Abstract
T2-family acidic endoribonucleases are represented in all genomes. A physiological role for RNase T2 has yet to be defined for metazoa. RNASET2 mutation in humans is linked with a leukoencephalopathy that arises in infancy characterized by cortical cysts and multifocal white matter lesions. We now show localization of RNASET2 within lysosomes. Further, we demonstrate that loss of rnaset2 in mutant zebrafish results in accumulation of undigested rRNA within lysosomes within neurons of the brain. Further, by using high field intensity magnetic resonance microimaging, we reveal white matter lesions in these animals comparable to those observed in RNASET2-deficient infants. This correlates with accumulation of Amyloid precursor protein and astrocytes at sites of neurodegeneration. Thus we conclude that familial cystic leukoencephalopathy is a lysosomal storage disorder in which rRNA is the best candidate for the noxious storage material.
T2 endoribonucleases are distributed throughout the kingdoms of life. They perform diverse biological roles in bacteria, fungi, and plants, including phosphate scavenging following nutritional stress, cleaving tRNA in response to oxidative stress, as well as mediating gametophyte self-incompatibility (reviewed in ref. 1). However, little is known about their functions in metazoa. Recently we uncovered that RNASET2 deficiency in man results in a leukoencephalopathy that arises in infancy characterized by cortical cysts and multifocal white matter lesions (2). Patients manifest with psychomotor impairment, micro- or normocephaly, spasticity, and epilepsy. However, neither the physiological role of RNASET2 nor the disease mechanism was apparent from this clinical presentation. Also, to our knowledge, no animal model has yet been generated to study this disease.
Of all the ribonuclease families, the RNase T2 family is the only one active at acidic pH (1) and consequently, the human RNASET2 was described as a putative lysosomal hydrolase (3, 4). Lysosomes are membrane bound organelles containing 60 or more hydrolytic enzymes active at an acidic pH and responsible for degrading all classes of macromolecules upon fusion of lysosomes with phagosomes and endosomes. Lysosomes play an essential role in autophagy and phagocytosis, being responsible for the degradation of molecules, whole organelles, and microorganisms and subsequent salvaging of metabolites (4). Approximately 50 genetic lysosomal diseases, collectively termed lysosomal storage disorders (LSDs), have now been recognized, which affect one in every 5,000 to 7,000 live births in the United States (5, 6). Inborn lysosomal defects result in disease through the accumulation of nondegraded macromolecules. The severity of disease symptoms depends generally on the nature of the accumulated substrate, giving a unique clinical and pathological diagnostic for each LSD. Generally, multiple organs are affected, leading to severe symptoms and premature death. CNS involvement is very common, with neuronal degeneration being apparent (7). Although the deficient gene and the nature of the accumulating product are known in the majority of LSDs, the events that lead from macromolecule accumulation to disease are still frequently unclear.
In zebrafish we have previously described two RNase T2 encoding paralogues: rnaset2l (ribonuclease T2 like, previously referred to as RNase Dre1), which is conserved only in fish, on chromosome 15, with 35% identity at the amino acid level to RNASET2; and rnaset2 (previously referred to as RNase Dre2) on chromosome 13, with 47% identity, that appears to be the orthologue of RNASET2 (8). In this study, we have identified zebrafish mutant in the rnaset2 gene and shown that these animals accumulate undigested rRNA in lysosomes within neurons of the brain. In adults this leads to neuronal degeneration that can be detected by both magnetic resonance microimaging (μMRI) and immunohistochemical analysis of Amyloid precursor protein (APP) and astrocyte accumulation. Thus we propose that the familial cystic leukoencephalopathy arising in RNASET2-deficient humans is a manifestation of an LSD in which rRNA is the best candidate for the noxious storage material.
Results and Discussion
Identification of an rnaset2 Mutation in Zebrafish.
