Abstract
The mTORC1 kinase promotes growth in response to growth factors, energy levels, and amino acids and its activity is often deregulated in disease. The Rag GTPases interact with mTORC1 and are proposed to activate it in response to amino acids by promoting mTORC1 translocation to a membrane-bound compartment that contains the mTORC1 activator Rheb. We show that amino acids induce the movement of mTORC1 to lysosomal membranes, where the Rag proteins reside. A complex encoded by the MAPKSP1, ROBLD3, and c11orf59 genes, which we term Ragulator, interacts with the Rag GTPases, recruits them to lysosomes, and is essential for mTORC1 activation. Constitutive targeting of mTORC1 to the lysosomal surface is sufficient to render the mTORC1 pathway amino acid insensitive and independent of Rag and Ragulator, but not Rheb, function. Thus, Rag-Ragulator mediated translocation of mTORC1 to lysosomal membranes is the key event in amino acid signaling to mTORC1.
Introduction
The multi-component kinase mTORC1 (mammalian target of rapamycin complex 1) regulates cell growth by coordinating upstream signals from growth factors, intracellular energy levels, and amino acid availability, and is deregulated in diseases such as cancer and diabetes (reviewed in (Guertin and Sabatini 2007)). The TSC1 and TSC2 proteins form a tumor suppressor complex that transmits growth factor and energy signals to mTORC1 by regulating the GTP-loading state of Rheb, a Ras-related GTP-binding protein. When bound to GTP, Rheb interacts with and activates mTORC1 (reviewed in (Laplante and Sabatini, 2009)) and appears to be necessary for the activation of mTORC1 by all signals, including amino acid availability. In contrast, TSC1-TSC2 is dispensable for the regulation of mTORC1 by amino acids and, in cells lacking TSC2, the mTORC1 pathway is sensitive to amino acid starvation but resistant to growth factor withdrawal (Roccio et al., 2006; Smith et al., 2005).
Recently, the Rag GTPases, which are also members of the Ras-family of GTP-binding proteins, were shown to be amino acid-specific regulators of the mTORC1 pathway (Kim et al., 2008; Sancak et al., 2008). Mammals express four Rag proteins—RagA, RagB, RagC, and RagD—that form heterodimers consisting of RagA or RagB with RagC or RagD. RagA and RagB, like RagC and RagD, are highly similar to each other and are functionally redundant (Hirose et al., 1998; Sancak et al., 2008; Schurmann et al., 1995; Sekiguchi et al., 2001). Rag heterodimers containing GTP-bound RagB interact with mTORC1, and amino acids induce the mTORC1-Rag interaction by promoting the loading of RagB with GTP, which enables it to directly interact with the raptor component of mTORC1 (Sancak et al., 2008). The activation of the mTORC1 pathway by amino acids correlates with the movement of mTORC1 from an undefined location to a compartment containing Rab7 (Sancak et al., 2008), a marker of both late endosomes and lysosomes (Chavrier et al., 1990; Luzio et al., 2007). How the Rag proteins regulate mTORC1 is unknown, but, in cells expressing a RagB mutant that is constitutively bound to GTP (RagBGTP), the mTORC1 pathway is insensitive to amino acid starvation and mTORC1 resides in the Rab7-positive compartment even in the absence of amino acids (Sancak et al., 2008). We previously proposed that amino acids promote the translocation of mTORC1—in a Rag-dependent fashion—to the surface of an endomembrane compartment where mTORC1 can find its well-known activator Rheb. Here, we show that the lysosomal surface is the compartment where the Rag proteins reside and to which mTORC1 moves in response to amino acids. We identify the trimeric Ragulator protein complex as a new component of the mTORC1 pathway that interacts with the Rag GTPases, is essential for localizing them and mTORC1 to the lysosomal surface, and is necessary for the activation of the mTORC1 pathway by amino acids. In addition, by expressing in cells a modified raptor protein that targets mTORC1 to the lysosomal surface, we provide evidence that supports our model of mTORC1 pathway activation by amino acids.
Results
Amino acids cause the translocation of mTORC1 to lysosomal membranes, where the Rag GTPases are already present
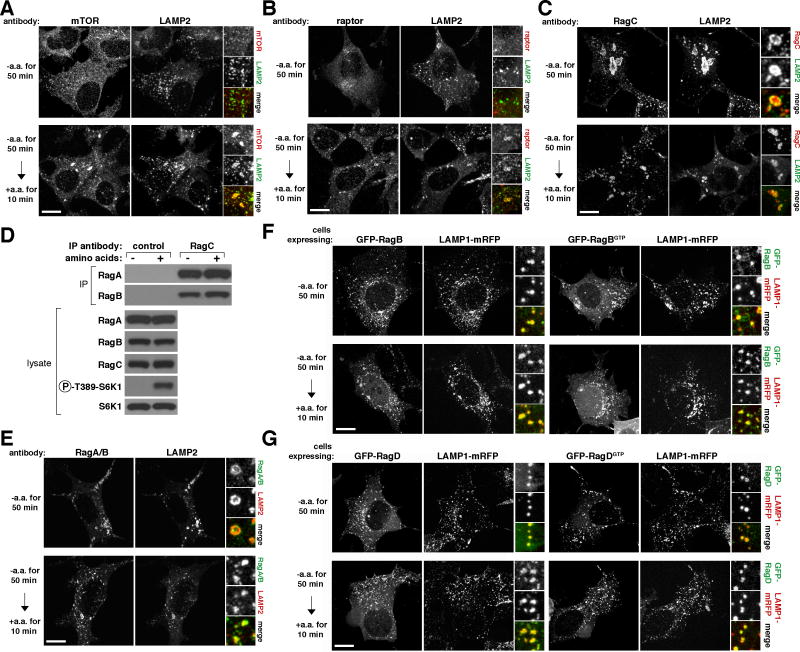
To better define the compartment to which mTORC1 moves upon amino acid stimulation, we co-stained human cells with antibodies to endogenous mTOR, raptor, or RagC as well as to various endomembrane markers (data not shown). This revealed that in the presence, but not in the absence, of amino acids mTOR and raptor co-localized with LAMP2 (Figures 1A and 1B), a well-characterized lysosomal marker (reviewed in (Eskelinen, 2006)). Amino acid stimulation also resulted in an appreciable increase in the average size of lysosomes, which, as determined by live cell imaging, was most likely caused by lysosome-lysosome fusion (R.Z., unpublished results). The amino acid-induced movement of mTOR to the LAMP2-positive compartment depends on the Rag GTPases as it was eliminated by the RNAi mediated co-knockdown of RagA and RagB (Figure S1A and S1B). Endogenous RagC also co-localized extensively with LAMP2, but, unlike mTORC1, this co-localization was unaffected by amino acid availability (Figure 1C). Consistent with amino acids not regulating the interaction between RagC and RagA or RagB (Figure 1D), an antibody that recognizes RagA and RagB stained lysosomes in both amino acid-starved and replete cells (Figure 1E). Lastly, GFP-tagged wild-type and GTP-bound mutants of RagB (RagBGTP) and RagD (RagDGTP) behaved identically to their endogenous counterparts (Figures 1F and 1G). Thus, amino acids stimulate the translocation of mTORC1 to the lysosomal surface, where the Rag GTPases reside irrespective of their GTP-loaded states or amino acid availability. Given that mTORC1 interacts with the Rag heterodimers in an amino acid-dependent fashion (Sancak et al., 2008), the mTORC1 and Rag localization data are consistent with the Rag GTPases serving as an amino acid-regulated docking site for mTORC1 on lysosomes.
Figure 1. mTORC1 localizes to lysosomal membranes in an amino acid-dependent fashion while the Rag GTPases are constitutively localized to the same compartment.
(A) Images of HEK-293T cells co-immunostained for lysosomal protein LAMP2 (green) and mTOR (red). Cells were starved of and restimulated with amino acids for the indicated times before processing and imaging.
(B) Images of HEK-293T cells co-immunostained for LAMP2 (green) and raptor (red) Cells were treated and processed as in (A).
(C) Images of HEK-293T cells co-immunostained for LAMP2 (green) and RagC (red). Cells were treated and processed as in (A).
(D) RagC interacts with RagA and RagB independently of amino acid availability. RagC-immunoprecipitates were prepared from HEK-293T cells starved or stimulated with amino acids as in (A), and immunoprecipitates and lysates were analyzed by immunoblotting for the indicated proteins.
(E) Images of HEK-293T cells co-immunostained for RagA/B (green) and LAMP2 (red). Cells were treated, processed, and imaged as in (A).
