Abstract
Elastic fibres are essential for normal physiology in numerous tissues, including arteries, lungs and skin. Fibulin-4 is an elastic-fibre-associated glycoprotein that is indispensable for elastic-fibre formation in mice. However, the mechanism by which fibulin-4 executes this function remains to be determined. Here, we established an in vitro functional assay system in which fibulin-4 was knocked down in human foreskin fibroblasts using siRNA (small interfering RNA) technology. With two different siRNAs, substantial knockdown of fibulin-4 was achieved, and this suppression was associated with impaired elastic-fibre formation by the fibroblasts. Real-time reverse transcription–PCR analysis showed that knockdown of fibulin-4 expression was accompanied by reduced expression of tropoelastin mRNA. Further analysis showed that this decrease was caused by transcriptional down-regulation of tropoelastin. This effect was selective, since the mRNA level of other elastic-fibre-associated proteins, including fibrillin-1, lysyl oxidase and lysyl oxidase-like-1, was not affected. Moreover, addition of conditioned medium from cultures of CHO (Chinese-hamster ovary) cells overexpressing fibulin-4 stimulated tropoelastin expression and elastic-fibre formation in cultures of Williams–Beuren-syndrome fibroblasts. Knocking down or knocking out fibulin-4 in mice led to a decrease in tropoelastin expression in the aorta. These results indicate that fibulin-4, considered as a structural protein, may also participate in regulating elastic-fibre formation in human cells through an unanticipated mechanism, namely the regulation of tropoelastin expression.
Keywords: elastic-fibre formation, elastin, extracellular-matrix protein, fibulin-4, human foreskin fibroblast, tropoelastin gene
INTRODUCTION
Elastic fibres are the major ECM (extracellular matrix) structures that provide tissues with elasticity throughout the body, especially in the major arteries, lungs and skin, where tissue elasticity is absolutely necessary for physiological function. Elastic fibres are composed of a central elastin core assembled on the scaffold of parallel microfibrils consisting of numerous glycoproteins [1,2]. Formation of elastic fibres (elastogenesis) is a developmentally regulated multi-step hierarchical assembly process of sequential interaction between elastin and its associated proteins [3]. In cells, EBP67 (67 kDa elastin-binding protein) binds to soluble tropoelastin and forms a complex with PPCA (protective protein–cathepsin A) and Neu1 (neuraminidase-1) to facilitate tropoelastin secretion. Consecutively, Neu1 catalyses removal of the terminal sialic acids from carbohydrate chains of microfibrillar glycoproteins and other adjacent matrix glycoconjugates, unmasking their penultimate galactosugars. The exposed galacto-sugars, in turn, interact with the galactolectin domain of the EBP, thereby inducing the release of transported tropoelastin molecules and facilitating their subsequent assembly into elastic fibres [4,5]. The released tropoelastin is deposited on microfibril scaffolds, cross-links to form polymers and assembles to make elastic fibres. To date, over 34 proteins have been found to be associated with elastic fibres [6]. Only a few of them have been shown to be structurally involved in elastogenesis. These include fibrillin-1 [7,8], fibrillin-2 [9], LOX (lysyl oxidase) [10,11], LOXL1 (LOX-like-1) [12], emilin-1[13], fibulin-4 [14] and fibulin-5 [15,16].
Fibulins are a family of extracellular glycoproteins that share the characteristics of a tandem array of cbEGF (calcium-binding epidermal growth factor) motifs, followed by a 120–140-aminoacid residue fibulin-type C-terminus [17–19]. Currently, seven genes have been identified in the fibulin family. Fibulin-1, 2, and -6 are larger in size than fibulin-3, -4, -5 and -7, containing additional anaphylatoxin (AT) motifs at their N-termini. Fibulin-3, -4 and -5 contain five cbEGF motifs preceded by a modified cbEGF domain at their N-termini. Because of the similar modular structure, fibulin-1 to -5 all show a rod structure with globules at one end (fibulin-3, -4 and -5) [20] or both ends (fibulin-1 and -2) [21,22] by rotary-shadowing electron microscopy.
The fibulins are widely distributed throughout the body and are more prominent in tissues rich in elastic fibres, such as aorta [20]. They bind to a large number of ECM proteins ranging from the basement-membrane proteins, proteoglycan aggregates, various microfibrils such as fibronectin microfibrils and fibrillin microfibrils, to elastic networks [6,17]. Indeed, all fibulins that have been analysed have been found to be associated with elastic fibres. Fibulin-1 is associated with the elastin core [23], fibulin-2 [24] and -4 [20] are associated with the microfibrils, and fibulin-2 [24] and -5 [20] are located at the interface between the elastin core and the microfibrils.
Despite the extensive similarity among the fibulins, only fibulin-4 and -5 have been implicated as regulators of elastic-fibre formation. Whereas fibulin-4-null mice fail to make elastic fibres [14], fibulin-5-null mice still assemble elastic fibres, although the fibres are abnormal [15,16]. On the other hand, the lack of fibulin-1 or -2 in mice does not affect the primary elastic-fibre formation [25,26]. Thus fibulin-4 appears to be the most critical member of this family of proteins in modulating elastic-fibre formation. However, the mechanisms underlying its involvement in elastogenesis remain unknown. The role of fibulin-4 in regulating elastic-fibre formation in human cells has not been determined.
Recently, extensive in vitro biochemical characterization has been carried out to compare fibulin-4 with fibulin-1, -2, -3 and -5 in their interactions with tropoelastin and fibrillin-1 [20,27]. Like fibulin-2 and -5, fibulin-4 binds to the N-terminus of fibrillin-1. The affinity of fibulin-4 for tropoelastin is higher than that of fibulin-1 and -3, but lower than that of fibulin-2 and -5. Consistent with this affinity pattern, the ability of fibulin-4 to facilitate binding of tropoelastin to fibrillin-1 is weaker than that of fibulin-2 and -5. These results suggest that, in common with other fibulins, fibulin-4 may be involved in regulating tropoelastin assembly into elastic fibres. However, the fact that only fibulin-4-null mice do not produce elastic fibres [14] indicates the additional novel functions of fibulin-4 to account for its unique and indispensable role(s) in elastic-fibre formation.
In the present study we sought to determine such putative roles for fibulin-4 in elastogenesis using an in vitro model of human cells. We established a functional assay system in which fibulin-4 was knocked down in HFFs (human foreskin fibroblasts) using siRNA (small interfering RNA) technology. We found that a decrease in fibulin-4 in these cells led to the down-regulation of tropoelastin expression, with impaired elastic-fibre formation. To our knowledge, this is the first demonstration of a novel mechanism by which fibulin-4 can also regulate elastic-fibre formation, namely by controlling tropoelastin expression.
EXPERIMENTAL
Cell culture
Fibroblasts isolated from dissected human perinatal foreskin tissues were maintained in DMEM (Dulbecco’s modified Eagle’s medium) (Invitrogen) containing 10 % (v/v) FBS (fetal bovine serum) (Atlas Biological) at 37 °C under a 5 % CO2 atmosphere. The fibroblasts were collected by the Cleveland MetroHealth Hospital Perinatal Clinical Research Center (supported by National Institutes of Health Research Center award RR-00080). The use of this unidentified, discarded tissue was deemed by both the MetroHealth and the Cleveland Clinic Institutional Review Board to be exempt from patient consent requirements. HEK-293T (human embryonic kidney 293T) cells (A.T.C.C.) were also maintained in DMEM containing 10 % FBS with antibiotic/antimycotic (Invitrogen). rASMCs (rat aortic smooth-muscle cells) were isolated and kindly provided by Dr Donald Jacobsen and Dr Olga Stenina (Cleveland Clinic, Cleveland, OH, U.S.A.). They were grown and maintained in DMEM/F12 medium containing 10 % FBS with antibiotic/antimycotic. The HPCE (human pigmented ciliary epithelial) cell line was generously provided by Dr Miguel Coca-Prados (Department of Ophthalmology and Visual Science, Yale University School of Medicine, New Haven, CT, U.S.A.). The cells were grown in DMEM containing 10 %FBS with the antibiotic/antimycotic.
Production of lentivirus carrying fibulin-4-specific shRNAs (short-hairpin RNAs)
Two fibulin-4-specific shRNAs were cloned into pLKO.1 vector (Open Biosystems). pL335 targets sequence 1007–1027 (GenBank® accession no. BC010456). pL337 targets sequence 765–787 (GenBank® accession no. BC010456). To produce the recombinant lentiviruses, the shRNA plasmid was co-transfected into HEK-293T cells with the packaging plasmid psPAX2 (Tronolab) [28] and the plasmid VSV-G (vesicular-stomatitis-virus envelope glycoprotein) kindly provided by Dr Andrei Gudkov (Cleveland Clinic, Cleveland, OH, U.S.A.) [29] using Lipofect-amine™ 2000 (Invitrogen) according to the manufacturer’s instructions. At 48 h after transfection, the infectious lentiviruses were harvested and filtered through 0.22-μm-pore-size filters (Millipore).
Lentivirus titering
HFFs were seeded in six-well plates at 1.125 × 105 cells/well. The cells were infected with the virus stock at dilutions from 1 × 10−1 to 1 × 10−6. At 24 h after infection, the medium was replaced with fresh medium containing 1 μg/ml puromycin (Sigma–Aldrich). Colonies formed after 12 days were stained with Crystal Violet and their numbers were counted.
Knocking down fibulin-4 in HFFs and in rASMCs
HFFs at passages 7–10 were seeded overnight at a density of 9.2 × 105 per 100-mm-diameter culture plate, infected with 1.0 × 107 plaque-forming units of shRNA lentiviruses, and incubated for 24 h at 37 °C under 5 % CO2. The lentiviral medium was replaced with fresh medium containing 1 μg/ml puromycin. After 48 h the culture was trypsinized and expanded for experiments. The fibulin-4 was also knocked down, following the same protocol, in highly elastogenic rASMCs (passages 7–10) that were seeded at a density of 5.83 × 105 per 60-mm-diameter culture plate.
Real-time RT–PCR (reverse transcription–PCR)
Total RNA from the cultured fibroblasts or mouse aorta was isolated using RNeasy kit (Qiagen) according to the manufacturer’s instruction. RT reactions were performed using Taqman Reverse Transcription Reagents (Applied Biosystems). qPCR (quantitative PCR) were performed using SYBR Green PCR Master Mix (Applied Biosystems) and carried out in 7000 Sequence Detection System (Applied Biosystems). Quantification of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA levels in the same cDNA samples was used as an internal control. Primers used are listed in Table 1.