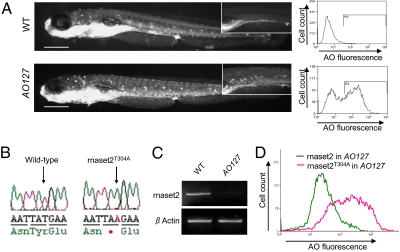
In a screen of N-ethyl-N-nitrosourea (ENU)-mutagenized zebrafish for altered acridine orange (AO) uptake, we identified mutant embryos with increased AO staining (Fig. 1A, compare WT with AO127 mutant). The gene affected in AO127 mutants was identified by positional cloning (Fig. S1A). Sequencing of a positional candidate zgc:113369, equivalent to rnaset2, which we previously described (8), identified a T-to-A transversion present only in mutant samples (T304A mutation; Fig. 1B). rnaset2 is expressed ubiquitously in zebrafish embryos and adults from conception (8). The mutation within codon 118 of the rnaset2 ORF creates a premature stop codon truncating the protein before its second conserved catalytic site, CAS II, resulting in loss of essential catalytic residues (Fig. S1B). Moreover, as shown by RT-PCR, this mutation results in a dramatic down-regulation of the rnaset2 messenger RNA (Fig. 1C), presumably through nonsense-mediated decay (9). Capped mRNA was produced from full-length WT rnaset2 cDNA and injected into one-cell stage AO127 mutant embryos. Flow cytometry analysis confirmed restoration of normal AO staining (Fig. 1D). In contrast, injection with the mutant version of rnaset2, rnaset2T304A, did not alter AO staining. Furthermore, knockdown of rnaset2 in AO127 heterozygotes using an antisense oligonucleotide induced increased AO staining (Fig. S2). Thus, we have identified a mutant harboring a null allele of rnaset2, an orthologue of RNASET2, and confirmed that this mutation is responsible for increased AO binding.
Fig. 1.
AO127 mutant animals are deficient in rnaset2. (A) AO127 mutants displayed a higher uptake of AO compared with WT embryos, quantifiable by flow cytometry. (Note the intense fluorescence common to both animals is a result of AO within the lumen of the pharynx and intestine, autofluorescence of residual yolk, and ready uptake of AO by olfactory neurons and hair cells within neuromasts.) (Scale bars: 200 μm.) (B) Sequencing of the gene zgc:113369, coding for the zebrafish orthologue of human RNASET2, identified a T-to-A transversion in the mutant sample. (C) RT-PCR indicates a dramatic down-regulation of rnaset2 mRNA in AO127 mutants compared with WT siblings. β-Actin serves as a control for RNA integrity and equal input cDNA. (D) Suppression of excess AO staining in AO127 mutants by injection of WT rnaset2 mRNA but not mutant mRNA.
RNASET2 Localizes to Lysosomes.
Although certain reports imply that RNase T2 molecules are localized to lysosomes (3, 4), others have suggested that they are secreted enzymes (1). To corroborate a lysosomal localization, immunofluorescence staining was performed on HEK 293 cells transfected with WT RNASET2. This revealed partial overlap in the subcellular distribution of RNASET2 with either a marker for the endoplasmic reticulum (ie, PDI) or Golgi apparatus, and significantly more overlap with a marker of late endosomes and lysosomes (LAMP-1; Fig. 2A). This is consistent with a protein that transits through the endoplasmic reticulum and Golgi apparatus to the lysosomes. In contrast, in cells transfected with RNASET2C184R, harboring a missense mutation as identified from a patient with cystic leukoencephalopathy (2), signal was retained within the endoplasmic reticulum (Fig. 2B), presumably as a result of activation of the misfolded protein response (10).
Fig. 2.
Subcellular localization of RNASET2. HEK293 cells were transfected with myc-epitope tagged WT RNASET2 (A) and the mutated version, RNASET2 C184R (B). Immunofluorescence for endoplasmic reticulum (PDI), Golgi, and late endosome (LAMP-1) markers show partial colocalization with WT RNASET2, suggesting that RNASET2 enters the secretory pathway. The mutated version colocalized completely with the PDI, suggesting ER retention. (Scale bar: 10 μm.)
rnaset2-Deficient Zebrafish Display Lysosomes Engorged with rRNA.
