Abstract
Argonaute and Piwi proteins are key players in the RNA silencing pathway, with the former interacting with micro-RNAs (miRNAs) and siRNAs, whereas the latter targets piwi-interacting RNAs (piRNAs) that are 2′-O-methylated (2′-OCH3) at their 3′ ends. Germline-specific piRNAs and Piwi proteins play a critical role in genome defense against transposable elements, thereby protecting the genome against transposon-induced defects in gametogenesis and fertility. Humans contain four Piwi family proteins designated Hiwi1, Hiwi2, Hiwi3, and Hili. We report on the structures of Hili-PAZ (Piwi/Argonaute/Zwille) domain in the free state and Hiwi1 PAZ domain bound to self-complementary 14-mer RNAs (12-bp + 2-nt overhang) containing 2′-OCH3 and 2′-OH at their 3′ ends. These structures explain the molecular basis underlying accommodation of the 2′-OCH3 group within a preformed Hiwi1 PAZ domain binding pocket, whose hydrophobic characteristics account for the preferential binding of 2′-OCH3 over 2′-OH 3′ ends. These results contrast with the more restricted binding pocket for the human Ago1 PAZ domain, which exhibits a reverse order, with preferential binding of 2′-OH over 2′-OCH3 3′ ends.
Small RNAs, which play a critical role in surveillance of the genome and transcriptome, impact on a variety of biological processes, including cell differentiation and proliferation, apoptosis, transposon silencing, and antiviral defense (1). These small RNAs vary in size between 20 and 30 nucleotides (nt) and are associated with Argonaute (Ago) family proteins. Amongst the small RNAs encoded in the genome, microRNAs (miRNAs) and small interfering RNAs (siRNAs) bind to the Ago subfamily, whereas Piwi-interacting RNAs (piRNAs) bind to the Piwi subfamily (2). Structure-function studies have provided detailed mechanistic insights into guide-strand mediated cleavage of target RNAs within Ago complexes (3). By contrast, there is no structural information to date on Piwi proteins or their component domains, either in the free or RNA-bound states. Progress in this area is important because piRNAs and Piwi proteins are germline specific and play a key role in protecting the genome against mobile genetic elements, thereby preventing transposon-induced defects in gametogenesis and fertility (4, 5).
The PAZ (Piwi/Argonaute/Zwille) domain was previously identified from structure-function studies as a RNA-binding module (6–8), with subsequent structure-function studies on RNA-bound complexes defining the principles underlying recognition of a 2-nt overhang at the 3′ end of double-stranded RNA (dsRNA) (9), as well as recognition of the 3′ end of single-stranded RNA (ssRNA) (10). In eukaryotes, the PAZ domain is only found in Ago (11, 12) and Piwi (13) subfamilies, as well as in the RNase III family ribonuclease Dicer (14, 15), all of which play essential roles in the RNA silencing pathway (16, 17).
Ago subfamily proteins constitute the core component of the RNA Induced Silencing Complex (18), and are loaded with Dicer-generated mature miRNAs. The passenger strand is cleaved by the nuclease activity of Ago, while the guide strand is retained through anchoring of its 5′ phosphate and 3′ ends within the Mid and PAZ domains respectively, of the Ago protein (19). Subsequent pairing of the guide strand with a complementary mRNA target, triggers silencing by either target cleavage or translational repression, with the structural basis underlying Ago-mediated nucleation, propagation, and cleavage of target RNA (3) reviewed recently (20). Introduction of a 2′-O-methyl group (2′-OCH3) at the 3′ end of RNA reduced the binding affinity for human Ago1 PAZ domain (9), consistent with human miRNAs not being 2′-O-methylated at that site.
Piwi subfamily proteins bind to a distinct class of small RNAs, called piRNAs (21–25). Generally, piRNAs are ssRNAs of 26- to 30-nt in length, with 5′ phosphate and 2′-O-methylated 3′ ends (26, 27), a process mediated by methyltransferase HEN1 (26, 28, 29). The four human Piwi proteins, designated Hiwi1, Hiwi2, Hiwi3, and Hili, exhibit significant sequence identity amongst their PAZ domains, but distinct differences with the human Ago1 PAZ domain (Fig. S1). Nevertheless, nearly all the residues involved in 3′-end overhang recognition in Ago1 PAZ domain (9, 10) are conserved in Piwi PAZ domains, particularly Tyr and His aromatic residues that line the binding pocket (see red triangles, Fig. S1). This observation raises an intriguing question regarding how the Piwi PAZ domain binding pocket accommodates the methyl group of 2′-OCH3 at the 3′ end of piRNAs, a modification that impedes binding to the Ago1 PAZ domain. A working postulate would be that the Piwi PAZ domain could bind the 3′-end 2-nt overhang in a manner similar to Ago1 PAZ domain (9), except for a wider binding pocket capable of accommodating the 2′-OCH3 of piRNAs. Importantly, there is a common 8- to 10-aa insertion between the β6 and β7 strands in the PAZ domains of all four Piwi PAZ subfamily members (designated Piwi PAZ-specific insertion) that is absent in Ago1 PAZ domain (Fig. S1) and could facilitate the generation of this more spacious binding pocket.