Table 1.
List of oligonucleotide primers used for real-time RT–PCR
| Species | Protein | Sequence (5′ → 3′) |
|---|---|---|
| Human | GAPDH-F | TCAACAGCGACACCCACTCC |
| GAPDH-R | TGAGGTCCACCACCCTGTTG | |
| Fibulin-4-F | ATGCGAGCAGCGCTGCTTCAA | |
| Fibulin-4-R | AGCCCCCATGGAAGTTGACAC | |
| Tropoelastin-F | GCTGACGCTGCTGCAGCCTA | |
| Tropoelastin-R | CAGCAAAAGCTCCACCTACA | |
| Tropoelastin pre-mRNA-F | CCAGCTACTTGGGAGGCTGA | |
| Tropoelastin pre-mRNA-R | CAGCAAAAGCTCCACCTACA | |
| LOX-F | GCAGATGTCAGAGATTATGATCA | |
| LOX-R | ATCGCCTGTGGTAGCCATAGT | |
| LOXL1-F | CAACTATGTGCAAGCATCCACT | |
| LOXL1-R: | GGTAATGCTGGTGGCAGCTGT | |
| Rat and mouse | Gapdh-F | CGGTGCTGAGTATGTCGTGGA |
| Gapdh-R | TGGCATGGACTGTGGTCATGA | |
| Rat | Fibulin-4-F | CACGGAATGCACAGATGGCTA |
| Fibulin-4-R | TGTCCACACAGCTCTCCTGTT | |
| Tropoelastin-F | TTAGGAGTCTCAACAGGTGCT | |
| Tropoelastin-R | CATTGGTATAGGGCAGTCCAT | |
| Mouse | Fibulin-4-F | CACGGAATGCACAGATGGCTA |
| Fibulin-4-R | CATCCACACAGCTCTCCTGTT | |
| Tropoelastin-F | CGGGTCTGACAGCGGTAGT | |
| Tropoelastin-R | CTCCAAGTCCTCCAGGACCT |
-F, forward; -R, reverse.
Data collection and analysis
All experiments using real-time RT–PCR were repeated at least three times with duplicate samples at each time and duplicate qPCRs for each sample. Relative RNA abundance was calculated using the Ct (threshold cycle value). All samples were corrected for total input RNA by normalizing to GAPDH mRNA. A ΔCt value was calculated by subtracting the Ct value of each gene from the Ct value of the internal control GAPDH. Relative RNA abundance to GAPDH mRNA (%GAPDH) was calculated by raising 2 to the power of ΔCt value. Relative RNA abundance to zero time, pLK or wild-type mouse aorta was calculated by dividing the %GAPDH of each sample by the %GAPDH from samples obtained at zero time or from pLK-infected cells or wild-type mouse aorta. Unless indicated otherwise, means ± S.D. were calculated from all data obtained. Student’s t test was used to calculate the P value. A P value of < 0.05 was considered significant.
mRNA stability
HFFs were seeded overnight in a six-well plate at 4.5 × 105 cells/well. Fresh medium containing 65 μM DRB (5,6-dichlorobenzimidazole riboside) (Sigma–Aldrich) was added to the culture (t=0). At the times indicated, the cells were washed twice with PBS, trypsinized to detach them, and used for real-time RT–PCR.
Pre-mRNA analysis
The abundance of tropoelastin pre-mRNA measured with real-time RT–PCR was used as an indicator of its transcriptional activity [30]. DNase-treated total RNA was isolated using RNeasy kits (Qiagen) according to the manufacturer’s instructions. RT reactions were performed using random hexamers. qPCRs were performed as described for real-time RT–PCR. The primers used (see Table 1) were designed to amplify sequences at an intron/exon junction.
Antibody production
To produce monoclonal antibodies against human fibulin-4, we first produced a recombinant N-terminal GST (glutathione transferase)–human fibulin-4 fusion protein. Using the template of a full-length human fibulin-4 cDNA (MHS1010-73567; Open Biosystems) and the primers h4fb169F-BamHI (5′-GGCCG-GATCCCAGGATTCTGAAGAGCCCGA-3′) and h4fb1416R-SalI (5′-GGCCGTCGACCGAAGGTGTAGGCCCCTACAAAG--3′), we used PCR to amplify the fibulin-4 cDNA without its natural leader sequence. The cDNA was in-frame and cloned into the BamHI/SalI sites of the vector pGEX4T-1 (Pharmacia Biotech) to make the recombinant construct. After sequence validation, the construct was transformed into Escherichia coli BL21 cells. After 4 h of 0.1 mM isopropyl β-D-1-thiogalactopyranoside induction, the GST–fibulin-4 protein was purified using the bulk and readipack GST purification modules (Amersham Pharmacia Biotech). The purified protein was injected into mice following a standard hybridoma fusion protocol for producing antibodies against human fibulin-4. The monoclonal antibody against fibulin-4 was made in the Hybridoma Core Facility, Lerner Research Institute, under the guidelines of an approved protocol from the institutional and national animal care committees. The specificity of the antibody against human fibulin-4 (5G11) was confirmed by Western-blot analysis. The 5G11 antibody recognized a single band on the Western-blot analysis of human fibroblast cellular extracts. The band intensity decreased significantly in the cellular extracts of fibroblast cultures in which fibulin-4 had been knocked down.
Preparation of conditioned medium from CHO (Chinese-hamster ovary) cells overexpressing human fibulin-4
A full-length human fibulin-4 cDNA expression construct was obtained by subcloning a PCR-amplified fibulin-4 cDNA without its natural leader sequence into the vector NT-pSecTag2 to produce a secretory protein with an N-terminal His6–c-myc epitope tag. The NT-pSecTag2 vector was modified from pSecTag2/HygroB (Invitrogen). To produce the modified vector, pSecTag2/HygroB was used as a template for PCR to include a His6 and c-myc epitope tag after the Igκ-chain leader sequence. PCR was performed using Taq DNA polymerase (Roche Diagnostics). The primers used were: NheI-F, 5′-GATCGCTAGCCACCATGGAGACAGACACA-3′; HindIII-Myc-His-R, 5′-GATCAAGCTTCAGATCCTCTTCTG-AGATGAGTTTTTGTTCATTGCTGGCGACGTCATGATGAT-GATGATGATGCGCGTCACCAGTGGAACCT-3′. Once amplified, the PCR fragment was digested with NheI and HindIII and subcloned into the corresponding sites in the pSecTag2/HygroB vector.
To produce an N-terminal His6–c-myc-tagged human fibulin-4 cDNA, human full-length fibulin-4 cDNA (Open Biosystems) was used as the template for PCR amplification with the following primers: h4f169F(KpnI), 5′-CTTCTC-CTGGTACCCAGGATTCTGAAGAGCCC GAC-3′; h4fb1419r-(XhoI-Stop), 5′-GTTTAAACCTCGAGTCAGAAGGTGTAGG-CCCCTA-3′.
PCR was performed using the Expand High Fidelity PCR system (Roche Diagnostics). The amplified PCR fragment was digested with KpnI and XhoI before subcloning into the corresponding sites in the NT-pSecTag2 vector. The resultant vector for human fibulin-4 with N-terminal His6–c-myc tag was further subcloned into pCDNA 3.1 (+) (Invitrogen) by digestion with, and ligation into, the BamHI and XhoI sites. This final fibulin-4 expression clone, pQC0701 (Figure 1), was transfected into CHO cells using Lipofectamine™ 2000 according to the manufacturer’s instructions. At 48 h after transfection, the medium was collected. The amount of fibulin-4 produced in the medium was estimated by an ELISA developed with our anti-fibulin-4 monoclonal antibody 5G11 and using our purified GST-fusion protein as a standard.
Figure 1. Partial amino acid sequence of the N-terminal His6–c-myc-tagged human fibulin-4 expressed by plasmid pQC0701.

The expressed protein contains an N-terminal Igκ-chain signal peptide (underlined sequence). The predicted signal peptide cleavage site is indicated by a filled triangle. The N-terminus of the cleaved polypeptide has a His6 tag and a c-myc epitope tag (italicized and bold). The predicted mature form of human fibulin-4 polypeptide was placed C-terminal to the myc epitope in parenthesis. Only eight amino acid residues from both the N-terminus and the C-terminus of human fibulin-4 are shown here.
Western-blot analysis
Conditioned medium (30 μl) from the pQC0701 transfected CHO cells or from the HFF culture was treated with 5 % (v/v) 2-mercaptoethanol or 100 mM DTT (dithiothreitol) and separated using SDS/7.5 %-(w/v)-PAGE. To prepare cell lysates from HFFs, the cells were cultured for 3 days after reaching confluency, rinsed with PBS, scraped off, pelleted and lysed with a buffer containing 50 mM Tris/HCl, pH 7.4, 150 mM NaCl, 1 % (v/v) Nonidet P40, 0.5 % (w/v) deoxycholate and 0.1 % (w/v) SDS in the presence of 2 mM PMSF and 1 × proteinase inhibitor cocktail (catalogue number, 11697498001; Roche Applied Science). After centrifugation for 10 min at 4 °C at 16 000 g, 50 μg of protein from the supernatant was mixed with the SDS loading buffer containing 100 mM DTT and separated on the acrylamide gel. Proteins on the gel were electrophoretically transferred on to an Immobilon-P membrane (Millipore) and probed with monoclonal antibody specific for human fibulin-4 (5G11). After incubation with the primary antibody, goat anti-mouse horseradish-peroxidase-conjugated IgG (Upstate Biotechnology) was added. Immune complexes were detected by Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences).