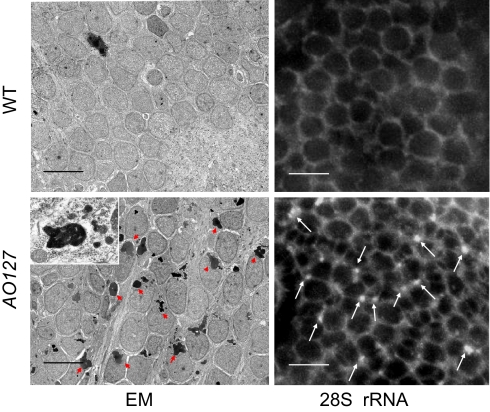
The localization of RNASET2 to lysosomes, combined with the increased AO staining observed in AO127 mutants and the fact that within cells AO preferentially stains lysosomes (11), led us to hypothesize the existence of enlarged or supernumerary lysosomes in our mutants. To address this, we examined sections of zebrafish embryos by using electron microscopy (EM) Indeed, enlarged electron-dense lysosomes were readily apparent in the brains of mutant embryos, whereas they were not apparent in WT siblings (Fig. 3). Further, in situ hybridization using a probe for 28S rRNA (12) revealed rRNA aggregates again specifically in AO127 mutants (Fig. 3; Fig. S3 shows control sense probe) presumably corresponding to the electron-dense material observed by EM. This suggests that the engorged lysosomes might be accumulating undigested rRNA. Enlarged lysosomes were not readily detected outside the central nervous system in AO127 mutant zebrafish embryos. Additionally, knockdown of RNASET2 in HEK 293 cells resulted in increased lysosome numbers but not the appearance of enlarged lysosomes (Fig. S4 A–D). Also, we failed to detect accumulation of rRNA in RNASET2-depleted HEK293 cells (Fig. S4E). We assume, therefore, that the increased AO uptake detected in most other cells within AO127 mutants was caused, as for HEK 293 cells, by increased lysosome numbers. Indeed, increased lysosome biogenesis has previously been documented as a compensatory mechanism when lysosome function is compromised (13).
Fig. 3.
Brain neurons of AO127 mutants contain lysosomes engorged with rRNA. Brain transverse sections from embryos 5 d after fertilization. EM revealed abundant electron-dense vesicles in the neurons of AO127 mutants (some of these are indicated by red arrowheads; Inset, electron-dense material is bounded by a limiting membrane). (Scale bars: 5 μm). In situ hybridization revealed accumulation of rRNA aggregates (white arrows) in the cytoplasm of AO127 mutant neurons. (Scale bars: 10 μm.)
rnaset2-Deficient Zebrafish Develop White Matter Lesions.
As previously described, the brains of RNASET2-deficient humans present a diagnostic pattern of brain abnormalities on MRI (2) (Fig. S5A). T2-weighted μMRI (14) was also used to analyze adult zebrafish. White matter lesions adjacent to brain ventricles were observed in all AO127 mutant animals analyzed (n = 7) and ventricles also appeared dilated; frequent focal white matter anomalies were scattered throughout brains (Fig. 4). Analysis of T2 relaxation time suggested that white matter lesions might be filled with fluid similar to CSF in ventricles (Fig. S5B). Moreover, immunohistochemical staining with an antibody recognizing APP indicated APP accumulation and possible axonal damage (15) specifically in mutant brains, particularly evident in white matter regions bordering ventricles (Fig. 4). Likewise, staining for GFAP revealed an increase of astrocytes around sites of putative damage (Fig. 4).
Fig. 4.
White matter anomalies in AO127 mutants. T2-weighted MR images of the brain of WT and mutant AO127 adult zebrafish. Sagittal images indicate the presence of a white matter anomaly (white arrow) in telencephalon of mutant, which was not seen in WT. Coronal images show the presence of lesions adjacent to the ventricles (white arrow). IHC was carried out on the same samples used for MRI. Axonal disruptions around ventricles were seen using APP as a marker, as well as a higher number of astrocytes using GFAP as a marker, suggesting the presence of damaged neurons and inflammation around the lesions. (Scale bars: 1 mm for MR sagittal images, 500 μm for coronal images, 100 μm for immunohistochemistry.)
Notwithstanding the neurodegeneration described, animals homozygous for the AO127 allele appear morphologically normal, reach a normal adult size and weight, are fertile, and do not display obvious gross motor defects. Clearly, this is in contrast to the debilitating psychomotor impairment observed in RNASET2-deficient humans. Previous RT-PCR and in situ hybridization analyses revealed extensive overlap between rnaset2l and rnaset2 expression in embryonic and adult tissues, with both isoforms being expressed in the central nervous system (8). Potentially, rnaset2l can partly compensate for rnaset2 deficiency. Indeed, by in gel RNase activity assay, we observed only partial reduction in RNase activity in the 20- to 30-kDa range at an acidic pH in protein extracts from AO127 mutant embryos compared with extracts from WT animals (Fig. S6A). RNase activity in this size range, although not affected by antisense oligonucleotide-mediated knockdown of rnaset2l alone, was completely abolished by depleting rnaset2l in an AO127 mutant background (Fig. S6B). Further, knockdown of rnaset2l by itself induced only a modest increase in AO staining in WT fish and failed to augment the increased AO staining observed in AO127 mutants (Fig. S6C). Thus, although rnaset2 is not an essential gene product in zebrafish and rnaset2l is likely to partly compensate for its deficiency, AO127 mutants nonetheless recapitulate aspects of the leukoencephalopathy observed in RNASET2-deficient humans, supporting their use as a preclinical animal model of this disease.