To address this question, we have solved crystal structures of Hili PAZ in the free state and Hiwi1 PAZ bound to a self-complementary 14-mer RNA (12-bp + 2-nt overhang) containing either a 2′-OCH3 or 2′-OH at its 3′ end. These structures identify a preformed PAZ-binding pocket capable of accommodating the 2′-OCH3 group and, based on the hydrophobic nature of the binding pocket, account for the higher binding affinity for 2′-OCH3 relative to its 2′-OH counterpart. A comparison of the structures of the Hiwi1 PAZ-RNA complex with the previously reported human Ago1 PAZ-RNA complex (9) highlights a more restrictive PAZ-binding pocket in the Ago1 complex, thereby accounting for the higher binding affinity for 2′-OH over 2′-OCH3 for this complex.
Results
Effect of RNA 3′-End 2′-O-Methylation on Binding to Piwi PAZ Domains.
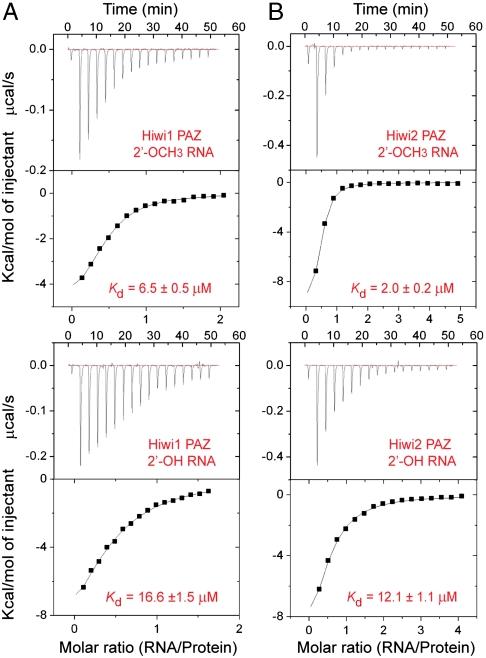
We measured apparent dissociation constants (Kd) for binding of RNA ligands containing 3′ ends that are either 2′-OCH3 or 2′-OH to the PAZ domains of human Piwi proteins by isothermal titration calorimetry (ITC) (Table 1). The 14-nt-long RNA used in these experiments forms a 12-nt self-complementary duplex with a U-U overhang at the 3′ end. The ITC binding data clearly establish that Hiwi1 PAZ (Fig. 1A), Hiwi2 PAZ (Fig. 1B), and Hili PAZ (Fig. S2) bind 2′-OCH3 RNA with 2.5- to 6-fold higher affinity compared to its 2′-OH counterpart. This trend is opposite from that observed for Ago1 PAZ domain, which binds 2′-OH RNA with higher affinity than its 2′-OCH3 counterpart (9).
Table 1.
ITC binding data for Piwi PAZ-RNA complex formation
| N, sites | Kd, μM | ΔH, Kcal/mol | ΔS, cal·mol-1· deg-1 | |
| Hiwi1 PAZ-RNA 14-mer 2′-OCH3 | 0.43 ± 0.01 | 6.5 ± 0.5 | −5.6 ± 0.2 | 5.04 |
| Hiwi1 PAZ-RNA 14-mer 2′-OH | 0.43 ± 0.03 | 16.6 ± 1.5 | −13.4 ± 1.2 | −23.1 |
| Hiwi2 PAZ-RNA 14-mer 2′-OCH3 | 0.39 ± 0.01 | 2 ± 0.2 | −10.9 ± 0.4 | −10.3 |
| Hiwi2 PAZ-RNA 14-mer 2′-OH | 0.45 ± 0.04 | 12.1 ± 1.1 | −14.3 ± 1.5 | −25.3 |
| Hili-PAZ-RNA 14-mer 2′-OCH3 | 0.59 ± 0.03 | 10 ± 0.5 | −19.0 ± 1.0 | −40.7 |
| Hili-PAZ-RNA 14-mer 2′-OH | 0.47 ± 0.16 | 33.9 ± 4.0 | −19.6 ± 7.1 | −45.4 |
| Hiwi1 PAZ (M381Y/379H)-RNA 14-mer 2′-OCH3 | 0.41 ± 0.02 | 9.5 ± 0.8 | −8.1 ± 0.5 | −4.2 |
| Hiwi1 PAZ (M381Y/379H)-RNA 14-mer 2′-OH | 0.52 ± 0.04 | 21.8 ± 2.0 | −12.8 ± 1.4 | −21.7 |
Fig. 1.
RNA-binding analyses on human Piwi PAZ domains. (A) ITC measurements for binding of Hiwi1 PAZ to self-complementary 14-mer RNA (12-bp + 2-nt overhang) containing 2′-OCH3 (Top) and 2′-OH (Bottom) at the 3′ end. The evaluated dissociation constants (Kd) are listed in the panels. (B) ITC measurements for binding of Hiwi2 PAZ to self-complementary 14-mer RNA containing 2′-OCH3 (Top) and 2′-OH (Bottom) at the 3′ end.