WBS (Williams–Beuren syndrome) fibroblast culture and treatment
With patient’s family consent and Institutional Ethics Committee approval from the Hospital for Sick Children in Toronto, fibroblasts were isolated from the skin biopsy as previously described [31,32] from a 2-year-old male patient with confirmed WBS chromosome deletion. WBS is characterized by a heterozygous deletion in chromosome 7 that harbours more than 20 genes, including elastin. [33,34]. These WBS fibroblasts displaying haploinsufficiency of the elastin gene were seeded in six-well plates (1 × 105 cells/well) and initially maintained in DMEM containing 10 %FBS until they reached confluency. The cultures were then incubated for 24 h in a medium consisting of equal volumes of DMEM containing 4 % FBS and CHO-cell conditioned medium that was harvested either from cultures of control CHO cells or CHO cells overexpressing human fibulin-4. The concentration of fibulin-4 in the medium tested was estimated to be 10 ng/ml by ELISA. At the end of the incubation period, the total RNA was extracted from all cultures and used for semi-quantitative RT–PCR to detect changes in elastin gene expression. The parallel cultures plated on coverslips were kept for additional 4 days and then fixed with ice-cold methanol and immunostained with a specific anti-elastin antibody as previously described [29].
Semi-quantitative RT–PCR
RT–PCRs (25 μl volume) were performed using OneStep RT–PCR kits (Qiagen) according to the manufacturer’s instructions and as described by Bunda et al. [35]. The RT step was performed for elastin and GAPDH reactions at 50 °C for 30 min, followed by 15 min at 95 °C. The elastin PCR (sense primer: 5′-GGTGCGGTGGTTCCTCAGCCTGG-3′; antisense primer: 5′-GGGCCTTGAGATACCCCAGTG-3′; designed to produce a 255 bp product) was performed under the following conditions: 25 cycles of 94 °C for 20 s, 63 °C for 20 s and 72 °C for 1 min, followed by a final extension for 10 min.
The GAPDH PCR (sense primer: 5′-TCCACCACCCTG-TTGCTGTAG-3′; antisense primer: 5′-GACCACAGTCCAAT-GCCATCACT-3′; designed to produce a 450 bp product) was performed under the following conditions: 21 cycles of 94 °C for 20 s, 58 °C for 30 s and 72 °C for 1 min, followed by a final extension at 72 °C for 10 min. Samples (5 μl) of the elastin and GAPDH PCR products from each reaction were separated on 1.2 %(w/v) agarose gels and stained with ethidium bromide. The amount of tropoelastin mRNA was normalized against the amount of GAPDH mRNA.
Immunostaining
Immunostaining of proteins of interest was performed as described by Bunda et al [35]. Cultured cells, grown on glass coverslips, were rinsed twice with PBS, fixed in cold methanol at − 20 °C for 30 min, blocked with 3 % (v/v) goat serum (Sigma–Aldrich) for 1 h at room temperature (22 °C), incubated for 2 h at room temperature with polyclonal antibodies against human elastin or fibrillin-1 (Elastin Product Company) or monoclonal antibody against human fibulin-4 (5G11), followed by a 1 h incubation at room temperature with either Alexa Fluor® 568–anti-rabbit IgG or with Alexa Fluor® 488–anti-mouse IgG (Invitrogen). The cultures were mounted in Vectashield mounting medium with DAPI (4′,6-diamidino-2-phenylindole) (Vector) and subjected to either confocal or fluorescence microscopy.
For florescence microscopy, the samples were examined with Olympus IX81 motorized inverted microscope that was mounted with Hamamatsu ORCA-ER digital camera and fitted out with an Olympus BH2-RFL-T3 100 W high-pressure mercury lamp burner. For confocal microscopy, the samples were examined with a Leica-Microsystems TCS-SP2 AOBS confocal microscope using an HCX PL APO 63 × -magnification/1.4-numerical-aperture oil-immersion lens. DAPI-stained nuclei were excited with the 351 nm line of an argon laser and emitted light was collected from 400 to 480 nm. Alexa Fluor® 488-labelled fibulin-4 was imaged using the 488 nm line of a krypton laser with emitted light collected from 500 to 550 nm. Alexa Fluor® 568-labelled elastin fibres or fibrillin-1 microfibrils were imaged using the 568 nm line of a krypton laser with emitted light collected from 575 to 630 nm. The amount of elastin in each image was measured using ImageProPlus 6.2 software (Media Cybernetics) to calculate the intensity of staining.
For immunostaining performed on the WBS cultures, the coverslips were blocked with 1 % goat serum, incubated for 1 h with polyclonal antibody against elastin, which also detects insoluble extracellular elastin (Elastin Product Company), followed by an 1 h incubation with goat anti-rabbit serum–FITC conjugated secondary antibody as described by Hinek et al. [36]. Nuclei were counterstained with propidium iodide. The pre-immune rabbit IgG and the secondary antibody alone were used as controls for immunostaining specificity. The cultures were mounted in Elvanol and examined with a Nikon Eclipse E1000 microscope with a Retiga EX cooled charge-coupled-device camera (Qimaging) and a video analysis system (Image-Pro Plus software; Media Cybernetics).
For immunostaining of mouse aortic tissues, embryonic day 13.5 (E13.5) embryos were isolated, fixed overnight at room temperature in Excel Plus (American Master Tech Scientific), dehydrated and embedded in paraffin. Sections of thickness 10 μm were de-paraffinized, rehydrated, blocked for 1 h with the freshly prepared 10 % goat serum and 1 % BSA in PBS, and incubated overnight at 4 °C with the polyclonal antibody against mouse tropoelastin PR385 (Elastin Products Company), followed by 1 h incubation at room temperature with Alexa Fluor® 488–anti-mouse IgG (Invitrogen). The samples were mounted in Vectashield mounting medium (Vector Laboratories) and analysed by fluorescence microscopy.
Mice
Mice were housed in the Biological Resources Unit of the Cleveland Clinic. Fibulin-4 knockout mice (fibulin-4−/−) were obtained from Deltagen. Fibulin-4-knockdown mice (fibulin-4R/R) were generated as previously described [37]. PCR was used for genotyping. Timed pregnant mice were killed by CO2 asphyxiation and used for E13.5 embryo isolation. Postnatal 10-day-old mice were killed by cervical dislocation and used to isolate aortas.
RESULTS
Knocking down fibulin-4 in HFFs impairs elastic-fibre formation
HFFs are one of the major elastogenic cells that are frequently used in studies of elastic-fibre formation. Since elastic-fibre formation in cell-culture systems varies significantly with the age of the donors and the tissue origin of the cells [38], we have established HFF cultures from foreskins of newborns which were highly elastogenic. Using immunostaining analyses for elastic fibres, we were able to detect fine fibres the next day after subculture if the cells contacted each other (Supplementary Figure S1, D1 at http://www.BiochemJ.org/bj/423/bj4230079add.htm). Considerably more elastic fibres could be detected by day 3 after subculture (Supplementary Figure S1, D3). By day 7, extensive elastic fibres could be visualized (Supplemenatry Figure S2A at http://www.BiochemJ.org/bj/423/bj4230079add.htm). The abundance of elastic fibres produced in the newborn foreskin fibroblasts was comparable with that produced in the hypertrophic scar fibroblasts (Supplementary Figure S2B) and much greater than that in adult skin fibroblasts (Supplementary Figure S2C). On day 3, even with cells at passage 10 or greater, we were able to detect elastic fibres in the cultures. These characteristics suggested that we had identified an excellent system for in vitro elastogenesis studies.
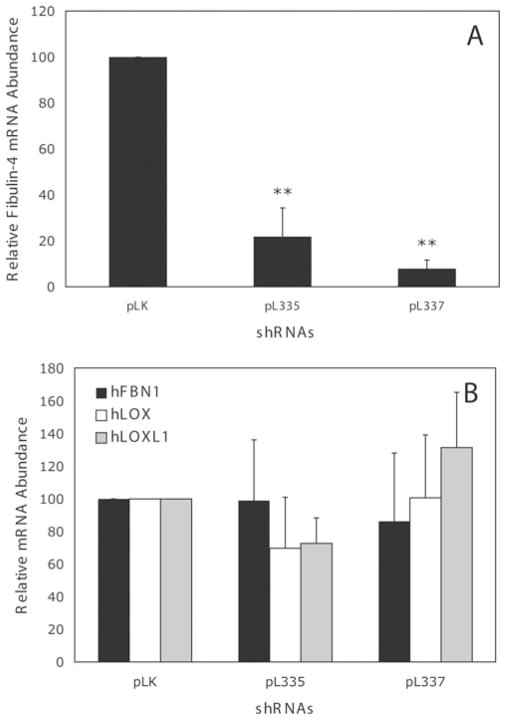
To determine the role of fibulin-4 in elastogenesis, we chose to knock down fibulin-4 in the HFF cultures using siRNA technology. We infected the fibroblasts with lentivirus expressing fibulin-4-specific shRNAs and analysed the effect on the fibulin-4 mRNA level using real-time RT–PCR. Compared with the amount of fibulin-4 mRNA produced in the control vector pLK virus-infected cells, HFFs infected with the fibulin-4-shRNA-producing viruses (pL335 and pL337) had a significantly decreased amounts of fibulin-4 mRNA (Figure 2A). Infecting the cells with pL335 or pL337 viruses resulted in an 80 and 90 % decrease respectively in fibulin-4 mRNA. However, no statistically significant difference (P > 0.05) in the mRNA level of several elastic-fibre-associated proteins, including fibrillin-1, LOX, and LOXL1 was evident among the cells infected with pLK, pL335 and pL337 (Figure 2B). Similarly, the GAPDH mRNA level in the cells, which was used as an internal control, remained the same in all cultures exposed to different viruses. These results suggest that fibulin-4 shRNAs specifically knocked down the expression of fibulin-4 mRNA in HFFs.
Figure 2. Relative mRNA abundance in fibroblasts infected with control pLK viruses or fibulin-4 shRNA viruses pL335 and pL337.
Infecting HFFs with pL335 or pL337 viruses leads to a significant decrease in fibulin-4 mRNA (A), with no statistically significant change in the mRNA level (B) of fibrillin-1 (hFBN1), lysyl oxidase (hLOX), and lysyl oxidase-like-1 (hLOXL1). Results are means ± S.D. for six to eight samples. **P < 0.05 compared with the control pLK.