(F) GFP-RagB and GFP-RagBGTP co-localize with co-expressed LAMP1-mRFP independently of amino acid availability. HEK-293T cells transfected with the indicated cDNAs were treated and processed as in (A).
(G) GFP-RagD and GFP-RagDGTP co-localize with co-expressed LAMP1-mRFP independently of amino acid availability. HEK-293T cells transfected with the indicated cDNAs were treated and processed as in (A). In all images, insets show selected fields that were magnified five times and their overlays. Scale bar is 10 μm.
See also Fig S1.
The translocation of mTORC1 to lysosomes does not depend on growth factors, Rheb, or mTORC1 activity
The movement of mTORC1 to lysosomes is a specific response to amino acids. In wild-type mouse embryonic fibroblasts (MEFs), amino acids promoted the translocation of mTORC1 to lysosomes even when cells were cultured in the absence of serum (Figure S1C), a condition in which mTORC1 signaling, as detected by phosphorylated S6K1, is not active (Figure S1D). Conversely, in the absence of amino acids, neither serum stimulation nor constitutive activation of Rheb caused by the loss of TSC2, led to the lysosomal translocation of mTORC1 (Figure S1C). In both wild-type and TSC2-null MEFs, RNAi-mediated suppression of Rheb1 expression inhibited mTORC1 activation by amino acids (Figure S1E), but did not interfere with the amino acid-induced movement of mTOR to lysosomes (Figure S1F). Thus, the amino acid-induced translocation of mTORC1 to the lysosomal surface occurs independently of mTORC1 activity and does not require TSC2, Rheb, or growth factors.
The trimeric Ragulator complex interacts with the Rag GTPases and co-localizes with them on lysosomal membranes
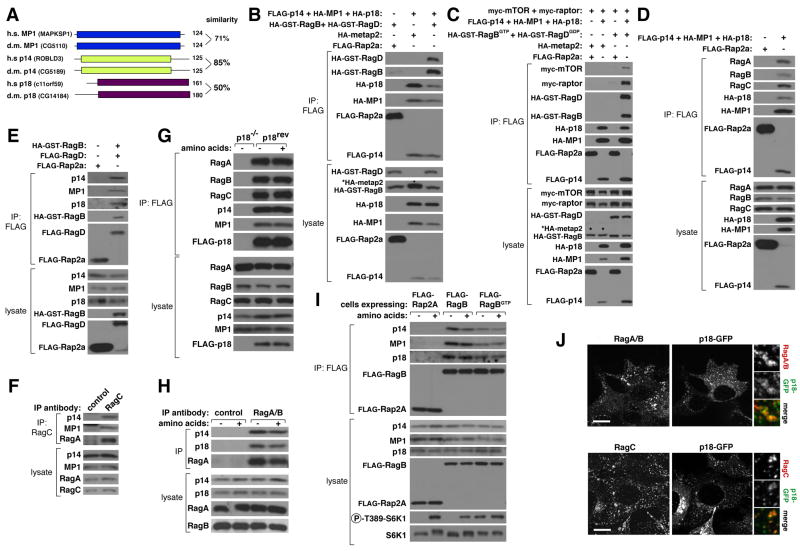
Inspection of the amino acid sequence of the Rag GTPases did not reveal any obvious lipid modification signals that might mediate Rag recruitment to lysosomal membranes. Thus, we pursued the possibility that unknown Rag-interacting proteins are needed to localize the Rag GTPases to lysosomes and play a role in mTORC1 signaling. To identify such proteins we used protein purification approaches that have led to the discovery of other mTOR pathway components (see supp. methods). Mass spectrometric analysis of anti-FLAG immunoprecipitates prepared from human HEK-293T cells stably expressing FLAG-RagB or FLAG-RagD, but not FLAG-Rap2a, consistently revealed the presence of proteins encoded by the MAPKSP1, ROBLD3, and c11orf59 genes (Figure 2A). Furthermore, the same proteins were also detected in immunoprecipitates of endogenous RagC but not control proteins like p53 or tubulin. Previous work indicates that these three small proteins interact with each other, localize to endosomes and lysosomes, and play positive roles in the MAPK pathway (Lunin et al., 2004; Nada et al., 2009; Schaeffer et al., 1998; Teis et al., 2006; Teis et al., 2002; Wunderlich et al., 2001). The proteins encoded by MAPKSP1, ROBLD3, and c11orf59 have been called MP1, p14, and p18, respectively, and we use these names throughout this study. For convenience and because MP1, p14, and p18 are Rag and mTORC1 regulators (see below) we refer to the trimeric complex as the ‘Ragulator’.
Figure 2. The trimeric Ragulator complex interacts and co-localizes with the Rag GTPases.
(A) Schematic amino acid sequence alignment of human MP1, p14, and p18 and their corresponding Drosophila orthologs.
(B) Recombinant epitope-tagged Ragulator co-immunoprecipitates recombinant RagB and RagD. Anti-FLAG immunoprecipitates were prepared from HEK-293T cells co-transfected with the indicated cDNAs in expression vectors and cell lysates and immunoprecipitates analyzed by immunoblotting for levels of indicated proteins. The * indicates the band corresponding to the metap2 protein as it has the same apparent molecular weight as HA-GST-RagB.
(C) Recombinant Ragulator co-immunoprecipitates mTORC1 when it is co-expressed with the GTP-bound mutant of RagB. HEK-293T cells were co-transfected with the indicated cDNAs in expression vectors and analyzed as in (B). The * indicates the bands corresponding to metap2 as it has the same apparent molecular weight as HA-GST-RagB.
(D) Recombinant Ragulator co-immunoprecipitates endogenous RagA, RagB, and RagC. HEK-293T cells were co-transfected with indicated cDNAs in expression vectors and anti-FLAG immunoprecipitates analyzed as in (B).
(E) Recombinant RagB-RagD heterodimers co-immunoprecipitate endogenous p14, MP1, and p18. HEK-293T cells were co-transfected with indicated cDNAs in expression vectors and anti-FLAG immunoprecipitates analyzed as in (B).
(F) Endogenous RagC co-immunoprecipitates endogenous p14 and MP1. Anti-RagC immunoprecipitates were prepared from HEK-293T cells and analyzed for the levels of the indicated proteins.
(G) Amino acids do not regulate the amounts of endogenous MP1, p14, RagA, or RagB that co-immunoprecipitate with recombinant p18. p18-null cells (p18−/−) or p18-null cells stably expressing FLAG-p18 (p18rev) were starved for amino acids for 50 min or starved and restimulated with amino acids for 10 min. After in-cell cross-linking, anti-FLAG immunoprecipitates were prepared from cell lysates and analyzed for the levels of the indicated proteins by immunoblotting.
(H) Amino acids do not affect the amounts of endogenous p14 and p18 that co-immunoprecipitate with endogenous RagA/B. HEK-293T cells were treated as in (G) and anti-RagA/B immunoprecipitates analyzed by immunoblotting for the indicated proteins.
(I) Endogenous Ragulator co-immunoprecipitates with FLAG-RagB independently of amino acid availability and GTP-loading of RagB. HEK-293T cells stably expressing FLAG-RagB or FLAG-RagBGTP were starved and restimulated with amino acids as in (G) and anti-FLAG immunoprecipitates analyzed for the levels of indicated proteins.
(J) The Rag GTPases co-localize with GFP-tagged p18. HEK-293T cells were transfected with a cDNA encoding p18-GFP, processed for immunostaining for endogenous RagA/B or RagC, and imaged for the RagA/B (red) or RagC (red) signal as well as for p18-GFP fluorescence (green). Note: not all cells express p18-GFP. In all images, insets show selected fields that were magnified five times and their overlays. Scale bar is 10 μm.
See also Fig S2.
Orthologues of MP1, p14, and p18 are readily detectable in vertebrates as well as in Drosophila (Figure 2A), but extensive database searches did not reveal any potential orthologues in budding or fission yeast. The amino acid sequences of MP1, p14, and p18 reveal little about their function and other than p14, which has a roadblock domain of unknown function (Koonin and Aravind, 2000), the proteins do not share sequence homology amongst themselves or with any other proteins in the databases besides their direct orthologues. In particular, they do not share any sequence similarity with the Ego1p or Ego3p, proteins, which interact with Gtr1p and Gtr2p (Dubouloz et al., 2005; Gao and Kaiser, 2006), the orthologues of the Rag proteins in budding yeast (Gao and Kaiser, 2006; Schurmann et al., 1995). The lysosomal localization of p18 requires its lipidation through N-terminal myristoylation and palmitoylation sites and p18 likely serves as a platform for keeping MP1 and p14 on the lysosomal surface (Nada et al., 2009).