In this study, we present a zebrafish model for a childhood-onset leukodystrophy. Furthermore, we demonstrate that familial cystic leukoencephalopathy is most likely an LSD in which rRNA accumulates in lysosomes in brain neurons. We hypothesize that RNase T2 is a factor in the specific ribosome recycling pathway termed “ribophagy,” previously described in yeast and through which ribosomes are targeted selectively to the vacuole to be salvaged (16). Significantly, absence of the RNase T2 encoded by rns2 in Arabidopsis also results in constitutive accumulation of undigested RNA in vacuoles, and rRNA acquires a longer half-life in rns2 mutants (17), so this seems to be a conserved housekeeping function for RNase T2 in multicellular organisms. Depletion of RNASET2 from HEK 293 cells did not result in the appearance of engorged lysosomes or accumulation of rRNA, but rather increased lysosome numbers, indicative of lysosomal dysfunction (13). Potentially, excess undigested rRNA could be “bailed” from these cells through exocytosis (18) and digested by secreted RNase A-like enzymes in the extracellular environment. The problem may be less easily resolved in neurons if rRNA levels were significantly higher or exocytosis by lysosomes limited.
Concerning the disease mechanism leading from RNase T2 deficiency to neurodegeneration, then, potentially, impaired ribophagy could compromise ribogenesis, reducing cell functionality/viability. Alternatively, as with other LSDs, macroautophagy might be impaired as an indirect consequence of lysosomal dysfunction, which in turn appears to harm cells (5, 19). Conversely, accumulated RNA may itself be cytotoxic, as mutations within another ribonuclease, RNASEH2—which neither targets rRNA nor is a lysosomal hydrolase—underlie a very similar leukoencephalopathy, Aicardi–Goutieres syndrome (20). Moreover, the resemblance of RNASET2 deficiency and Aicardi–Goutieres syndrome to in utero CMV infection (2, 20) implies that accumulation of certain RNA species may be recognized by the organism as nonself and thereby elicit an inflammatory reaction. This in turn may be a potential cause of the disease symptoms. Indeed, another causative mutation in Aicardi–Goutieres syndrome affects the SAMHD1 gene, which encodes a regulator of the innate immune system (21). None of the disease mechanisms suggested here are, however, mutually exclusive.
Future Perspectives.
rnaset2-deficient zebrafish could serve as preclinical animal models for further elucidating disease pathophysiology linking RNA accumulation with axonal degeneration and white matter alterations. The early manifestation of the phenotype in transparent zebrafish embryos as well as easy detection by AO staining suggests that therapeutic targets could be readily validated in this system. To assess disease course as well as the impact of any candidate therapeutics, longitudinal studies in adult fish could be performed using μMRI on living fish as previously described (14).
Experimental Procedures
Detailed methods are included in SI Experimental Procedures.
Animal Husbandry, Mutagenesis, and Selection of AO127 Mutants.
Zebrafish were raised and maintained at the University of Manchester Biological Services Unit as described elsewhere (22). ENU mutagenesis was performed on both AB and TL males as previously described (23). Day 5 postfertilization embryos were stained with 3 μg/μL of AO (Sigma) in embryo water for 15 min then washed three times in embryo water. After anesthesia with tricaine (Sigma), AO staining was visualized using a Zeiss stereo Lumar V12 fluorescent microscope fitted with filter set Lumar 38 and images were acquired using an AxioCam MRm camera and processed using AxioVision software (version 4.8; Image Associates). Subsequently, embryos were dissociated in trypsin and AO content was measured by flow cytometry using a Beckman Coulter Cyan ADP and data analyzed using Summit software (Dako).
Positional Cloning.
We followed a previously described detailed protocol (24) (also available from http://zon.tchlab.org/). SI Experimental Procedures includes further details on positional cloning.
RNA Expression and Knockdown in Zebrafish.