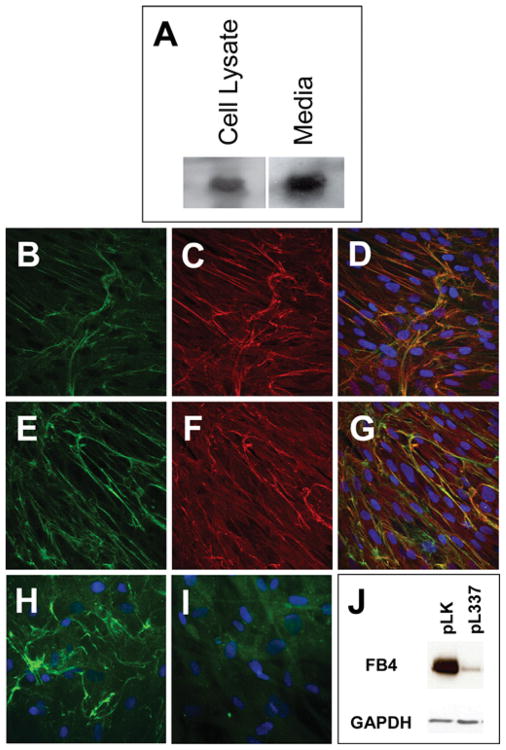
To confirm that fibulin-4 shRNAs also knocked down fibulin-4 protein in HFFs, we performed Western-blot analysis of the cultured HFF cells using antibodies specific for fibulin-4. As shown in Figure 3(A), fibulin-4 was present in the cultured medium as well as in the cell lysate. To determine whether fibulin-4 was present in the ECM, we performed immunostaining assay of the HFF culture. Figures 3(B) and 3(E) show that fibulin-4-specific antibody decorated fibrous structures in the culture. Double staining with antibodies against either elastin (Figure 3C) or fibrillin-1 (Figure 3F) showed that fibulin-4 co-localized in ECM with the elastic fibres (Figure 3D) and the fibrillin microfibrils (Figure 3G). In the presence of pL337 virus, fibulin-4 staining was significantly decreased (Figure 3I). This decrease was confirmed by Western blots of the cultured HFF cells (Figure 3J). Taken together, these results demonstrate that knocking down fibulin-4 decreased not only fibulin-4 mRNA, but also protein deposition in the ECM.
Figure 3. Characterization of fibulin-4 produced in HFF cells.
(A) Western-blot analysis of fibulin-4 in HFF cell lysate and cultured medium. (B–G) Confocal images of passage-10 HFF cultures immunostained with antibodies against human fibulin-4 (B, E), human elastin (C) and human fibrillin-1 (F). Co-localization of fibulin-4 with elastin and fibrillin-1 are shown in the merged images, (D) and (G) respectively. (H, I) Fluorescent images of HFF cultures infected with pLK virus (H) or pL337 virus (I) and immunostained with fibulin-4 antibody. (J) Western-blot analysis of fibulin-4 (FB4) in HFF cells infected with the control pLK lentivirus or the fibulin-4 shRNA-producing lentivirus pL337. Note that pL337 infection led to the abrogation of fibulin-4 staining in the ECM (I) and a significant decrease in the amount of fibulin-4 in the HFF cells (J).
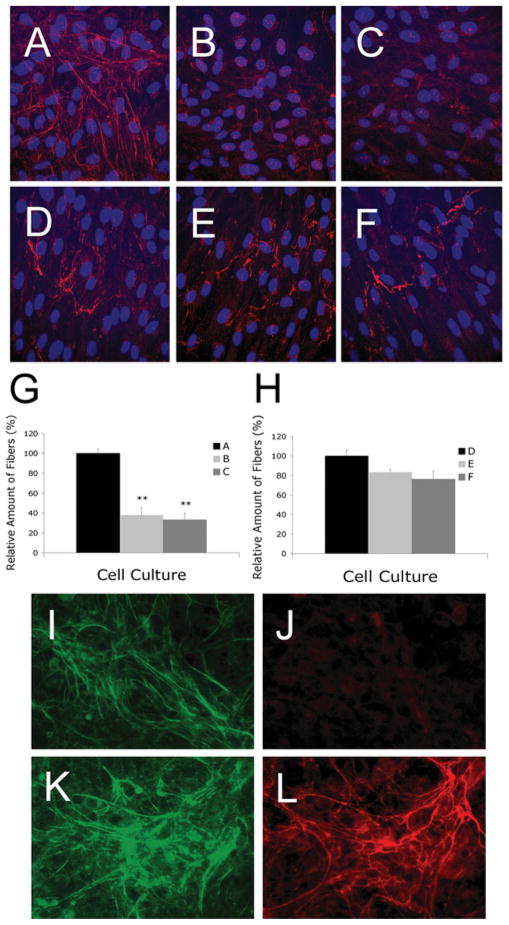
To determine the effects of knocking down fibulin-4 on elastic-fibre formation in HFFs, we performed immunostaining. Using an antibody raised to human elastin, an extensive network of elastic fibres was detected in the 3-day-old culture of fibroblasts infected with the control pLK virus (Figure 4A). However, only sparse elastic fibres were detected in cultures infected with shRNAs, pL335 (Figure 4B) or pL337 (Figure 4C). Quantification of immunodetectable elastin showed at least a 3-fold decrease in its contents in the cultures infected with fibulin-4 shRNA viruses (Figure 4G). To confirm the effects were specific for elastic fibres, we performed the following immunostaining experiments. First, we stained the cultures with antibodies against fibrillin-1, one of the major components of microfibrils. No statistically significant difference in the microfibril networks were present in cultures infected with all three viruses (Figures 4D–4F and 4H). Secondly, we stained immortalized HPCE cells with antibodies against elastin, fibrillin-1 and fibulin-4. HPCE cells expressed fibrillin-1 and fibulin-4, but not tropoelastin [39]. Fibulin-4 and fibrillin-1 antibodies stained their respective target proteins, whereas anti-elastin antibody failed to stain any fibres in the cultures (Figures 4I–4L). Thus the antibodies used in the present study were specific and did not cross-react with each other. Furthermore, the deposition of another matrix protein (fibronectin) that is not associated with elastic fibres was not affected by fibulin-4 knockdown (results not shown). These results suggest that down-regulation of fibulin-4 specifically impairs elastic-fibre formation.
Figure 4. Representative images of HFFs immunostained with antibodies against elastin (A, B and C) or fibrillin-1 (D, E and F) and HPCE cells immunostained with antibodies against fibulin-4 (I, K), elastin (J) or fibrillin-1 (L).
HFF cells were seeded on to glass coverslips in a 12-well plate with 1.8 × 105 cells/well and were allowed to grow for 3 days before immunostaining. Fibroblasts infected with pLK control viruses (A) produced extensive elastic fibres, whereas fibroblasts infected with fibulin-4 shRNA viruses pL335 (B) and pL337(C) produced significantly fewer elastic fibres. No statistically significant difference in the production of fibrillin-1 microfibrils was present among fibroblast cultures infected with pLK (D), pL335 (E) and pL337 (F). (G) Quantification of elastic fibres taken from at least four fields of multiple cultures represented in (A), (B) and (C). (H) Quantification of microfibrils taken from four fields of multiple cultures represented in (D), (E) and (F). **P < 0.05 compared with the respective control pLK. (I–L) HPCE cells were seeded on to glass coverslips in a 12-well plate with 4 × 105 cells/well and were allowed to grow for 7 days before immunostaining. The cultures produced significant amounts of fibulin-4 (I, K) and fibrillin-1 (L). However, no elastic fibres were detectable (J).
Decrease in tropoelastin mRNA in fibulin-4-deficient HFFs and rASMCs
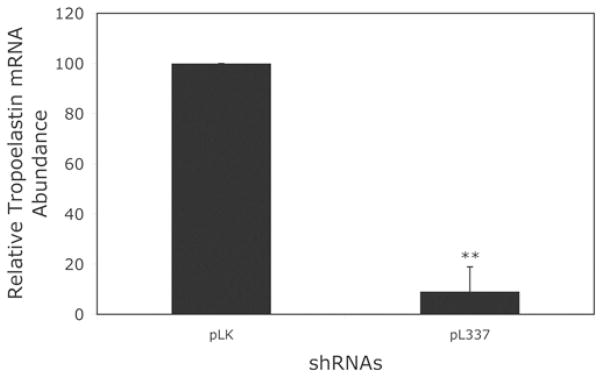
Tropoelastin is the soluble precursor of elastin polymers. To determine whether knocking down fibulin-4 affects tropoelastin expression, we performed a real-time RT–PCR analysis of tropoelastin mRNA in HFF cultures exposed to pL335 or the control pLK virus. Figure 5 shows that exposing these cells to pL335 resulted in a significant decrease in tropoelastin mRNA levels. Likewise, infection of parallel cultures with another fibulin-4 shRNA virus pL337 targeting a different site on fibulin-4 mRNA also resulted in a significant decrease in the tropoelastin mRNA level (Figure 5). Moreover, three base mutations to inactivate pL337 shRNA against fibulin-4 were associated with a significant decrease in its anti-elastogenic effect. Since the target sequence for pL337 is exactly the same between the human and the rat fibulin-4, we also tested the effects of pL337 on rat tropoelastin expression. We observed a significant decrease in the tropoelastin mRNA level in cultured rASMCs upon fibulin-4 knockdown with the pL337 shRNA (Figure 6). Taken together, these results suggest that knocking down fibulin-4 leads to down-regulation of tropoelastin mRNA in cultured HFFs and ASMCs.
Figure 5. Relative tropoelastin mRNA abundance in fibroblasts infected with control pLK viruses or fibulin-4 shRNA viruses pL335 and pL337.
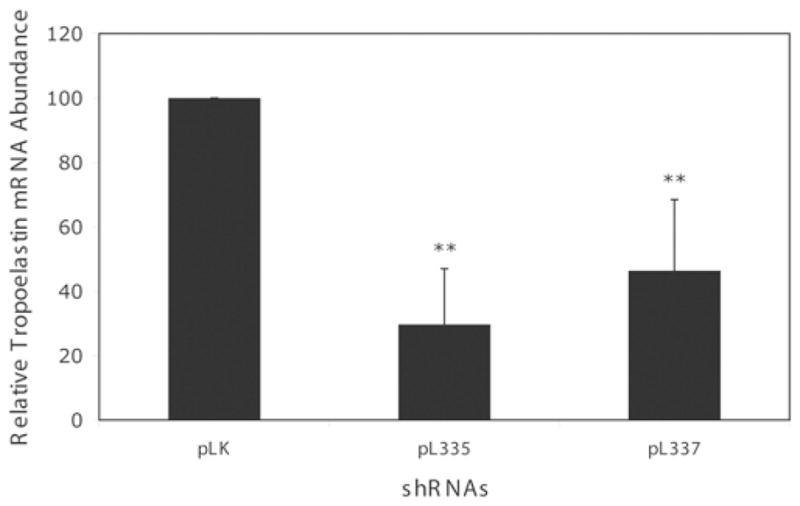
Infecting HFFs with either pL335 or pL337 viruses led to a significant decrease in tropoelastin mRNA. Results are means±S.D. for six to eight samples. **P < 0.05 compared with the control pLK. The results are based on eight samples from three experiments. Some variability in the magnitude of tropoelastin mRNA expression was observed and later traced to the batches of FBS used.