In humans a mutation that leads to a partial reduction in the expression of p14 causes a pronounced growth defect so that individuals carrying the mutation are below the third percentile in age-adjusted height (Bohn et al., 2006). Furthermore, mice engineered to lack either p14 or p18 die around embryonic day 7–8 and exhibit severe growth retardation (Nada et al., 2009; Teis et al., 2006). Given the major role of the mTORC1 pathway in growth control, these loss of function phenotypes were of interest to us.
As an initial step in verifying our mass spectrometric identification of MP1, p14, and p18 as Rag-interacting proteins, we co-expressed them along with RagB and RagD in HEK-293T cells and found that the Ragulator, but not the control Rap2A protein, co-immunoprecipitated both Rag GTPases but not the metap2 protein that has the same molecular weight as tagged RagB (Figure 2B). Furthermore, when co-expressed with a RagB mutant (RagBGTP) that binds constitutively to GTP, the Ragulator co-immunoprecipitated the mTORC1 components raptor and mTOR (Figure 2C), consistent with the GTP-loading of RagB promoting the interaction of the Rag heterodimers with mTORC1 (Sancak et al., 2008). Furthermore, endogenous RagA, RagB, and RagC co-purified with recombinant Ragulator (Figure 2D) and endogenous Ragulator components co-purified with the recombinant RagB-RagD heterodimer (Figure 2E). Lastly, endogenous p14 and MP1 were present in immunoprecipitates prepared with an antibody directed against endogenous RagC that readily co-immunoprecipitates RagA (Figure 2F).
Amino acids did not appreciably regulate the interaction of recombinant p18 with endogenous p14, MP1 or the Rag GTPases (Figure 2G). Similarly, amino acids did not affect the interaction of endogenous Ragulator with endogenous Rag A/B (Figure 2H). The amounts of p14, p18, and MP1 that co-immunoprecipitated with the GTP-bound RagB mutant (RagBGTP) were slightly less than with wild-type RagB (Figure 2I). Because mTORC1 pathway activity is high in cells expressing RagBGTP (Sancak et al., 2008) the reduced Ragulator-Rag interaction in these cells may reflect a compensatory mechanism to reduce mTORC1 activity. To test if the Rag GTPases interact with one or more Ragulator components directly, we performed in vitro binding assays between purified RagB-RagD heterodimers and individual Ragulator proteins. p18 interacted with RagB-RagD in vitro, but not with the Rap2a control protein (Figure S2A). In contrast, we did not detect a direct interaction between either p14 or MP1 and the Rag GTPases (data not shown), suggesting that p18 is the principal Rag-binding subunit of the Ragulator. Lastly, within HEK-293T cells, GFP-tagged p18 co-localized with endogenous RagA/B and RagC (Figure 2J). Collectively, these results show that the Ragulator interacts with the Rag GTPases and that a super-complex consisting of Ragulator, a Rag heterodimer, and mTORC1 can exist within cells.
Ragulator localizes the Rag proteins to the lysosomal surface and is necessary for the amino acid-dependent recruitment of mTORC1
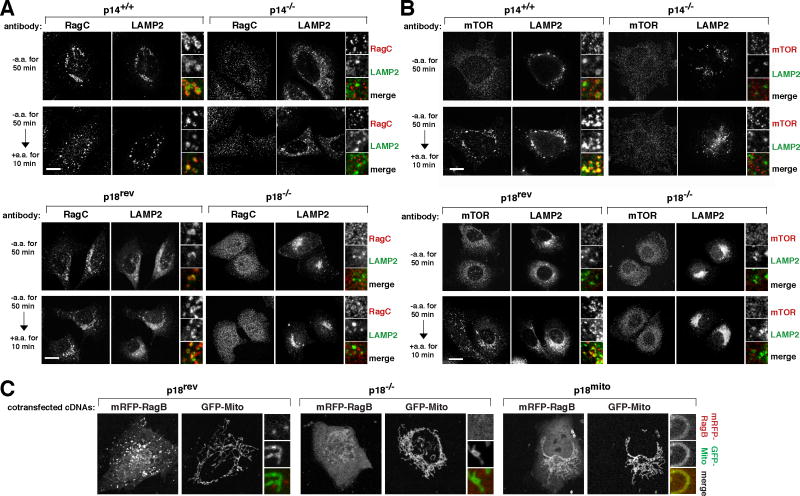
Because the Rag GTPases interact with Ragulator and given the function of p18 in localizing MP1 and p14 to lysosomes (Nada et al., 2009), it seemed possible that the Ragulator is necessary for localizing the Rag proteins to the lysosomal surface. Indeed, in cells lacking p14 or p18 (Nada et al., 2009; Teis et al., 2006), endogenous RagC was localized in small puncta throughout the cytoplasm of the cells rather than to lysosomes (Figure 3A), the morphology of which was not obviously affected by the loss of either protein. In contrast, in p14+/+ cells or p18-null cells reconstituted with wild-type p18 (p18rev), RagC constitutively co-localized with the LAMP2 lysosomal marker (Figure 3A). Analogous results were obtained in HEK-293T cells with an RNAi-mediated reduction in MP1 expression (Figure S3A). Consistent with the essential role of the Rag proteins in the translocation of mTORC1 to the lysosomal surface (Figure S1), in cells lacking p14 or p18 or in HEK-293T cells with p14, p18, or MP1 knockdowns, amino acids failed to induce lysosomal recruitment of mTOR, which was found throughout the cytoplasm in both amino acid starved and stimulated cells (Figures 3B, S3B, and S3D). Thus, all Ragulator subunits are required for lysosomal targeting of the Rag GTPases and mTORC1.
Figure 3. The Ragulator is necessary to localize the Rag GTPases and mTORC1 to lysosomal membranes.
(A) Images of p14-null or p18-null cells or their respective controls co-immunostained for RagC (red) and LAMP2 (green). Cells were starved of and restimulated with amino acids for the indicated times before processing for the immunofluorescence assay and imaging.
(B) Images of p14-null or p18-null cells or their respective controls co-immunostained for mTOR (red) and LAMP2 (green). Cells were treated and processed as in (A).
(C) Co-localization of mRFP-RagB (red) with GFP-Mito (green) in cells expressing mitochondrially-localized p18. p18-null cells (p18−/−), or p18-null cells expressing wild type p18 (p18rev) or mitochondrially-localized p18 (p18mito), were transiently transfected with the indicated cDNAs in expression plasmids and imaged. In all images, insets show selected fields that were magnified five times and their overlays. Scale bar is 10 μm.
See also Fig S3.
To determine if Ragulator is sufficient to control the intracellular localization of the Rag proteins, it was necessary to target Ragulator to a location that is distinct from the lysosomal surface. As p18 binds both p14 and MP1 and is necessary for targeting them to the lysosomal surface (Nada et al., 2009), we chose to manipulate the intracellular localization of p18. To accomplish this we generated a variant of p18, called p18mito, which lacks its N-terminal lipidation sites but is fused at its C-terminus to the transmembrane region of OMP25, which is sufficient to target heterologous proteins to the mitochondrial surface (Nemoto and De Camilli, 1999). When expressed in p18-null cells, p18mito was associated with mitochondria as verified by co-localization with the established mitochondrial protein Cytochrome c (Figure S3E). Remarkably, in the p18-null cells expressing p18mito, RFP-tagged RagB co-localized with the mitochondrial marker GFP-mito (Figure 3C). In contrast, RFP-RagB did not co-localize with GFP-mito in p18-null cells (p18−/−) or p18rev cells, and instead was present in a cytoplasmic or lysosomal pattern, respectively (Figure 3C). In cell expressing p18mito, mTORC1 activity remained very low and mTOR was not recruited to the mitochondria (Figure S3E and S3F), likely because the mitochondrial surface does not contain the machinery necessary to load the Rag GTPases with the appropriate nucleotides. These results indicate that the location of p18 is sufficient to define that of the Rag proteins and are consistent with Ragulator serving as a constitutive docking site on lysosomes for the Rag heterodimers, which, in amino acid-replete cells, have an analogous function for mTORC1.