Capped mRNA encoding rnaset2 was transcribed using the T7 mMessage mMachine kit (Ambion) and approximately 1 nL was injected into one to four cell stage mutant embryos using a PLI-90 Pico-Injector (Harvard Apparatus). Morpholino-modified antisense oligonucleotides (MOs) targeting rnaset2l and rnaset2 as well as a standard control MO were obtained from GeneTools and 1 nL of a 300 nM stock was injected into one to four cell stage embryos. AO binding was analyzed at 3 d after fertilization by flow cytometry as described earlier.
Immunofluorescence.
HEK 293 cells growing on glass coverslips were stably transfected with plasmid expressing WT or C184R mutant RNASET2 using the Effectene transfection reagent (Qiagen). Immunostaining was performed using standard methods and a polyclonal rabbit RNASET2-antiserum (generated by conventional methods [Seramun]; dilution 1:2,000). PDI (ER marker; Abcam), 58K Golgi protein (Abcam), and LAMP-1 (lysosome marker; BD Biosciences) antibodies were applied to detect subcellular colocalization. Alexa 488- and Cy3-conjugated anti-rabbit and anti-mouse secondary antibodies were used (Molecular Probes and Jackson ImmunoResearch; diluted 1:1,000). The cells were embedded in ProLongGold mounting medium with DAPI (Invitrogen). Fluorescence signals were detected by using an Axio-Imager M.1 fluorescence microscope (Zeiss).
HEK 293 Culture and RNASET2 Knockdown.
HEK 293 cells, cultured in DMEM, supplemented with 10% FBS, 2 mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and 400 μg/mL Geneticin at 37 °C and 5% CO2, were transfected with 800 ng of control or RNASET2-targeting miRNA plasmid using Fugene HD (Roche) transfection reagent as instructed by the manufacturer. (SI Experimental Procedures provides vector details). Western blotting with the rabbit polyclonal RNASET2 anti-serum described earlier was used to demonstrate RNASET2 protein knockdown. In parallel, cells were stained with 5 μM of the intravital dye LysoTracker Green DND-26 (Invitrogen) and analyzed immediately by flow cytometry using a Beckman Coulter Cyan ADP and data analyzed by using Summit software (Dako). Cells flow-sorted for mCherry fluorescence were fixed and prepared for EM analysis.
Transmission EM.
Transmission EM (TEM) was carried out in the EM facility of the Faculty of Life Sciences, University of Manchester. Embryos were sedated, fixed, and processed as previously described (25). The protocol followed for HEK 293 cells is detailed in SI Experimental Procedures. TEM observations were carried out using a FEI Tecnai12 BioTwin microscope and photographs were taken using a 4 × 4 k CCD camera (Eagle; FEI Company) equipped with Kodak 4489 film. Images were acquired using an Imacon scanner and FlexColor software.
In Situ Hybridization for 28S rRNA.
The in situ hybridization protocol was adapted from Rouquette et al. (12) and performed on frozen sections using a 28S rRNA Cy3 tagged antisense oligodeoxynucleotide probe (Invitrogen; SI Experimental Procedures). Images were collected on a Leica TCS SP5 AOBS inverted confocal using a 40× objective. When acquiring 3D optical stacks, the confocal software was used to determine the optimal number of Z sections. Only the maximum intensity projections of these 3D stacks are shown.
MRI.
MRI studies of the human brain were performed using a 3T MRI system (Magnetom Trio; Siemens) with eight-channel phased-array head coils. Appropriate informed consent was obtained from the patients’ parents. MRI was performed on adult zebrafish as previously described (14) using a 400 MHz (9.4 T) vertical bore system and a transmit/receive birdcage radiofrequency coil with an inner diameter of 10 mm and a 1 Tm−1 gradient insert from Bruker. SI Experimental Procedures provides further details on MRI.
Paraffin Sections and Immunostaining.
Paraffin-embedded tissue sections (5 μm) were used for immunohistochemistry (IHC). The primary antibodies used were mouse anti-APP (1:500 dilution; Zymed) and mouse anti-GFAP (1:300 dilution; Dako), incubated overnight at 4 °C. The secondary antibody used was Vectastain Biotinylated horse anti–pan-Ig antibody. Following incubation with the secondary antibody, the sections were incubated with the ABC kit (Vectorlabs) and stained with DAB (Vectorlabs) before being mounted with Pertex. Images were captured using a Zeiss Axioplan microscope and Axiocam MR camera, and processed using Axiovision software.