Figure 6. Relative tropoelastin mRNA abundance in rASMCs infected with control pLK viruses or fibulin-4 shRNA virus pL337.
Infecting rASMCs with pL337 virus leads to a significant decrease in tropoelastin mRNA, as analysed by real-time RT–PCR. Results are means ± S.D. for eight samples. **P < 0.05 compared with the control pLK.
Down-regulation of tropoelastin expression in the aorta of fibulin-4-knockdown and fibulin-4-knockout mice
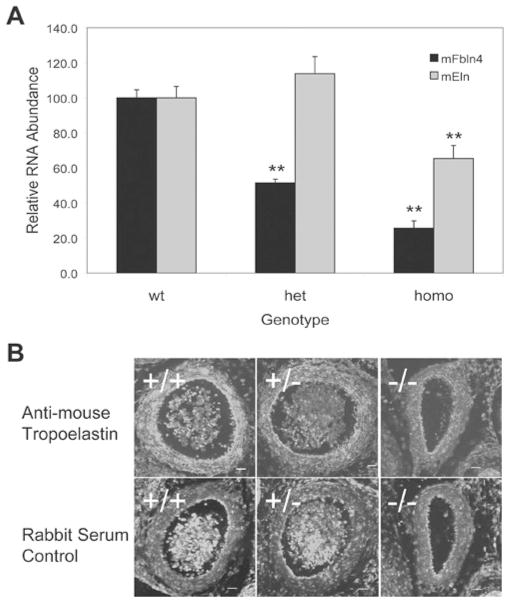
We have previously reported that down-regulation of fibulin-4 in mice, knockout (fibulin-4−/−) or knockdown (fibulin-4R/R), significantly attenuates elastic-fibre formation [14,37]. Depending on the dosage of fibulin-4 expressed in the mice, various defects in the aortic elastic laminae were manifest, ranging from disorganization in fibulin-4+/R mice and fragmentation in fibulin-4R/R mice to the complete absence in fibulin-4−/− mice. In addition to the defective elastic laminae, histological analysis of 10-day-old fibulin-4R/R mice revealed an increase in ASMC proliferation. Since knocking down tropoelastin in mice also induced the proliferation of ASMCs, we decided to determine if tropoelastin mRNA level deceases in the aorta of fibulin-4R/R mice. Real-time RT–PCR analysis of the aortic RNAs showed that the amount of tropoelastin mRNA in the fibulin-4R/R mouse aorta did decrease to approx. 50 % of the amount present in the wild-type aorta (Figure 7A).
Figure 7. Decreased tropoelastin expression in the aorta of fibulin-4-knockdown and fibulin-4-knockout mice.
(A) Real-time RT–PCR analysis of fibulin-4 (mFbln4) and tropoelastin (mEln) mRNA in the aorta of 10-day-old fibulin-4-knock-down mice. Results are means ± S.E.M. for nine to eleven samples. **P < 0.05 compared with the wild-type (wt) littermates. het, heterozygous; homo, homozygous. (B) Immunostaining images of E13.5 aorta isolated from fibulin-4-knockout mice. Top panel, stained with tropoelastin antibodies; Bottom panel, stained with control rabbit serum. The experiment was repeated at least three times with consistent results.
Unlike fibulin-4R/R mice, fibulin-4−/− mice die by P2 (postnatal day 2). Using lung RNAs from the P1 mice, our Northern-blot analysis did not show a decrease in the tropoelastin mRNA level in the fibulin-4−/− mice [14]. To clarify the effects of fibulin-4 knockout on tropoelastin expression, we performed immunostaining on the aorta from fibulin-4+/+, fibulin-4+/− and fibulin-4−/− mice with antibodies against tropoelastin, which identify the precursor tropoelastin and not the insoluble elastic laminae. In a previous study we showed that E13.5 was the earliest developmental stage at which elastic laminae were detectable in mice [14]. At this stage, elastic laminae started to form only under the endothelial cell layer and not between the smooth-muscle cell layers. From E14.5 and thereafter, as the concentric elastic laminae start to form between the smooth-muscle cell layers, the level of immunostaining would not reveal the extent of tropoelastin expression because of the lack of the antibody’s reactivity with insoluble elastin. Therefore we chose E13.5 to perform the immunostaining. As shown in Figure 7(B), immunostaining of the E13.5 mouse aorta with antibodies against mouse tropoelastin decorated most ASMC layers in the fibulin-4+/+ mice. The staining was significantly less robust in the fibulin-4−/− aorta. Compared with that of fibulin-4+/+ aorta, the staining of fibulin4+/− and fibulin-4−/− aorta decreased approx. 50 and 80 % respectively. Taken together, these in vivo results support our in vitro findings that fibulin-4 plays a role in regulating tropoelastin expression.
Down-regulation of tropoelastin transcriptional activity in fibulin-4-deficient fibroblasts
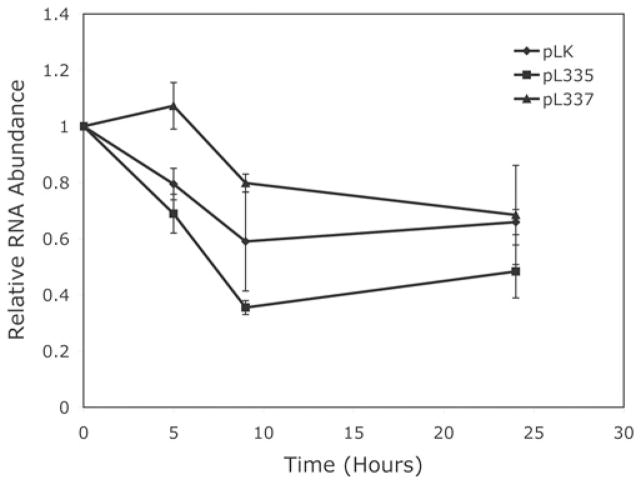
Steady-state mRNA levels are controlled both by transcriptional activity and post-transcriptional mRNA stability. To determine whether the decrease in tropoelastin mRNA in the fibulin-4-knockdown cells was caused by a decrease in mRNA stability, we treated HFFs with the RNA polymerase II-specific inhibitor DRB (65 μM) [40] after infection with the shRNA lentivirus and compared the rates of tropoelastin mRNA degradation. Results of the time-course assay shown in Figure 8 indicate that, in the presence of DRB, tropoelastin mRNA decreased gradually in the cultures infected with either the control viruses (pLK) or the fibulin-4 shRNA viruses (pL335 or pL337). The rate of tropoelastin mRNA degradation was not significantly different among the cultures. These results suggest that the decrease in tropoelastin mRNA level in the fibulin-4-shRNA-treated cells was not caused by an acceleration of mRNA degradation.
Figure 8. Changes over time of relative tropoelastin mRNA abundance in fibroblasts infected with control pLK virus or fibulin-4 shRNA viruses pL335 and pL337.
The rate of tropoelastin mRNA degradation in pL335- or pL337-infected fibroblasts was not significantly different from that in pLK-infected cells as determined by Student’s t test. Results are means ± S.D. for five to eight samples.
To determine whether knocking down fibulin-4 would affect tropoelastin transcription, we performed real-time RT–PCR analysis to determine tropoelastin pre-mRNA levels. The abundance of tropoelastin pre-mRNA has been used as an indicator of tropoelastin transcriptional activity [30]. Analysis of tropoelastin pre-mRNA in fibroblasts infected with either pL335 or pL337 viruses showed a significant decrease in message levels as compared with that of cells infected with control pLK viruses (Figure 9). Taken together, the above-presented results indicate that knocking down fibulin-4 leads to a down-regulation of tropoelastin transcription.
Figure 9. Relative amount of tropoelastin pre-mRNA in fibroblasts infected with control pLK viruses or fibulin-4 shRNA viruses pL335 and pL337.
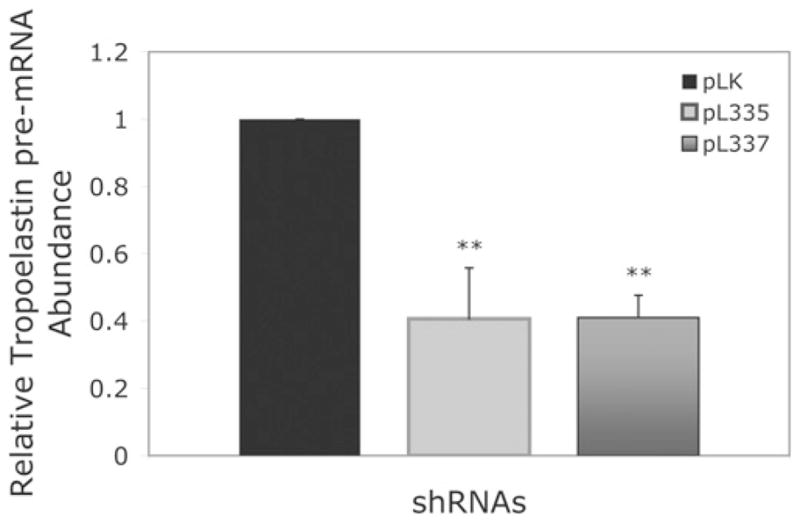
Fibroblasts infected with pL335 or pL337 produced a significant decreased amount of tropoelastin pre-mRNA compared with that in the pLK-infected cells, indicating that the transcriptional activity of tropoelastin significantly decreased in the pL335- or pL337-infected cells as compared with that in the pLK-infected cells. Results are means ± S.D. for four or five samples. **P < 0.05 compared with the control pLK.