Ragulator is necessary for TORC1 activation by amino acids in mammalian and Drosophila cells
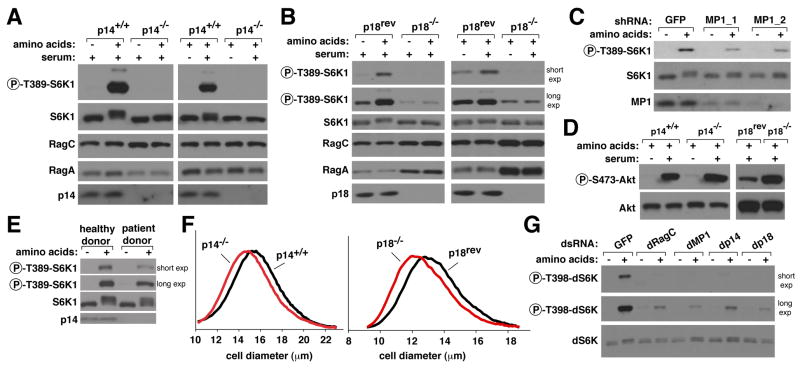
We employed the cells lacking p14 or p18 to determine if Ragulator is necessary for mTORC1 activation by amino acids. Strikingly, in both p14- and p18-null cells, but not in control cells, amino acids were incapable of activating the mTORC1 pathway as detected by the phosphorylation of S6K1 (Figures 4A and 4B) and 4E-BP1 (Figure S4A). Similarly, cells derived from patients with a homozygous mutation in the p14 gene that causes a reduction in p14 expression (Bohn et al., 2006) showed a defect in amino acid-induced mTORC1 activation compared to cells derived from a healthy donor (Figure 4E). In addition, autophagy, a process normally inhibited by mTORC1, was activated in p14-null cells, as detected by an increase compared to in control cells in the size and number of GFP-LC3-II puncta (Figures S4B and S4C). mTORC1 activity was also suppressed in HEK-293T cells with RNAi-induced reductions in p14, p18, or MP1 levels (Figures 4C and S3C). Consistent with the known requirement of amino acids and Rag function for growth factors to activate mTORC1 (Sancak et al., 2008), serum was also incapable of activating the mTORC1 pathway in cells null for p14 or p18 (Figures 4A and 4B). In contrast, no defect was observed in the level of S473 phosphorylation of Akt (Figure 4D). In fact, Akt phosphoylation was slightly higher in the p14-null and p18-null cells than in controls cells, which likely results from the lack of the well-appreciated inhibitory input from mTORC1 to the PI3K pathway in these cells (reviewed in (Manning, 2004)). As mTORC2 is the growth factor-regulated S473 kinase of Akt (Sarbassov et al., 2005), these results also indicate that the Ragulator does not play a detectable positive role in mTORC2 signaling. Interestingly, in the p18-null cells the expression of RagA and RagC was higher than in control cells (Figure 4B), suggesting that feedback signals in these cells may be trying to overcome the defect in mTORC1 activity by boosting Rag expression or that Ragulator also negatively controls Rag GTPase levels. Consistent with p18, p14, and MP1 forming a complex, the expression or stability of the Ragulator proteins seems to be co-regulated because in cells that lack p14, p18 protein levels are also reduced, and, similarly, in cells that lack p18, p14 protein levels are also low (Figure S2B). A well-known function of the mTORC1 pathway is the positive regulation of cell growth, so that inhibition of the pathway leads to a reduction in cell size (reviewed in (Laplante and Sabatini, 2009)). Consistent with Ragulator being a positive component of the mTORC1 pathway, the p14- and p18-null cells were smaller in size than their respective controls (Figure 4F).
Figure 4. Ragulator-null and -depleted cells are highly deficient in the activation of mTORC1 signaling by amino acids.
(A) p14 is necessary for the activation of the mTORC1 pathway by amino acids and serum. p14-null or control cells were starved of amino acids or serum for 50 minutes, or starved and re-stimulated with amino acids or serum for 10 minutes. Immunoblot analyses were used to measure the levels of the indicated proteins and phosphorylation states.
(B) p18 is necessary for the activation of the mTORC1 pathway by amino acids and serum. p18-null or control cells were treated and analyzed as in (A).
(C) Partial knockdown of MP1 blunts mTORC1 pathway activation by amino acids. HEK-293T cells expressing a control shRNA or two distinct shRNAs targeting MP1 were starved for amino acids for 50 minutes, or starved and stimulated with amino acids for 10 minutes and analyzed as in (A).
(D) p14 and p18 are not necessary for mTORC2 pathway activity. p14-null or control cells were starved for serum, or starved and then re-stimulated with serum as in (A). p18-null or control cells were grown in complete media. Cell lysates were prepared and analyzed by immunobloting for the levels of Akt1 and Akt phosphorylation at the S473 site phosphorylated by mTORC2.
(E) Decreased p14 expression impairs amino acid-induced mTORC1 activation in human cells. Cells derived from patients with lower p14 expression or healthy individuals were treated and analyzed as in (A).
(F) Cells lacking Ragulator are smaller than control cells. Cell size distributions of p14-null or p18-null cells are overlaid with those from corresponding control cells.
(G) Ragulator function is conserved in Drosophila cells. Drosophila S2 cells were transfected with a control dsRNA, or dsRNAs targeting dRagC, dMP1, dp14, or dp18, starved of amino acids for 90 minutes, or starved and restimulated with amino acids for 30 minutes. Levels of indicated proteins and phosphorylation states were analyzed by immunobloting.
See also Fig S4.
Many components of the TORC1 pathway, such as the Rag proteins, have conserved roles in mammalian and Drosophila cells (Kim et al., 2008; Sancak et al., 2008). RNAi-inducing dsRNAs that target the Drosophila orthologues of MP1 (CG5110), p14 (CG5189), and p18 (CG14184) were as effective at blocking amino acid-stimulated activation of dTORC1 in Drosophila S2 cells as dsRNAs targeting dRagC (Figure 4G). Our loss of function experiments indicate that Ragulator is a component of the TORC1 pathway that, like the Rag GTPases, is essential for amino acids to activate TORC1 signaling in mammalian and Drosophila cells.
Forced targeting of mTORC1 to the lysosomal surface eliminates the amino acid sensitivity of the mTORC1 pathway
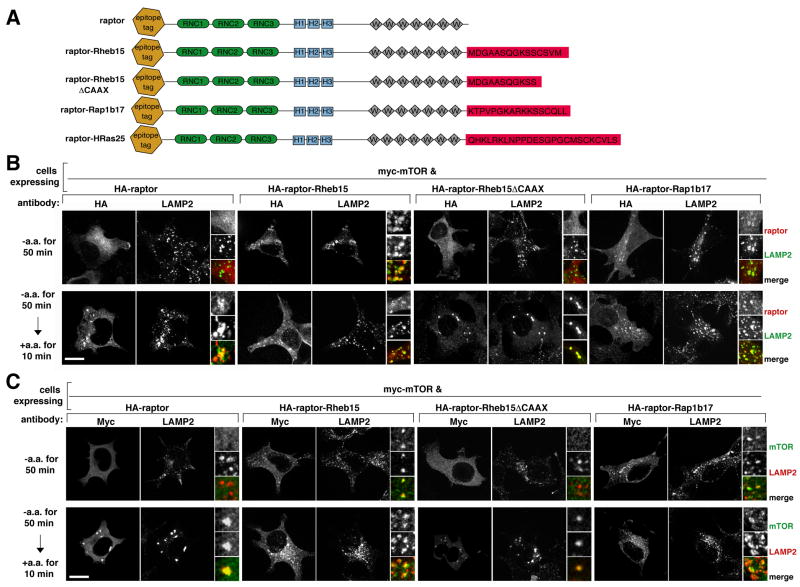
The findings we have presented so far are consistent with the amino acid-induced movement of mTORC1 to the lysosomal surface being necessary for the activation of mTORC1 by amino acids. To test if the placement of mTORC1 on lysosomal membranes is sufficient to mimic the amino acid input to mTORC1, it was necessary to force mTORC1 onto these membranes in the absence of amino acids. To accomplish this we expressed in HEK-293T cells modified raptor proteins that consist of epitope-tagged raptor fused to the intracellular targeting signals of Rheb1 or Rap1b, small GTPases that localize, in part, to the lysosomal surface (Pizon et al., 1994; Saito et al., 2005; Sancak et al., 2008). Because the targeting signals of these proteins are in their C-terminal tails, we added the last 15 or 17 amino acids of Rheb1 or Rap1b, respectively, to the C-terminus of raptor (Figure 5A). For simplicity, we refer to these fusion proteins as raptor-Rheb15 and raptor-Rap1b17. As a control we generated a raptor fusion protein that lacks the CAAX box of the Rheb1 targeting signal (raptor-Rheb15ΔCAAX) and so cannot associate with membranes (Buerger et al., 2006; Clark et al., 1997; Takahashi et al., 2005).
