup and analysis and Larbi Amazit for providing pSG5-SRC-1 WT and pSG5-SRC-1 mutant constructions. This work was supported by NIH grants 1-PO1-DK59820, HD07857, NURSA/DK62434 and PO1DK58398. J.V. Sagen is funded by grants from the Norwegian Cancer Society, University of Bergen, Det regionale samarbeidsorganet (Helse Vest RHF and University of Bergen), and the Norwegian Society of Endocrinology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Koves TR, Stevens R, Millington D, Newgard CB. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat Med. 2004;10:268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- Burgess SC, He T, Yan Z, Lindner J, Sherry AD, Malloy CR, Browning JD, Magnuson MA. Cytosolic phosphoenolpyruvate carboxykinase does not solely control the rate of hepatic gluconeogenesis in the intact mouse liver. Cell metabolism. 2007;5:313–320. doi: 10.1016/j.cmet.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill GF., Jr. Fuel metabolism in starvation. Annu Rev Nutr. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra AR, Louet JF, Saha P, An J, Demayo F, Xu J, York B, Karpen S, Finegold M, Moore D, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke's disease. Science. 2008;322:1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A, Louet JF, Lagouge M, Lerin C, Antal MC, Meziane H, Schoonjans K, Puigserver P, O'Malley BW, Auwerx J. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1{alpha}. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17187–17192. doi: 10.1073/pnas.0808207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington GJ, Wang N, Hanson RW. C/EBP alpha: a critical regulator of genes governing integrative metabolic processes. Curr Opin Genet Dev. 1995;5:565–570. doi: 10.1016/0959-437x(95)80024-7. [DOI] [PubMed] [Google Scholar]
- Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell metabolism. 2008;8:186–200. doi: 10.1016/j.cmet.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S. Control of mitochondrial beta-oxidation flux. Prog Lipid Res. 2002;41:197–239. doi: 10.1016/s0163-7827(01)00024-8. [DOI] [PubMed] [Google Scholar]
- Ferrara CT, Wang P, Neto EC, Stevens RD, Bain JR, Wenner BR, Ilkayeva OR, Keller MP, Blasiole DA, Kendziorski C, et al. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet. 2008;4:e1000034. doi: 10.1371/journal.pgen.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster DW. The role of the carnitine system in human metabolism. Ann N Y Acad Sci. 2004;1033:1–16. doi: 10.1196/annals.1320.001. [DOI] [PubMed] [Google Scholar]
- Garcia-Cazorla A, Rabier D, Touati G, Chadefaux-Vekemans B, Marsac C, de Lonlay P, Saudubray JM. Pyruvate carboxylase deficiency: metabolic characteristics and new neurological aspects. Ann Neurol. 2006;59:121–127. doi: 10.1002/ana.20709. [DOI] [PubMed] [Google Scholar]
- Granner D, Pilkis S. The genes of hepatic glucose metabolism. The Journal of biological chemistry. 1990;265:10173–10176. [PubMed] [Google Scholar]
- Groen AK, van Roermund CW, Vervoorn RC, Tager JM. Control of gluconeogenesis in rat liver cells. Flux control coefficients of the enzymes in the gluconeogenic pathway in the absence and presence of glucagon. The Biochemical journal. 1986;237:379–389. doi: 10.1042/bj2370379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Inoue J, Lambert G, Yim SH, Gonzalez FJ. Disruption of hepatic C/EBPalpha results in impaired glucose tolerance and age-dependent hepatosteatosis. The Journal of biological chemistry. 2004;279:44740–44748. doi: 10.1074/jbc.M405177200. [DOI] [PubMed] [Google Scholar]
- Jeong JW, Kwak I, Lee KY, White LD, Wang XP, Brunicardi FC, O'Malley BW, DeMayo FJ. The genomic analysis of the impact of steroid receptor coactivators ablation on hepatic metabolism. Molecular endocrinology (Baltimore, Md. 2006;20:1138–1152. doi: 10.1210/me.2005-0407. [DOI] [PubMed] [Google Scholar]
- Jungas RL, Halperin ML, Brosnan JT. Quantitative analysis of amino acid oxidation and related gluconeogenesis in humans. Physiol Rev. 1992;72:419–448. doi: 10.1152/physrev.1992.72.2.419. [DOI] [PubMed] [Google Scholar]
- Le Lay J, Tuteja G, White P, Dhir R, Ahima R, Kaestner KH. CRTC2 (TORC2) contributes to the transcriptional response to fasting in the liver but is not required for the maintenance of glucose homeostasis. Cell metabolism. 2009;10:55–62. doi: 10.1016/j.cmet.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:672–687. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell metabolism. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Louet JF, Coste A, Amazit L, Tannour-Louet M, Wu RC, Tsai SY, Tsai MJ, Auwerx J, O'Malley BW. Oncogenic steroid receptor coactivator-3 is a key regulator of the white adipogenic program. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17868–17873. doi: 10.1073/pnas.0608711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louet JF, O'Malley BW. Coregulators in adipogenesis: what could we learn from the SRC (p160) coactivator family? Cell Cycle. 2007;6:2448–2452. doi: 10.4161/cc.6.20.4777. [DOI] [PubMed] [Google Scholar]