Fibulin-4 stimulates tropoelastin expression and elastic-fibre formation in WBS skin fibroblasts
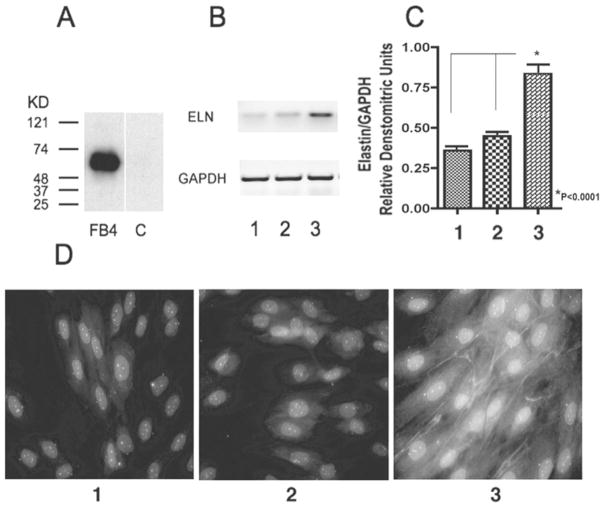
To determine whether exogenous fibulin-4 could affect elastogenesis, we tested elastin gene expression and elastic-fibre formation in cultures of skin fibroblasts derived from WBS patients characterized with haploinsufficiency of the elastin gene [32]. We found that WBS fibroblasts maintained for 5 days, either in DMEM containing 2 %FBS or in the medium consisting of equal volumes of DMEM and conditioned medium from cultures of control CHO cells, did not form elastic fibres (Figures 10D, panel 1 and 10D, panel 2). By contrast, the parallel cultures of WBS fibroblasts maintained in medium consisting of equal volumes of DMEM and conditioned medium from cultures of CHO cells overexpressing fibulin-4 (final concn. of fibulin-4: 10 ng/ml) demonstrated a significant formation of immunodetectable elastic fibres (Figure 10D, panel 3).
Figure 10. Effects of fibulin-4 on the elastogenesis of WBS fibroblasts.
(A) Western-blot analysis of fibulin-4 in CHO-cell conditioned medium. Fibulin-4 was detected only in the conditioned medium (lane FB4) of fibulin-4-overexpressing cells and not in the conditioned medium (lane C) of control cells. (B, C) Expression of tropoelastin and GAPDH mRNAs in the WBS fibroblast cultures in DMEM with 2 % FBS (lane 1), in conditioned medium from control CHO cells mixed with equal volume of DMEM with 2 % FBS (lane 2) or in conditioned medium from CHO cells overexpressing fibulin-4 cells mixed with equal volume of DMEM with 2 % FBS (lane 3). Fibroblasts exposed to fibulin-4-overexpressing conditioned medium demonstrated a significantly higher level of tropoelastin mRNA. The experiment was repeated three times. **P < 0.0001 compared with lane 1 or lane 2. (D) WBS fibroblasts maintained for 5 days either in DMEM containing 2 % FBS or in the medium consisting of equal volumes of DMEM and conditioned medium from control CHO cell cultures did not form elastic fibres (D, panel 1) and (D, panel 2). In contrast, the parallel cultures of WBS fibroblasts maintained in medium consisting of equal volumes of DMEM and conditioned medium from cultures of CHO cells overexpressing fibulin-4 (final concn. of fibulin-4: 10 ng/ml) demonstrated a significant formation of immunodetectable elastic fibres (D, panel 3).
Semi-quantitative RT–PCR analysis demonstrated a significant increase in levels of tropoelastin mRNA in 24-h-old cultures of WBS fibroblasts exposed to conditioned medium containing fibulin-4 as compared with both control groups (Figures 10B and 10C). These results further support the stimulatory role of fibulin-4 in elastogenesis, which in WBS fibroblasts affected the existing ELN (human elastin gene) allele.
DISCUSSION
Our studies were aimed to determine whether fibulin-4 also regulates elastogenesis in human cells, and, if so, to elucidate the mechanism underlying such regulation. Accordingly, we applied siRNA technology to knock down fibulin-4 in human foreskin fibroblasts. Using this approach, we have demonstrated that fibulin-4 is required for elastic-fibre formation in human cells. Down-regulation of fibulin-4 in the fibroblasts results in a decrease in tropoelastin mRNA level. Consistent with these in vitro results, fibulin-4 deficiency in mice also led to a decrease in tropoelastin expression. As the expression of tropoelastin in many in vivo and in vitro systems, including human skin fibroblasts, is primarily controlled by the abundance of tropoelastin mRNA [41–44], these results suggest that a decrease in fibulin-4 causes down-regulation of tropoelastin gene expression and impaired elastic-fibre formation. In this respect, fibulin-4 differs from two other elastic-fibre-associated proteins, MAGP1 (microfibril-associated glycoprotein-1) and fibrillin-2, which when knocked down lead to a significant impairment in elastic-fibre formation [45], but not to a primary decrease in tropoelastin mRNA levels. Furthermore, as independent support for these observations, the conditioned medium from CHO cells overexpressing fibulin-4 stimulated elastic-fibre formation and an increase in tropoelastin expression by WBS skin fibroblasts, which are inefficient in making elastic fibres. By contrast, it has been previously shown that addition of fibulin-5 to cultured skin fibroblasts enhances elastic-fibre formation, but does so without affecting tropoelastin mRNA levels [46].
How fibulin-4 executes this unique function in regulating tropoelastin expression is presently unknown. Knocking down fibulin-4 in mice enhances TGF-β (transforming growth factor-β) signalling [37], and TGF-β plays pleiotropic roles in regulating ECM structure and composition [47–50]. Cellular overproliferation tends to accompany tropoelastin deficiency [51,52]. Thus, as a future study, it would be interesting to investigate whether fibulin-4 regulates tropoelastin expression directly or indirectly through regulatory factors such as TGF-β. It should be noted that, in addition to being secreted to the ECM, fibulin-4 may be present intracellularly as a shorter isoform [53]. Functional co-ordination between the intracellular fibulin-4 and the extracellular fibulin-4 might be required to regulate tropoelatstin mRNA. We were unable to stimulate tropoelastin production by addition of extracellular fibulin-4 to fibulin-4-knockdown cells (results not shown) and requisite balance between intracelullar and extracellular fibulin-4 may explain why such experiments were not successful.
The fibulin-4 gene is located on chromosome 11q13 [53,54], a region associated with several pathologies, including inherited retinopathies and disorders of bone homoeostasis [55–58]. This is also a region of frequent amplification, translocations and other rearrangements that are associated with tumorigenesis. Indeed, fibulin-4 was initially identified as a mutant p53-binding protein [59]. Its potential oncogenic roles have been implicated in promoting cell growth and in its aberrant expression in several human colon tumours [53]. As a member of the fibulin family that is closely associated with elastic fibres, the critical role of fibulin-4 in elastic-fibre formation was first demonstrated in fibulin-4-knockout mice that fail to make elastic fibres [14]. Subsequent report on fibulin-4-knockdown mice suggested the perturbation of TGF-β signalling associated with the significant defects in elastic-fibre formation [37]. Recently, mutations in fibulin-4 have been identified as a cause of cutis laxa phenotype, a connective-tissue disorder in which the skin becomes inelastic [60,61]. Interestingly, impaired elastin deposition or decreased tropoelastin expression have also been mechanistically linked with tumour progression (Costello syndrome [62,63]) and with heightened proliferation of vascular smooth-muscle cells leading to arterial occlusions (WBS and supravalvular aortic stenosis [51,64]). These studies together show the potential multifunctional roles that fibulin-4 may play. The finding that is reported in the present paper further reveals a previously unappreciated role of fibulin-4, namely the regulation of tropoelastin expression during elastic-fibre formation.
In summary, the results presented here provide the first experimental evidence that fibulin-4, regarded as a structural protein of the extracellular matrix, should also be considered as a regulatory factor required for normal elastin gene expression in human fibroblasts. This newly discovered functional mechanism suggests potential pathophysiological links between the modulation of fibulin-4 levels and diseases characterized with impaired elastic-fibre formation.
Supplementary Material
Acknowledgments
We thank Dr Barry Starcher (The University of Texas Health Science Centre at Tyler) and Dr Alana Major (Cleveland Clinic) for sharing their expertise in analysing elastic fibres in cultured HFFs. We thank Lori Mavrakis (Cleveland Clinic) for help in establishing cell cultures and Yiqing Jiang (Cleveland Clinic) for technical assistance. We thank Earl Poptic for making the monoclonal antibody against human fibulin-4. The confocal images were captured and analysed by Dr Judy Drazba of the Imaging Core of the Lerner Research Institute, Cleveland Clinic, Cleveland, OH, U.S.A. Human foreskin tissues were harvested through the Birthing Services Department at the Cleveland Clinic and the Perinatal Clinical Research Center at the Cleveland MetroHealth Hospital.
FUNDING
This work was supported by the American Heart Association [grant number 0630193N (to Q. C.)]; the American Health Assistance Foundation [grant number M2006-020 (to Q. C.)]; the Lijf en Leven Foundation, The Netherlands (to J. E.); the Canadian Institute of Health research [grant number PG 13920 (to A. H.)]; the Heart and Stroke Foundation of Ontario [grant number NA 5435 (to A. H.)]; the National Institutes of Health [grant numbers HL17964 (to E. F. P.), RR-00080 (Research Center Award to P. E. D.)].
Abbreviations
- cbEGF
calcium-binding epidermal growth factor
- CHO
Chinese-hamster ovary
- Ct
threshold cycle value
- DAPI
4′,6-diamidino-2-phenylindole
- DMEM
Dulbecco’s modified Eagle’s medium
- DRB
5,6-dichlorobenzimidazole riboside
- DTT
dithiothreitol
- E13.5 (etc.)
embryonic day 13.5 (etc.)
- EBP67
67 kDa elastin-binding protein
- ECM
extracellular matrix
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GST
glutathione transferase
- HEK-293T
human embryonic kidney 293T
- HFF
human foreskin fibroblast
- HPCE
human pigmented ciliary epithelial
- LOX
lysyl oxidase
- LOXL1
LOX-like-1
- Neu1
neuraminidase-1
- P2 (etc.)
postnatal day 2 (etc.)