Figure 5. In cells expressing raptor variants fused to the targeting signals of Rheb1 or Rap1b, mTORC1 localizes to lysosomal membranes in an amino acid-independent fashion.
(A) Schematic of raptor fusion proteins that target mTORC1 to lysosomal membranes (raptor-Rheb15; raptor-Rap1b17) or to the plasma membrane (Raptor-HRas25) as well as proteins used as controls (wild-type raptor; raptor-Rheb15 CAAX).
(B) Images of amino acid starved or replete cells expressing lysosomally-targeted or control HA-tagged raptor proteins and co-immunostained for the HA epitope (red) and endogenous LAMP2 (green). HEK-293T cells were transfected with the indicated cDNAs, starved of and restimulated with amino acids for the indicated times, and processed in the immunofluorescence assay.
(C) Images of amino acid starved or replete cells co-expressing myc-mTOR and the indicated raptor fusion proteins and co-immunostained for the myc epitope (green) and endogenous LAMP2 (red). HEK-293T cells were co-transfected with the indicated cDNAs and treated and processed as in (B). In all images, insets show selected fields that were magnified five times and their overlays. Scale bar is 10 μm.
When expressed in cells together with myc-mTOR, raptor-Rheb15 and raptor-Rap1b17 localized to lysosomes in the presence or absence of amino acids, as judged by co-staining with LAMP2 (Figure 5B). In contrast, raptor-Rheb15ΔCAAX behaved like wild-type raptor and localized to lysosomes only upon amino acid stimulation (Figure 5B). In all cases the localization of the co-expressed myc-mTOR mirrored that of the wild-type or altered forms of raptor, indicating that C-terminal modifications of raptor do not perturb its interaction with mTOR (Figure 5C), which was confirmed in co-immunoprecipitation experiments (Figure S5A).
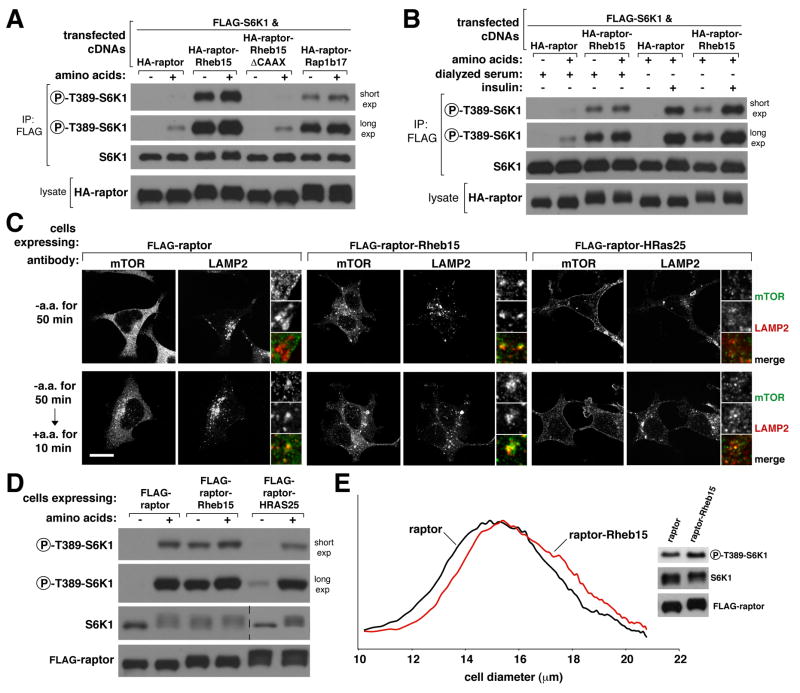
Remarkably, transient expression of raptor-Rheb15 or raptor-Rap1b17 in HEK-293T cells was sufficient to render the mTORC1 pathway, as judged by the phosphorylation of S6K1, resistant to amino acid starvation (Figure 6A). In contrast, the expression of wild-type raptor or raptor-Rheb15ΔCAAX did not affect the amino acid sensitivity of the pathway (Figure 6A). In HEK-293E cells, the expression of raptor-Rheb15 made S6K1 phosphorylation insensitive to amino acid starvation, but did not affect its regulation by insulin (Figure 6B). Thus, lysosomal targeting of mTORC1 can substitute for the amino acid, but not growth factor, input to mTORC1. This is consistent with previous work showing that growth factors signal to mTORC1 in large part through the TSC1-TSC2-Rheb axis (reviewed in (Laplante and Sabatini, 2009)), and not through the Rag GTPases (Sancak et al., 2008).
Figure 6. Constitutive association of raptor with lysosomal membranes, but not the plasma membrane, is sufficient to make the mTORC1 pathway insensitive to amino acid starvation.
(A) The mTORC1 pathway is not sensitive to amino acid starvation in cells that express lysosomally-targeted but not control raptor proteins. HEK-293T cells were co-transfected with the indicated cDNA expression plasmids and starved of amino acids for 50 minutes or starved and restimulated with amino acids for 10 minutes. Cell lysates and anti-FLAG-S6K1 immunoprecipitates were analyzed by immunobloting for the levels of the indicated proteins and phosphorylation states.
(B) The mTORC1 pathway is sensitive to serum starvation and insulin stimulation in cells that express lysosomally-targeted as well as control raptor proteins. HEK-293E cells were co-transfected with the indicated cDNA expression plasmids, starved of amino acids for 50 minutes or starved and restimulated with amino acids for 10 minutes. Duplicate cultures were starved of serum for 50 minutes or starved and stimulated with insulin for 10 minutes. Cell lysates and anti-FLAG-S6K1 immunoprecipitates were analyzed by immunobloting for the levels of the indicated proteins and phosphorylation states.
(C) Images of cells stably expressing FLAG-raptor, FLAG-raptor-Rheb15, or FLAG-raptor-HRas25 and co-immunostained for endogenous mTOR (green) and endogenous LAMP2 (red). HEK-293T cells stably expressing the indicated proteins were treated as in (A) for the indicated times before processing in the immunofluorescence assay. In all images, insets show selected fields that were magnified five times and their overlays. Scale bar is 10 μm.
(D) Targeting of mTORC1 to the lysosomal but not the plasma membrane makes the mTORC1 pathway insensitive to amino acid starvation. HEK-293T cells stably expressing FLAG-raptor, FLAG-raptor-Rheb15, or FLAG-raptor-HRas25 were treated as in (A) and analyzed by immunoblotting for the levels of the indicated proteins and phosphorylation states.
(E) Targeting of mTORC1 to the lysosomal membrane increases cell size and pathway activity in cells under normal growth conditions. Cell size distributions of cells that stably express FLAG-raptor or FLAG-raptor-rheb15 as well as immunoblot analyses of the mTORC1 pathway in the same cells.
To verify the effects of lysosomally-targeted mTORC1 in a more physiological setting than that achieved by transient cDNA expression, we generated HEK-293T cell lines stably expressing FLAG-tagged raptor-Rheb15 or wild-type raptor. In cells expressing the lysosomally-targeted but not wild-type raptor, mTOR was always associated with lysosomes, irrespective of amino acids (Figure 6C). As with the transient expression of raptor-Rheb15, its stable expression rendered the mTORC1 pathway fully resistant to amino acid starvation (Figure 6D). Furthermore, under normal growth conditions these cells had an increase in mTORC1 activity and were larger than controls (Figure 6E).
We next examined if the targeting of mTORC1 to membranes other than lysosomal membranes could also eliminate the amino acid sensitivity of the mTORC1 pathway. This was not the case because although the stable expression of a raptor variant consisting of raptor fused to the last 25 amino acids of H-Ras (raptor-HRas25) (Figures 5A and S5B) was sufficient to target a fraction of cellular mTOR to the plasma membrane (Figure 6C), it did not render the mTORC1 pathway resistant to amino acid starvation (Figure 6D).