- Magnuson MA, She P, Shiota M. Gene-altered mice and metabolic flux control. The Journal of biological chemistry. 2003;278:32485–32488. doi: 10.1074/jbc.R300020200. [DOI] [PubMed] [Google Scholar]
- Martinez-Jimenez CP, Gomez-Lechon MJ, Castell JV, Jover R. Underexpressed coactivators PGC1alpha and SRC1 impair hepatocyte nuclear factor 4 alpha function and promote dedifferentiation in human hepatoma cells. The Journal of biological chemistry. 2006;281:29840–29849. doi: 10.1074/jbc.M604046200. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell metabolism. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Newgard CB. Regulation of Glucose Metabolism in the Liver. In: DeFronzo RA, Ferrannini E, Kenn H, Zimmet P, editors. International Textbook of Diabetes. Third Edition John Wiley & Sons; Chichester: 2004. pp. 253–275. [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell metabolism. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley BW, Qin J, Lanz RB. Cracking the coregulator codes. Curr Opin Cell Biol. 2008;20:310–315. doi: 10.1016/j.ceb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Park EA, Gurney AL, Nizielski SE, Hakimi P, Cao Z, Moorman A, Hanson RW. Relative roles of CCAAT/enhancer-binding protein beta and cAMP regulatory element-binding protein in controlling transcription of the gene for phosphoenolpyruvate carboxykinase (GTP). The Journal of biological chemistry. 1993;268:613–619. [PubMed] [Google Scholar]
- Park EA, Roesler WJ, Liu J, Klemm DJ, Gurney AL, Thatcher JD, Shuman J, Friedman A, Hanson RW. The role of the CCAAT/enhancer-binding protein in the transcriptional regulation of the gene for phosphoenolpyruvate carboxykinase (GTP). Molecular and cellular biology. 1990;10:6264–6272. doi: 10.1128/mcb.10.12.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Gehin M, Annicotte J, Rocchi S, Champy MF, O'Malley BW, Chambon P, Auwerx J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden M, Petersen KF, Shulman GI. Nuclear magnetic resonance studies of hepatic glucose metabolism in humans. Recent Prog Horm Res. 2001;56:219–237. doi: 10.1210/rp.56.1.219. [DOI] [PubMed] [Google Scholar]
- Roesler WJ, Park EA, McFie PJ. Characterization of CCAAT/enhancer-binding protein alpha as a cyclic AMP-responsive nuclear regulator. The Journal of biological chemistry. 1998;273:14950–14957. doi: 10.1074/jbc.273.24.14950. [DOI] [PubMed] [Google Scholar]
- Ronnebaum SM, Ilkayeva O, Burgess SC, Joseph JW, Lu D, Stevens RD, Becker TC, Sherry AD, Newgard CB, Jensen MV. A pyruvate cycling pathway involving cytosolic NADP-dependent isocitrate dehydrogenase regulates glucose-stimulated insulin secretion. The Journal of biological chemistry. 2006;281:30593–30602. doi: 10.1074/jbc.M511908200. [DOI] [PubMed] [Google Scholar]
- Sekine K, Chen YR, Kojima N, Ogata K, Fukamizu A, Miyajima A. Foxo1 links insulin signaling to C/EBPalpha and regulates gluconeogenesis during liver development. Embo J. 2007;26:3607–3615. doi: 10.1038/sj.emboj.7601784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey KB, Storey JM. Tribute to P. L. Lutz: putting life on ‘pause’--molecular regulation of hypometabolism. J Exp Biol. 2007;210:1700–1714. doi: 10.1242/jeb.02716. [DOI] [PubMed] [Google Scholar]
- Timchenko N, Wilson DR, Taylor LR, Abdelsayed S, Wilde M, Sawadogo M, Darlington GJ. Autoregulation of the human C/EBP alpha gene by stimulation of upstream stimulatory factor binding. Molecular and cellular biology. 1995;15:1192–1202. doi: 10.1128/mcb.15.3.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Iakova P, Wilde M, Awad S, Timchenko NA. Liver tumors escape negative control of proliferation via PI3K/Akt-mediated block of C/EBP alpha growth inhibitory activity. Genes Dev. 2004;18:912–925. doi: 10.1101/gad.1183304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- Yamada T, Kawano H, Sekine K, Matsumoto T, Fukuda T, Azuma Y, Itaka K, Chung UI, Chambon P, Nakamura K, et al. SRC-1 is necessary for skeletal responses to sex hormones in both males and females. J Bone Miner Res. 2004;19:1452–1461. doi: 10.1359/JBMR.040515. [DOI] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.