- PPCA
protective protein–cathepsin A
- qPCR
quantitative PCR
- (r)ASMC
(rat) aortic smooth-muscle cell
- RT–PCR
reverse transcription–PCR
- shRNA
short-hairpin RNA
- siRNA
small interfering RNA
- TGF-β
transforming growth factor-β
- WBS
Williams–Beuren syndrome
Footnotes
AUTHOR CONTRIBUTION
Qiuyun Chen directed the overall study, performed the experiments to obtain data, analysed data and prepared the manuscript for publication. Teng Zhang performed experiments to obtain data and analysed data during revision. Joseph Roshetsky performed experiments to obtain data and analysed data. Zhufeng Ouyang performed experiments to obtain data and analysed data. Jeroren Essers provided fibulin-4-knockdown transgenic mice and suggestions for the analysis. Chun Fan performed experiments to obtain data and analysed the data during revision. Qing Wang undertook project planning, troubleshooting and manuscript preparation during revision. Aleksander Hinek provided expert advice on elastic-fibre analysis, contributed data and was involved in manuscript editing. Edward Plow undertook overall project planning, troubleshooting and manuscript preparation. Paul DiCorleto undertook overall project planning, troubleshooting and manuscript preparation.
References
- 1.Mecham R, Davis E. Elastic fibre structure and assembly. In: Yurchenco P, Birk D, Mecham R, editors. Extracellular Matrix Assembly and Structure. Academic Press; San Diego: 1994. pp. 281–314. [Google Scholar]
- 2.Vrhovski B, Weiss AS. Biochemistry of tropoelastin. Eur J Biochem. 1998;258:1–18. doi: 10.1046/j.1432-1327.1998.2580001.x. [DOI] [PubMed] [Google Scholar]
- 3.Kielty CM, Sherratt MJ, Shuttleworth CA. Elastic fibres. J Cell Sci. 2002;115:2817–2828. doi: 10.1242/jcs.115.14.2817. [DOI] [PubMed] [Google Scholar]
- 4.Hinek A, Pshezhetsky AV, von Itzstein M, Starcher B. Lysosomal sialidase (neuraminidase-1) is targeted to the cell surface in a multiprotein complex that facilitates elastic fibre assembly. J Biol Chem. 2006;281:3698–3710. doi: 10.1074/jbc.M508736200. [DOI] [PubMed] [Google Scholar]
- 5.Hinek A. Nature and the multiple functions of the 67-kD elastin-/laminin binding protein. Cell Adhes Commun. 1994;2:185–193. doi: 10.3109/15419069409004436. [DOI] [PubMed] [Google Scholar]
- 6.Kielty CM. Elastic fibres in health and disease. Expert Rev Mol Med. 2006;8:1–23. doi: 10.1017/S146239940600007X. [DOI] [PubMed] [Google Scholar]
- 7.Pereira L, Andrikopoulos K, Tian J, Lee SY, Keene DR, Ono R, Reinhardt DP, Sakai LY, Biery NJ, Bunton T, et al. Targetting of the gene encoding fibrillin-1 recapitulates the vascular aspect of Marfan syndrome. Nat Genet. 1997;17:218–222. doi: 10.1038/ng1097-218. [DOI] [PubMed] [Google Scholar]
- 8.Carta L, Pereira L, Arteaga-Solis E, Lee-Arteaga SY, Lenart B, Starcher B, Merkel CA, Sukoyan M, Kerkis A, Hazeki N, et al. Fibrillins 1 and 2 perform partially overlapping functions during aortic development. J Biol Chem. 2006;281:8016–8023. doi: 10.1074/jbc.M511599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhry SS, Gazzard J, Baldock C, Dixon J, Rock MJ, Skinner GC, Steel KP, Kielty CM, Dixon MJ. Mutation of the gene encoding fibrillin-2 results in syndactyly in mice. Hum Mol Genet. 2001;10:835–843. doi: 10.1093/hmg/10.8.835. [DOI] [PubMed] [Google Scholar]
- 10.Maki JM, Rasanen J, Tikkanen H, Sormunen R, Makikallio K, Kivirikko KI, Soininen R. Inactivation of the lysyl oxidase gene Lox leads to aortic aneurysms, cardiovascular dysfunction, and perinatal death in mice. Circulation. 2002;106:2503–2509. doi: 10.1161/01.cir.0000038109.84500.1e. [DOI] [PubMed] [Google Scholar]
- 11.Hornstra IK, Birge S, Starcher B, Bailey AJ, Mecham RP, Shapiro SD. Lysyl oxidase is required for vascular and diaphragmatic development in mice. J Biol Chem. 2003;278:14387–14393. doi: 10.1074/jbc.M210144200. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Zhao Y, Gao J, Pawlyk B, Starcher B, Spencer JA, Yanagisawa H, Zuo J, Li T. Elastic fibre homeostasis requires lysyl oxidase-like 1 protein. Nat Genet. 2004;36:178–182. doi: 10.1038/ng1297. [DOI] [PubMed] [Google Scholar]
- 13.Zanetti M, Braghetta P, Sabatelli P, Mura I, Doliana R, Colombatti A, Volpin D, Bonaldo P, Bressan GM. EMILIN-1 deficiency induces elastogenesis and vascular cell defects. Mol Cell Biol. 2004;24:638–650. doi: 10.1128/MCB.24.2.638-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLaughlin PJ, Chen Q, Horiguchi M, Starcher BC, Stanton JB, Broekelmann TJ, Marmorstein AD, McKay B, Mecham R, Nakamura T, Marmorstein LY. Targeted disruption of fibulin-4 abolishes elastogenesis and causes perinatal lethality in mice. Mol Cell Biol. 2006;26:1700–1709. doi: 10.1128/MCB.26.5.1700-1709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura T, Lozano PR, Ikeda Y, Iwanaga Y, Hinek A, Minamisawa S, Cheng CF, Kobuke K, Dalton N, Takada Y, et al. Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature. 2002;415:171–175. doi: 10.1038/415171a. [DOI] [PubMed] [Google Scholar]
- 16.Yanagisawa H, Davis EC, Starcher BC, Ouchi T, Yanagisawa M, Richardson JA, Olson EN. Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature. 2002;415:168–171. doi: 10.1038/415168a. [DOI] [PubMed] [Google Scholar]
- 17.Timpl R, Sasaki T, Kostka G, Chu ML. Fibulins: a versatile family of extracellular matrix proteins. Nat Rev Mol Cell Biol. 2003;4:479–489. doi: 10.1038/nrm1130. [DOI] [PubMed] [Google Scholar]
- 18.Argraves WS, Greene LM, Cooley MA, Gallagher WM. Fibulins: physiological and disease perspectives. EMBO Rep. 2003;4:1127–1131. doi: 10.1038/sj.embor.7400033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu ML, Tsuda T. Fibulins in development and heritable disease. Birth Defects Res C Embryo Today. 2004;72:25–36. doi: 10.1002/bdrc.20003. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi N, Kostka G, Garbe JH, Keene DR, Bachinger HP, Hanisch FG, Markova D, Tsuda T, Timpl R, Chu ML, Sasaki T. A comparative analysis of the fibulin protein family. Biochemical characterization, binding interactions, and tissue localization. J Biol Chem. 2007;282:11805–11816. doi: 10.1074/jbc.M611029200. [DOI] [PubMed] [Google Scholar]
- 21.Sasaki T, Kostka G, Gohring W, Wiedemann H, Mann K, Chu ML, Timpl R. Structural characterization of two variants of fibulin-1 that differ in nidogen affinity. J Mol Biol. 1995;245:241–250. doi: 10.1006/jmbi.1994.0020. [DOI] [PubMed] [Google Scholar]
- 22.Pan TC, Sasaki T, Zhang RZ, Fassler R, Timpl R, Chu ML. Structure and expression of fibulin-2, a novel extracellular matrix protein with multiple EGF-like repeats and consensus motifs for calcium binding. J Cell Biol. 1993;123:1269–1277. doi: 10.1083/jcb.123.5.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roark EF, Keene DR, Haudenschild CC, Godyna S, Little CD, Argraves WS. The association of human fibulin-1 with elastic fibres: an immunohistological, ultrastructural, and RNA study. J Histochem Cytochem. 1995;43:401–411. doi: 10.1177/43.4.7534784. [DOI] [PubMed] [Google Scholar]
- 24.Reinhardt DP, Sasaki T, Dzamba BJ, Keene DR, Chu ML, Gohring W, Timpl R, Sakai LY. Fibrillin-1 and fibulin-2 interact and are colocalized in some tissues. J Biol Chem. 1996;271:19489–19496. doi: 10.1074/jbc.271.32.19489. [DOI] [PubMed] [Google Scholar]
- 25.Kostka G, Giltay R, Bloch W, Addicks K, Timpl R, Fassler R, Chu ML. Perinatal lethality and endothelial cell abnormalities in several vessel compartments of fibulin-1-deficient mice. Mol Cell Biol. 2001;21:7025–7034. doi: 10.1128/MCB.21.20.7025-7034.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sicot FX, Tsuda T, Markova D, Klement JF, Arita M, Zhang RZ, Pan TC, Mecham RP, Birk DE, Chu ML. Fibulin-2 is dispensable for mouse development and elastic fibre formation. Mol Cell Biol. 2008;28:1061–1067. doi: 10.1128/MCB.01876-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Hallous E, Sasaki T, Hubmacher D, Getie M, Tiedemann K, Brinckmann J, Batge B, Davis EC, Reinhardt DP. Fibrillin-1 interactions with fibulins depend on the first hybrid domain and provide an adaptor function to tropoelastin. J Biol Chem. 2007;282:8935–8946. doi: 10.1074/jbc.M608204200. [DOI] [PubMed] [Google Scholar]
- 28.Arrighi JF, Pion M, Wiznerowicz M, Geijtenbeek TB, Garcia E, Abraham S, Leuba F, Dutoit V, Ducrey-Rundquist O, van Kooyk Y, et al. Lentivirus-mediated RNA interference of DC-SIGN expression inhibits human immunodeficiency virus transmission from dendritic cells to T cells. J Virol. 2004;78:10848–10855. doi: 10.1128/JVI.78.20.10848-10855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singhi AD, Kondratov RV, Neznanov N, Chernov MV, Gudkov AV. Selection–subtraction approach (SSA): a universal genetic screening technique that enables negative selection. Proc Natl Acad Sci USA. 2004;101:9327–9332. doi: 10.1073/pnas.0403080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swee MH, Parks WC, Pierce RA. Developmental regulation of elastin production. Expression of tropoelastin pre-mRNA persists after down-regulation of steady-state mRNA levels. J Biol Chem. 1995;270:14899–14906. doi: 10.1074/jbc.270.25.14899. [DOI] [PubMed] [Google Scholar]