Forced targeting of mTORC1 to the lysosomal surface eliminates the requirement in mTORC1 signaling for Rag and Ragulator, but not Rheb, function
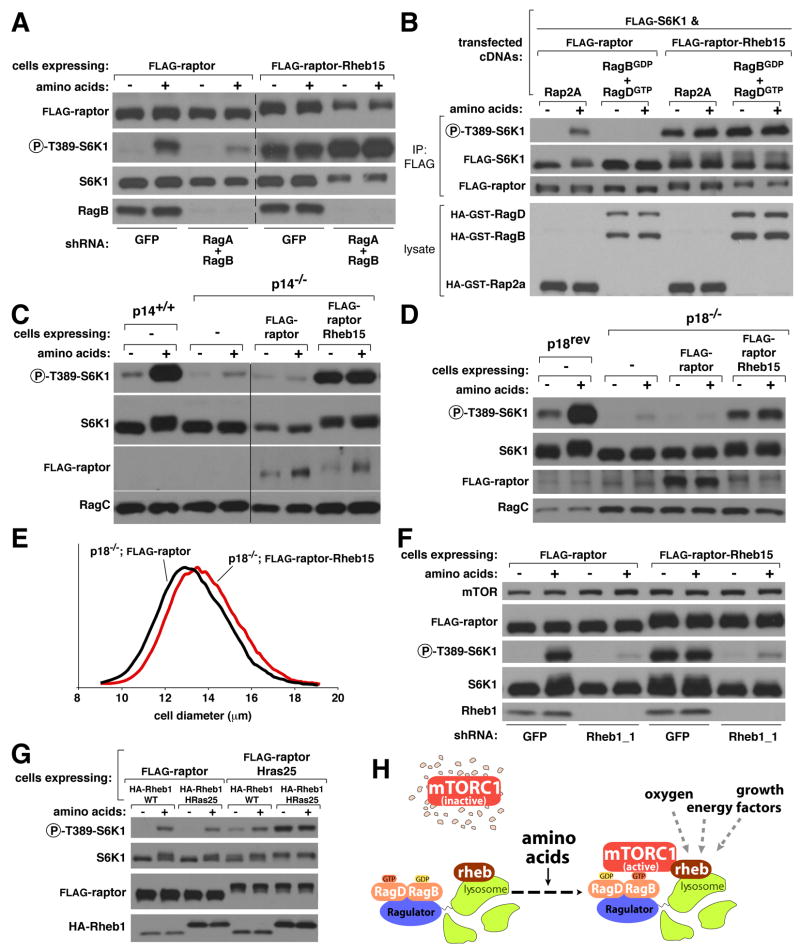
The ability to constitutively localize mTORC1 to lysosomal membranes enabled us to probe in more detail the role of the Rag and Rheb GTPases, as well as Ragulator, in the activation of mTORC1 by amino acids. We hypothesized that if the major role of the Rag GTPases is to allow mTORC1 to localize to lysosomes, then in cells that express raptor-Rheb15, mTORC1 activity should be independent of Rag function. Indeed, while in control cells the RNAi-mediated knockdown of both RagA and RagB strongly blunted the activation of mTORC1 by amino acids, it did not reduce the amino acid-insensitive mTORC1 activity observed in raptor-Rheb15 expressing cells (Figure 7A). As an additional approach to inhibit Rag function, we exploited the fact that co-expression of a GDP-bound RagB mutant (RagBGDP) and a GTP-bound RagD mutant (RagDGTP) eliminates mTORC1 pathway activity within cells (Kim et al., 2008; Sancak et al., 2008). Expression of RagBGDP-RagDGTP completely prevented mTORC1 activation by amino acids in control cells, but had no effect on the amino acid-insensitive mTORC1 activity of cells expressing raptor-Rheb15 (Figure 7B).
Figure 7. Targeting of mTORC1 to the lysosomal surface makes the activity of the mTORC1 pathway independent of Rag and Ragulator, but not, Rheb function.
(A) In cells that express FLAG-raptor-Rheb15, mTORC1 pathway activity is independent of Rag GTPase function. Lysates of HEK-293T cells expressing FLAG-raptor or FLAG-raptor-Rheb15 were analyzed by immunobloting for the indicated proteins and phosphorylation states after disruption of Rag function by RNAi-mediated co-knockdown of RagA and RagB. Cells were starved of amino acids for 50 minutes or starved and restimulated with amino acids for 10 minutes before lysis.
(B) In cells that express FLAG-raptor-Rheb15, mTORC1 pathway activity is independent of Rag GTPase function. Lysates of HEK-293T cells expressing FLAG-raptor or FLAG-raptor-Rheb15 were analyzed as in (A) after disruption of Rag function by expression of the dominant negative RagBGDP-RagDGTP heterodimer. Cells were treated and processed as in (A).
(C) Stable expression of FLAG-raptor-Rheb15 but not FLAG-raptor in p14-null cells is sufficient to reactivate the mTORC1 pathway and make it insensitive to amino acid starvation. Cells stably expressing the indicated proteins were treated and analyzed as in (A).
(D) Stable expression of FLAG-raptor-Rheb15 but not FLAG-raptor in p18-null cells is sufficient to reactivate the mTORC1 pathway and make it insensitive to amino acid starvation. Cells stably expressing the indicated proteins were treated and analyzed as in (A).
(E) In p18-null cells expression of raptor-Rheb15, but not wild-type raptor, increases cell size. Cell size distributions of p18-null cells that stably express FLAG-raptor or FLAG-raptor-Rheb15.
(F) In cells that express FLAG-raptor-Rheb15, the activity of the mTORC1 pathway is still Rheb-dependent. Lysates of HEK-293T cells that stably express FLAG-raptor or FLAG-raptor-Rheb15 were analyzed by immunobloting for the indicated proteins and phosphorylation states after disruption of Rheb function by an RNAi-mediated knockdown of Rheb1. Cells were treated as in (A).
(G) Co-expression of plasma membrane-targeted raptor and plasma membrane-targeted Rheb1 renders the mTORC1 pathway insensitive to amino acid starvation. HEK-293T cells stably expressing the indicated proteins were treated and analyzed as in (A).
(H) Model for amino-acid induced mTORC1 activation. In the absence of amino acids, mTORC1 cannot associate with the endomembrane system, and has no access to its activator Rheb. In the presence of amino acids, the Rag GTPases, which are tethered to the lysosomal surface by the Ragulator, serve as a docking site for mTORC1, allowing mTORC1 to associate with endomembranes and thus encounter and become activated by Rheb.
See also Fig S5.
If the main function of Ragulator in the mTORC1 pathway is to localize the Rag GTPases to the lysosomes then it should be possible to reactivate the mTORC1 pathway in Ragulator-null cells by expressing raptor-Rheb15. Remarkably, the stable expression of raptor-Rheb15, but not wild-type raptor, in p14- or p18-null cells reactivated mTORC1 signaling and made it insensitive to amino acid deprivation (Figure 7C and 7D). Furthermore, expression of raptor-Rheb15 in the p18-null cells was sufficient to increase their size (Figure 7E). In contrast to the results observed with the Rag GTPases and Ragulator, RNAi-mediated suppression of Rheb1 blocked amino acid-induced mTORC1 activation in cells expressing raptor-Rheb15 to the same extent as it did in control cells (Figure 7F).
To test whether the presence of mTORC1 and Rheb on the same membrane compartment is sufficient to render the mTORC1 pathway insensitive to amino acid levels, we generated cells in which mTORC1 and Rheb are both present on the plasma membrane. To accomplish this we prepared a Rheb1 variant, called Rheb1-HRas25, that localizes to the plasma-membrane (Figure S5C) because it contains the C-terminal 25 amino acids of H-Ras instead of the normal Rheb1 localization signal. When Rheb1-HRas25 was stably co-expressed with raptor-HRas25, but not wild-type raptor, the mTORC1 pathway became insensitive to amino acid starvation (Figure 7G). Importantly, mTORC1 signaling remained amino acid-sensitive in cells in which either Rheb or mTORC1, but not both, was targeted to the plasma membrane (Figure 7G).
Discussion
Our findings, together with previous work showing that Rheb is required for amino acids to activate the mTORC1 pathway (Roccio et al., 2006;Smith et al., 2005) and can localize to late endosomes/lysosomes (Saito et al., 2005; Sancak et al., 2008), is consistent with a model in which amino acids induce mTORC1 to associate with the endomembrane system of the cell and thus allow it to encounter its activator Rheb. In this model the essential role of the Ragulator-Rag complex is to serve as an amino acid-regulated docking site for mTORC1 on lysosomal membranes (see schematic in Figure 7H). The proposed link between the Rag and Rheb GTPases in the regulation of the mTORC1 pathway provides an explanation for why activation of mTORC1 occurs only when activators of both Rheb (e.g., growth factors and energy) and the Rags (i.e., amino acids) are available. For technical reasons (Buerger et al., 2006; Sancak et al., 2008), it has not been possible to determine the intracellular localization of endogenous Rheb and work using overexpressed GFP-tagged Rheb1 has placed it on various endomembrane compartments, including endosomes and lysosomes (Buerger et al., 2006; Saito et al., 2005; Sancak et al., 2008; Takahashi et al., 2005). Our results suggests that at some point in its life cycle Rheb must traverse the lysosomal surface in order to encounter mTORC1 and so in our model we have chosen to place Rheb on this compartment (Figure 7H). However, at any given time only a small fraction of cellular Rheb may actually be on the lysosomal surface or, alternatively, some of the mTORC1 within the cell may move to a non-lysosomal endomembrane compartment that also contains Rheb. These issues will only be answered once a definitive location for endogenous Rheb can be determined.