Supplementary Material
Acknowledgments
We are indebted to Barry McColl, Stuart Allan, Paul Shore, Martin Lowe, and Philip Woodman for reagents and advice (all University of Manchester, Manchester, UK). In addition, we thank staff of the University of Manchester Biological Services Unit for animal caretaking, Ann Warhurst in the histology facility, Mike Jackson in the flow cytometry facility, and Samantha Forbes in the Electron Microscopy facility. This work was supported by Grants C11876/A4495 from Cancer Research UK (to A.H.), Centre for Medical Systems Biology (to A.A.); Deutsche Forschungsgemeinschaft Grant GA354/6-1 (to J.G. and M.H.); the Bundesministerium für Bildung and Forschung through the German Leukodystrophy Network (M.H.); and the Research Program, Faculty of Medicine, Georg August University Göttingen (M.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.J.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009811107/-/DCSupplemental.
References
- 1.Luhtala N, Parker R. T2 Family ribonucleases: Ancient enzymes with diverse roles. Trends Biochem Sci. 2010;35:253–259. doi: 10.1016/j.tibs.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henneke M, et al. RNASET2-deficient cystic leukoencephalopathy resembles congenital cytomegalovirus brain infection. Nat Genet. 2009;41:773–775. doi: 10.1038/ng.398. [DOI] [PubMed] [Google Scholar]
- 3.Campomenosi P, et al. Characterization of RNASET2, the first human member of the Rh/T2/S family of glycoproteins. Arch Biochem Biophys. 2006;449:17–26. doi: 10.1016/j.abb.2006.02.022. [DOI] [PubMed] [Google Scholar]
- 4.Lübke T, Lobel P, Sleat DE. Proteomics of the lysosome. Biochim Biophys Acta. 2009;1793:625–635. doi: 10.1016/j.bbamcr.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballabio A, Gieselmann V. Lysosomal disorders: From storage to cellular damage. Biochim Biophys Acta. 2009;1793:684–696. doi: 10.1016/j.bbamcr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nat Rev Mol Cell Biol. 2004;5:554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- 7.Walkley SU. Pathogenic cascades in lysosomal disease—why so complex? J Inherit Metab Dis. 2009;32:181–189. doi: 10.1007/s10545-008-1040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillwig MS, et al. Zebrafish RNase T2 genes and the evolution of secretory ribonucleases in animals. BMC Evol Biol. 2009;9:170. doi: 10.1186/1471-2148-9-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 10.Anelli T, Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zelenin AV. Fluorescence microscopy of lysosomes and related structures in living cells. Nature. 1966;212:425–426. doi: 10.1038/212425a0. [DOI] [PubMed] [Google Scholar]
- 12.Rouquette J, Choesmel V, Gleizes PE. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science (New York) 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 14.Kabli S, Alia A, Spaink HP, Verbeek FJ, De Groot HJ. Magnetic resonance microscopy of the adult zebrafish. Zebrafish. 2006;3:431–439. doi: 10.1089/zeb.2006.3.431. [DOI] [PubMed] [Google Scholar]
- 15.Suenaga T, et al. Bundles of amyloid precursor protein-immunoreactive axons in human cerebrovascular white matter lesions. Acta Neuropathol. 1994;87:450–455. doi: 10.1007/BF00294171. [DOI] [PubMed] [Google Scholar]
- 16.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 17.Hillwig MS, et al. RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1009809108. 10.1073/pnas.1009809108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luzio JP, Pryor PR, Bright NA. Lysosomes: Fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 19.Settembre C, Fraldi A, Rubinsztein DC, Ballabio A. Lysosomal storage diseases as disorders of autophagy. Autophagy. 2008;4:113–114. doi: 10.4161/auto.5227. [DOI] [PubMed] [Google Scholar]
- 20.Crow YJ, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 21.Rice GI, et al. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) Eugene, OR: Univ of Oregon Press; 2000. [Google Scholar]
- 23.van Eeden FJ, Granato M, Odenthal J, Haffter P. Developmental mutant screens in the zebrafish. Methods Cell Biol. 1999;60:21–41. doi: 10.1016/s0091-679x(08)61892-0. [DOI] [PubMed] [Google Scholar]
- 24.Bahary N, et al. The Zon laboratory guide to positional cloning in zebrafish. Methods Cell Biol. 2004;77:305–329. doi: 10.1016/s0091-679x(04)77017-x. [DOI] [PubMed] [Google Scholar]
- 25.Michailidou C, et al. Dissecting the roles of Raf- and PI3K-signalling pathways in melanoma formation and progression in a zebrafish model. Dis Model Mech. 2009;2:399–411. doi: 10.1242/dmm.001149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.