- 31.Hinek A, Braun KR, Liu K, Wang Y, Wight TN. Retrovirally mediated overexpression of versican v3 reverses impaired elastogenesis and heightened proliferation exhibited by fibroblasts from Costello syndrome and Hurler disease patients. Am J Pathol. 2004;164:119–131. doi: 10.1016/S0002-9440(10)63103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urban Z, Riazi S, Seidl TL, Katahira J, Smoot LB, Chitayat D, Boyd CD, Hinek A. Connection between elastin haploinsufficiency and increased cell proliferation in patients with supravalvular aortic stenosis and Williams–Beuren syndrome. Am J Hum Genet. 2002;71:30–44. doi: 10.1086/341035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francke U. Williams–Beuren syndrome: genes and mechanisms. Hum Mol Genet. 1999;8:1947–1954. doi: 10.1093/hmg/8.10.1947. [DOI] [PubMed] [Google Scholar]
- 34.Merla G, Ucla C, Guipponi M, Reymond A. Identification of additional transcripts in the Williams–Beuren syndrome critical region. Hum Genet. 2002;110:429–438. doi: 10.1007/s00439-002-0710-x. [DOI] [PubMed] [Google Scholar]
- 35.Bunda S, Kaviani N, Hinek A. Fluctuations of intracellular iron modulate elastin production. J Biol Chem. 2005;280:2341–2351. doi: 10.1074/jbc.M409897200. [DOI] [PubMed] [Google Scholar]
- 36.Hinek A, Zhang S, Smith AC, Callahan JW. Impaired elastic-fibre assembly by fibroblasts from patients with either Morquio B disease or infantile GM1-gangliosidosis is linked to deficiency in the 67-kD spliced variant of β-galactosidase. Am J Hum Genet. 2000;67:23–36. doi: 10.1086/302968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanada K, Vermeij M, Garinis GA, de Waard MC, Kunen MG, Myers L, Maas A, Duncker DJ, Meijers C, Dietz HC, et al. Perturbations of vascular homeostasis and aortic valve abnormalities in fibulin-4 deficient mice. Circ Res. 2007;100:738–746. doi: 10.1161/01.RES.0000260181.19449.95. [DOI] [PubMed] [Google Scholar]
- 38.Mecham RP. Methods in elastic tissue biology: elastin isolation and purification. Methods. 2008;45:32–41. doi: 10.1016/j.ymeth.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robb BW, Wachi H, Schaub T, Mecham RP, Davis EC. Characterization of an in vitro model of elastic fibre assembly. Mol Biol Cell. 1999;10:3595–3605. doi: 10.1091/mbc.10.11.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kahari VM, Olsen DR, Rhudy RW, Carrillo P, Chen YQ, Uitto J. Transforming growth factor-β up-regulates elastin gene expression in human skin fibroblasts. Evidence for post-transcriptional modulation. Lab Invest. 1992;66:580–588. [PubMed] [Google Scholar]
- 41.Davidson JM, Smith K, Shibahara S, Tolstoshev P, Crystal RG. Regulation of elastin synthesis in developing sheep nuchal ligament by elastin mRNA levels. J Biol Chem. 1982;257:747–754. [PubMed] [Google Scholar]
- 42.Burnett W, Finnigan-Bunick A, Yoon K, Rosenbloom J. Analysis of elastin gene expression in the developing chick aorta using cloned elastin cDNA. J Biol Chem. 1982;257:1569–1572. [PubMed] [Google Scholar]
- 43.Parks WC, Secrist H, Wu LC, Mecham RP. Developmental regulation of tropoelastin isoforms. J Biol Chem. 1988;263:4416–4423. [PubMed] [Google Scholar]
- 44.Sephel GC, Sturrock A, Giro MG, Davidson JM. Increased elastin production by progeria skin fibroblasts is controlled by the steady-state levels of elastin mRNA. J Invest Dermatol. 1988;90:643–647. doi: 10.1111/1523-1747.ep12560800. [DOI] [PubMed] [Google Scholar]
- 45.Tsuruga E, Yajima T, Irie K. Microfibril-associated glycoprotein-1 and fibrillin-2 are associated with tropoelastin deposition in vitro. Int J Biochem Cell Biol. 2005;37:120–129. doi: 10.1016/j.biocel.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Hirai M, Horiguchi M, Ohbayashi T, Kita T, Chien KR, Nakamura T. Latent TGF-β-binding protein 2 binds to DANCE/fibulin-5 and regulates elastic fibre assembly. EMBO J. 2007;26:3283–3295. doi: 10.1038/sj.emboj.7601768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts AB, Heine UI, Flanders KC, Sporn MB. Transforming growth factor-β. Major role in regulation of extracellular matrix. Ann NY Acad Sci. 1990;580:225–232. doi: 10.1111/j.1749-6632.1990.tb17931.x. [DOI] [PubMed] [Google Scholar]
- 48.Jones JA, Spinale FG, Ikonomidis JS. Transforming growth factor-β signaling in thoracic aortic aneurysm development: a paradox in pathogenesis. J Vasc Res. 2009;46:119–137. doi: 10.1159/000151766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Larco JE, Todaro GJ. Growth factors from murine sarcoma virus-transformed cells. Proc Natl Acad Sci USA. 1978;75:4001–4005. doi: 10.1073/pnas.75.8.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts AB, Frolik CA, Anzano MA, Sporn MB. Transforming growth factors from neoplastic and nonneoplastic tissues. Fed Proc Fed Am Soc Exp Biol. 1983;42:2621–2626. [PubMed] [Google Scholar]
- 51.Li DY, Brooke B, Davis EC, Mecham RP, Sorensen LK, Boak BB, Eichwald E, Keating MT. Elastin is an essential determinant of arterial morphogenesis. Nature. 1998;393:276–280. doi: 10.1038/30522. [DOI] [PubMed] [Google Scholar]
- 52.Mecham RP, Lange G, Madaras J, Starcher B. Elastin synthesis by ligamentum nuchae fibroblasts: effects of culture conditions and extracellular matrix on elastin production. J Cell Biol. 1981;90:332–338. doi: 10.1083/jcb.90.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gallagher WM, Greene LM, Ryan MP, Sierra V, Berger A, Laurent-Puig P, Conseiller E. Human fibulin-4: analysis of its biosynthetic processing and mRNA expression in normal and tumour tissues. FEBS Lett. 2001;489:59–66. doi: 10.1016/s0014-5793(00)02389-9. [DOI] [PubMed] [Google Scholar]
- 54.Katsanis N, Venable S, Smith JR, Lupski JR. Isolation of a paralog of the Doyne honeycomb retinal dystrophy gene from the multiple retinopathy critical region on 11q13. Hum Genet. 2000;106:66–72. doi: 10.1007/s004390051011. [DOI] [PubMed] [Google Scholar]
- 55.Gong Y, Vikkula M, Boon L, Liu J, Beighton P, Ramesar R, Peltonen L, Somer H, Hirose T, Dallapiccola B, et al. Osteoporosis-pseudoglioma syndrome, a disorder affecting skeletal strength and vision, is assigned to chromosome region 11q12–13. Am J Hum Genet. 1996;59:146–151. [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson ML, Gong G, Kimberling W, Recker SM, Kimmel DB, Recker RB. Linkage of a gene causing high bone mass to human chromosome 11 (11q12–13) Am J Hum Genet. 1997;60:1326–1332. doi: 10.1086/515470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stone EM, Nichols BE, Streb LM, Kimura AE, Sheffield VC. Genetic linkage of vitelliform macular degeneration (Best’s disease) to chromosome 11q13. Nat Genet. 1992;1:246–250. doi: 10.1038/ng0792-246. [DOI] [PubMed] [Google Scholar]
- 58.Li Y, Muller B, Fuhrmann C, van Nouhuys CE, Laqua H, Humphries P, Schwinger E, Gal A. The autosomal dominant familial exudative vitreoretinopathy locus maps on 11q and is closely linked to D11S533. Am J Hum Genet. 1992;51:749–754. [PMC free article] [PubMed] [Google Scholar]
- 59.Gallagher WM, Argentini M, Sierra V, Bracco L, Debussche L, Conseiller E. MBP1: a novel mutant p53-specific protein partner with oncogenic properties. Oncogene. 1999;18:3608–3616. doi: 10.1038/sj.onc.1202937. [DOI] [PubMed] [Google Scholar]
- 60.Hucthagowder V, Sausgruber N, Kim KH, Angle B, Marmorstein LY, Urban Z. Fibulin-4: a novel gene for an autosomal recessive cutis laxa syndrome. Am J Hum Genet. 2006;78:1075–1080. doi: 10.1086/504304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dasouki M, Markova D, Garola R, Sasaki T, Charbonneau NL, Sakai LY, Chu ML. Compound heterozygous mutations in fibulin-4 causing neonatal lethal pulmonary artery occlusion, aortic aneurysm, arachnodactyly and mild cutis laxa. Am J Med Genet A. 2007;143:2635–2641. doi: 10.1002/ajmg.a.31980. [DOI] [PubMed] [Google Scholar]
- 62.Hinek A, Smith AC, Cutiongco EM, Callahan JW, Gripp KW, Weksberg R. Decreased elastin deposition and high proliferation of fibroblasts from Costello syndrome are related to functional deficiency in the 67-kD elastin-binding protein. Am J Hum Genet. 2000;66:859–872. doi: 10.1086/302829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tatano Y, Takahashi T, Tsuji D, Takeuchi N, Tsuta K, Takada G, Ohsawa M, Sakuraba H, Itoh K. Significant decrease in tropoelastin gene expression in fibroblasts from a Japanese Costello syndrome patient with impaired elastogenesis and enhanced proliferation. J Biochem (Tokyo) 2006;140:193–200. doi: 10.1093/jb/mvj146. [DOI] [PubMed] [Google Scholar]
- 64.Urban Z, Michels VV, Thibodeau SN, Davis EC, Bonnefont JP, Munnich A, Eyskens B, Gewillig M, Devriendt K, Boyd CD. Isolated supravalvular aortic stenosis: functional haploinsufficiency of the elastin gene as a result of nonsense-mediated decay. Hum Genet. 2000;106:577–588. doi: 10.1007/s004390000285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.