The trimeric p14, p18, and MP1 protein complex, which we call Ragulator, is a Rag-interacting complex that is essential for amino acid signaling to mTORC1 and represents an additional critical component of the TORC1 signaling pathway in mammals and flies. p18 directly interacts with the Rag GTPases (Figure S2A) as well as with p14 and MP1 (Nada et al., 2009) and so may serve as a scaffold to bring the Rag GTPases and MP1-p14 next to each other. In vitro we have not detected a direct interaction between the Rag GTPases and either MP1 or p14, but both proteins are, like p18, necessary for localizing the Rag GTPases to the lysosomal surface. p14 is required to maintain normal p18 expression levels (Figure S2B), suggesting that within cells p14 and MP1 form a crucial part of the Ragulator structure. Given the non-specific nature of the p14 and p18 names, in the future it may be best to rename these proteins, perhaps to names that reflect their essential roles in the mTORC1 pathway.
The location of the Rag GTPases, the Ragulator, and mTORC1 on the lysosomal surface implicates this organelle as the site of a yet to be discovered sensing system that signals amino acid availability to the Ragulator-Rag complex. The lysosomal location of the amino acid sensing branch of the mTORC1 pathway is consistent with increasing evidence that lysosomes, and their yeast counterparts, vacuoles, are at the nexus of amino acid metabolism within cells. Lysosomes are a major site of protein degradation and amino acid recycling and vacuoles store amino acids at high concentrations (reviewed in (Li and Kane, 2009)). Thus, mTORC1 and its regulators may reside on the lysosomal surface so as to sense a currently unknown aspect of lysosomal function that reflects the intracellular pools of amino acids.
It is interesting to consider the differences and similarities between the still poorly understood amino acid signaling mechanisms employed by the mTORC1 and yeast TORC1 pathways. Consistent with previous work in mammalian cells (Sancak et al., 2008), the Gtr1p-Gtr2p heterodimer that is orthologous to RagA/B-RagC/D, interacts with yeast TORC1 when Gtr1p is GTP-loaded (Binda et al., 2009). TORC1 and the Gtr proteins are located on the surface of the vacuole (Berchtold and Walther, 2009; Binda et al., 2009), the yeast equivalent of lysosomes, but, unlike in mammals, yeast TORC1 does not leave the vacuolar surface upon amino acid deprivation although amino acids do control the interaction of TORC1 with Gtr1p-Gtr2p (Binda et al., 2009). This finding suggests that there must exist a distinct mechanism for retaining TORC1 at the vacuolar surface and that in yeast the interaction between TORC1 and Gtr1p-Gtr2p serves other purposes besides controlling the intracellular location of TORC1. In contrast, our current work argues that in mammals the main role of the Rag GTPase and the associated Ragulator complex is to control the association of mTORC1 with the cellular endomembrane system, in particular, lysosomes. Rheb, which is essential for the activation of mTORC1 by all upstream signals, does not appear to be part of the TORC1 pathway in yeast (reviewed in (Berchtold and Walther, 2009)). As we suggest that the Rag-dependent and amino acid-regulated translocation of mTORC1 to the lysosomal surface may ultimately be a mechanism for controlling the access of mTORC1 to Rheb, the absence of Rheb in the yeast TORC1 pathway may make regulation of TORC1 localization unnecessary. That known Rag- and Gtr-interacting proteins share no sequence homology also suggests that the mechanisms through which the Rag and Gtr GTPases regulate mTORC1 and yeast TORC1, respectively, have diverged. Although it is clear that the Ragulator and EGO complexes both control the intracellular localization of the Rag (this paper) and Gtr (Gao and Kaiser, 2006) GTPases, respectively, whether these complexes have additional functions remains to be determined.
Previous studies suggest that MP1-p14-p18 complex plays an adaptor role in the MAP Kinase (MAPK) pathway (reviewed in (Dard and Matthias, 2006)) and our current findings do not contradict these results. However, considering the very strong inhibition of the mTORC1 pathway that occurs in cells lacking p14 or p18, it seems possible that some of the impairment in MAPK signaling observed in those cells reflects an altered feedback signaling from Akt to the MAPK pathway. For example, in Ragulator-null cells, Akt is slightly activated, almost certainly because the well-known inhibitory signal from mTORC1 to PI3K is absent. As Akt suppresses MAPK signaling by phosphorylating and inhibiting Raf (Zimmermann and Moelling, 1999), it is conceivable that the activation of Akt that occurs in Ragulator-null cells could account, at least in part, for the inhibition of MAPK signaling that has been observed in these cells.
Mice lacking either p14 or p18 die around embryonic day 7.5–8 and have obvious growth defects (Nada et al., 2009; Teis et al., 2006). We would not be surprised if, when generated, mice lacking the Rag proteins die at around the same age and present similar defects. On the other hand, mice lacking the core mTORC1 component raptor die earlier (before embryonic day 6.5) than p14- and p18-null mice (Guertin et al., 2006). This may be expected because although loss of p14 or p18 completely blocks mTORC1 activation by amino acids, cells lacking the Ragulator proteins are likely to retain a low residual level of mTORC1 activity that may be sufficient to support development further than in embryos completely lacking mTORC1 function. Lastly, our results suggest that the strong growth retardation observed in humans with a mutation that reduces p14 expression (Bohn et al., 2006), is a result of partial suppression of the mTORC1 pathway. If this turns out to be the case, it would represent the first human example of a loss of function mutation in a positive component of the mTORC1 pathway.
Materials and Methods
Cell Lines and Tissue Culture
HEK-293E cells; HEK-293T cells; TSC2+/+, TSC2−/−, p14+/+, and p14−/− MEFs were cultured in DMEM with 10% IFS. p18rev, p18mito, and p18−/− cells were cultured in DMEM with 10% FBS. HEK-293E and HEK-293T cells express E1a and SV40 large T antigen, respectively. In HEK-293E, but not HEK-293T, cells the mTORC1 pathway is strongly regulated by serum and insulin (Sancak et al., 2007). TSC2−/−, p53−/− and TSC2+/+, p53−/− MEFs were kindly provided by Dr. David Kwiatkowski (Harvard Medical School). The HEK-293E cell line was kindly provided by Dr. John Blenis (Harvard Medical School). p14−/− and control MEFs were kindly provided by Dr. Lukas A. Huber (Innsbruck Medical University) and described in (Teis et al., 2006). p18−/− cells are epithelial in nature and p18rev cells are p18−/− cells in which wild-type p18 has been re-expressed (Nada et al., 2009). Patient-derived cells with a homozygous mutation in the ROBLD3 (p14) gene 3′ untranslated region and control healthy donor-derived cells were kindly provided by Dr. Christoph Klein (Universität München) and have been described in (Bohn et al., 2006)
Amino Acid and Serum Starvation and Stimulation of Cells
Serum and/or amino acid starvation of HEK-293T cells, HEK-293E cells, p14-null and control cells, p18-null and control cells, MEFs, patient-derived and healthy donor-derived cells were performed essentially as described (Sancak et al., 2008). Serum was dialyzed against PBS in dialysis cassettes (Thermo Scientific) having a 3,500 molecular weight cut off.
Preparation of Cell Lysates and Immunoprecipitations
Cell lysate preparation, cell lysis and immunoprecipitations were done as described in supplemental materials and methods.
For co-transfection experiments, 2 million HEK-293T or HEK-293E cells were plated in 10 cm culture dishes. 24 hours later, cells were transfected with the indicated plasmids as follows: 50 ng or 1500 ng myc-mTOR in pRK5; 20 ng or 500 ng HA-, myc- or FLAG-Raptor in pRK5 or pLJM1 with or without the targeting signals; 100 ng HA-GST-Rap2a in pRK5; 100 ng HA-GST-Rheb1 in pRK5; 100 ng HA-GST-RagB in pRK5, 100 ng HA-GST-RagD in pRK5; 1 ng FLAG-S6K1 in pRK7; 50 ng or 600 ng HA- or FLAG-p14 in pRK5; 75 ng or 600 ng HA-MP1 in pRK5; 50 ng or 800 ng HA-p18 in pRK5. The total amount of plasmid DNA in each transfection was normalized to 2 μg using empty pRK5.
Cell Size Determinations
To measure cell size, 2 million HEK-293T cells or 200,000 of other cell types were plated into 10 cm culture dishes. 24 hours later the cells were harvested by trypsinization in a 4 ml volume and diluted 1:20 with counting solution (Isoton II Diluent, Beckman Coulter). Cell diameters were determined using a particle size counter (Coulter Z2, Beckman Coulter) running Coulter Z2 AccuComp software.
Mammalian Lentiviral shRNAs and cDNAs
Lentiviral shRNAs targeting human Rheb1, RagB, and RagC have been described (Sancak et al., 2008). Lentiviral shRNAs targeting mouse Rheb1 and human p14 were obtained from Sigma-Aldrich. Lentiviral shRNAs targeting the mRNA for human MP1 and human p18 were cloned into pLKO.1 vector as described (Sarbassov et al., 2005). The target sequences are provided in supplemental materials and methods.
Virus generation and infection was done as previously described (Sancak et al., 2008).
Raptor was cloned into the AgeI and BamHI sites of a modified pLKO.1 vector (pLJM1) (Sancak et al., 2008) with or without the Rheb1, Rap1b and HRas targeting signals or cloned into the pRK5 vector with or without the same localization signals. After sequence verification, pLJM1 based plasmids were used in transient cDNA transfections or to produce lentivirus needed to generate cell lines stably expressing these proteins. pRK5 based plasmids were also used for transient transfection experiments. The p18mito expression plasmid was generated by cloning a mutant p18 with amino acids 2-5 changed to alanines into a modified version of the pLKO.1 vector that added, to the C-terminus of p18, the mitochondrial localization signal of OMP25 protein. This plasmid was used in transient cDNA transfections or to produce lentivirus needed to generate stable cell lines. HA-Rheb1 and HA-Rheb1-HRas25 were cloned into pLJM5, a derivative of pLJM1 carrying a hygromycin instead of puromycin resistance gene. The vectors were used as above for lentivirus production.
Immunofluorescence Assays
50,000 HEK-293T cells or 20,000 of other cell types were plated on fibronectin coated glass coverslips in 12-well tissue culture plates. 24 hours later, the slides were rinsed with PBS once and fixed for 15 minutes with 4% paraformaldehyde in PBS warmed to 37°C. The slides were rinsed twice with PBS and cells were permeabilized with 0.05% Triton X-100 in PBS for 30 seconds. After rinsing twice with PBS, the slides were incubated with primary antibody in 5% Normal Donkey Serum for 2 hours at room temperature, rinsed four times with PBS and incubated with secondary antibodies produced in donkey (diluted 1:1000 in 5% Normal Donkey Serum) for one hour at room temperature in the dark, washed four times with PBS. Slides were mounted on glass coverslips using Vectashield (Vector Laboratories) and imaged.
Transient transfections for immunofluorescence assays were performed as described in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank all members of the Sabatini Lab for helpful suggestions and Eric Spooner for the mass spectrometric analysis of samples. We thank Dr. Lukas A. Huber for providing p14-null and control cells and Dr. Christoph Klein for providing patient-derived and healthy donor-derived cells. This work was supported by grants from the NIH (CA103866 and AI47389) and Department of Defense (W81XWH-07-0448) to D.M.S., awards from the W.M. Keck Foundation and LAM Foundation to D.M.S, and fellowship support from the LAM Foundation and from the Jane Coffin Childs Memorial Fund for Medical Research to R.Z. D.M.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berchtold D, Walther TC. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol Biol Cell. 2009:1565–1575. doi: 10.1091/mbc.E08-10-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda M, Péli-Gulli M, Bonfils G, Panchaud N, Urban J, Sturgill T, Loewith R, De Virgilio C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Bohn G, Allroth A, Brandes G, Thiel J, Glocker E, Schäffer AA, Rathinam C, Taub N, Teis D, Zeidler C, et al. A novel human primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nat Med. 2006;13:38–45. doi: 10.1038/nm1528. [DOI] [PubMed] [Google Scholar]
- Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem Biophys Res Commun. 2006;344:869–880. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- Chavrier P, Parton RG, Hauri HP, Simons K, Zerial M. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990;62:317–329. doi: 10.1016/0092-8674(90)90369-p. [DOI] [PubMed] [Google Scholar]
- Clark GJ, Kinch MS, Rogers-Graham K, Sebti SM, Hamilton AD, Der CJ. The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J Biol Chem. 1997;272:10608–10615. doi: 10.1074/jbc.272.16.10608. [DOI] [PubMed] [Google Scholar]
- Dard N, Matthias P. Scaffold proteins in MAP kinase signaling: more than simple passive activating platforms. BioEssays. 2006;28:146–156. doi: 10.1002/bies.20351. [DOI] [PubMed] [Google Scholar]
- Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Eskelinen EL. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol Aspects Med. 2006;27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Gao M, Kaiser C. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Hirose E, Nakashima N, Sekiguchi T, Nishimoto T. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J Cell Sci. 1998;111:11–21. doi: 10.1242/jcs.111.1.11. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:395–345. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Aravind L. Dynein light chains of the Roadblock/LC7 group belong to an ancient protein superfamily implicated in NTPase regulation. Curr Biol. 2000;10:774–776. doi: 10.1016/s0960-9822(00)00774-0. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SC, Kane PM. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim Biophys Acta. 2009;1793:650–663. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunin VV, Munger C, Wagner J, Ye Z, Cygler M, Sacher M. The Structure of the MAPK Scaffold, MP1, Bound to Its Partner, p14. J Biol Chem. 2004;279:23422–23430. doi: 10.1074/jbc.M401648200. [DOI] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- Manning BD. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J Cell Biol. 2004;167:399–403. doi: 10.1083/jcb.200408161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada S, Hondo A, Kasai A, Koike M, Saito K, Uchiyama Y, Okada M. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK ERK pathway to late endosomes. EMBO J. 2009;28:477–489. doi: 10.1038/emboj.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y, De Camilli P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 1999;18:2991–3006. doi: 10.1093/emboj/18.11.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizon V, Desjardins M, Bucci C, Parton RG, Zerial M. Association of Rap1a and Rap1b proteins with late endocytic/phagocytic compartments and Rap2a with the Golgi complex. J Cell Sci. 1994;107:1661–1670. doi: 10.1242/jcs.107.6.1661. [DOI] [PubMed] [Google Scholar]
- Roccio M, Bos JL, Zwartkruis FJT. Regulation of the small GTPase Rheb by amino acids. Oncogene. 2006;25:657–664. doi: 10.1038/sj.onc.1209106. [DOI] [PubMed] [Google Scholar]
- Saito K, Araki Y, Kontani K, Nishina H, Katada T. Novel Role of the Small GTPase Rheb: Its Implication in Endocytic Pathway Independent of the Activation of Mammalian Target of Rapamycin. J Biochem. 2005;137:429–430. doi: 10.1093/jb/mvi046. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and Regulation of Akt/PKB by the Rictor-mTOR Complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Schaeffer HJ, Catling AD, Eblen ST, Collier LS, Krauss A, Weber MJ. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- Schurmann A, Brauers A, Maßmann S, Becker W, Joost H. Cloning of a Novel Family of Mammalian GTP-binding Proteins (RagA, RagBs, RagBl) with Remote Similarity to the Ras-related GTPases. J Biol Chem. 1995;270:28982–28988. doi: 10.1074/jbc.270.48.28982. [DOI] [PubMed] [Google Scholar]
- Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Nakagawa M, Young SG, SY Differential Membrane Localization of ERas and Rheb, Two Ras-related Proteins Involved in the Phosphatidylinositol 3-Kinase/mTOR Pathway. J Biol Chem. 2005;280:32768–32774. doi: 10.1074/jbc.M506280200. [DOI] [PubMed] [Google Scholar]
- Teis D, Taub N, Kurzbauer R, Hilber D, Araujo ME, Erlacher M, Offterdinger M, Villunger A, Geley S, Bohn G, et al. p14 MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J Cell Biol. 2006;175:861–868. doi: 10.1083/jcb.200607025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Wunderlich W, Huber LA. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell. 2002;3:803–814. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- Wunderlich W, Fialka I, Teis D, Alpi A, Pfeifer A, Parton RG, Lottspeich F, Huber LA. A Novel 14-Kilodalton Protein Interacts with the Mitogen-activated Protein Kinase Scaffold MP1 on a Late Endosomal/Lysosomal Compartment. J Cell Biol. 2001;152:765–776. doi: 10.1083/jcb.152.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann S, Moelling K. Phosphorylation and Regulation of Raf by Akt (Protein Kinase B) Science. 1999;286:1741–1744. doi: 10.1126/science.286.5445.1741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.