Structure of a Hiwi1 PAZ Domain Bound to 3′-End 2′-O-Methylated RNA.
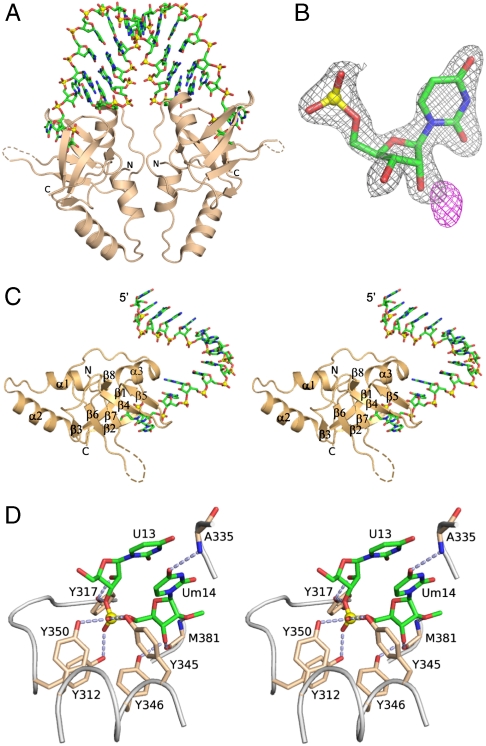
We next attempted to grow crystals of PAZ domains of Hiwi1/2/3 and Hili proteins bound to self-complementary duplexes of varying length containing 2-nt 3′-overhangs with 2′-OCH3 modifications at their 3′ ends. We were able to successfully grow crystals for a Hiwi1 PAZ construct (amino acids 271–391) bound to a self-complementary GCGAAUAUUCGCUUm 14-mer sequence that diffracted to 2.90 Å. This complex (space group P4122, Table S1) is composed of Hiwi1 PAZ domains bound to U-Um 2-nt overhangs (Um stands for the 3′-terminal U modified by a 2′-OCH3 group) at either end of the duplex (Fig. 2A). We introduced a Met site (chose V361M mutant following sequence alignment of Piwi PAZ domains, Fig. S1) in the Hiwi1 PAZ sequence, which not only improved the phasing power [following selenomethionine (SeMet) incorporation], but also dramatically improved the resolution of the complex to 2.1 Å (Table S1). The V361M mutant is located 8 Å from the Hiwi1 PAZ-binding pocket and had no impact on its conformation (Fig. S3). The methyl group of 2′-OCH3 at the 3′ end can be readily detected in the electron density map of the Hiwi1 PAZ-RNA complex (Fig. 2B). The 2Fo - Fc map (black) is contoured at 1.8σ and Fo - Fc map (magenta) is contoured at 2.0σ in Fig. 2B.
Fig. 2.
Crystal structure of the Hiwi1 PAZ-RNA complex. (A) Ribbon representation of the structure of the Hiwi1 PAZ-RNA complex. The RNA is a self-complementary 14-mer (12-bp + 2-nt overhang) sequence containing 2′-OCH3 at its 3′ end. The complex contains Hiwi1 PAZ modules bound to the 2-nt overhangs at either end of the RNA duplex. (B) Experimental electron density (Fo - Fc map) observed for the methyl group of 2′-OCH3 of Um in the Hiwi1-PAZ-RNA complex. Fo - Fc map is contoured at 2.0σ and shown in magenta. Electron density (2Fo - Fc map) corresponding to terminal nucleotide Um without methyl group (green) is contoured at 1.8σ and shown in black. (C) Stereo view of the structure of the Hiwi1 PAZ-RNA complex containing one PAZ domain bound to the overhang-containing strand, corresponding to the 1∶1 PAZ∶ssRNA stoichiometry seen in the asymmetric unit in the crystal. (D) Stereo view of intermolecular interactions between 2-nt U13-Um14 overhang (in green) with labeled amino acid side chains (in biscuit) lining the Hiwi1 PAZ-binding pocket. Intermolecular hydrogen-bonds are indicated by dashed silver lines.
The RNA duplex conformation in the Hiwi1 PAZ-RNA complex superpositions reasonably well over most of its duplex length with an A-form RNA duplex, except toward the 3′-overhang ends (rmsd of 2.6 Å over all atoms), as shown in Fig. S4. The PAZ domains bound at either end of the RNA duplex are brought into close proximity, such that crystal packing interactions between them could introduce strain into the conformation of the bound RNA.
A view of one symmetrical half of the complex composed of the overhang-containing strand inserting its 3′ end into the Hiwi1 PAZ domain is shown in stereo in Fig. 2C, with the α-helices and β-strands labeled in this figure. The RNA can be readily traced in the complex and the 2Fo - Fc map (contoured at 1.2σ) is plotted in Fig. S5. Details of intermolecular contacts between the 2-nt U-Um overhang that is 2′-OCH3 modified at its 3′ end and aromatic residues lining the Hiwi1 PAZ-binding pocket are shown in stereo in Fig. 2D. The overhang nucleotides U13 and Um14 retain a stacked A-helical conformation and are inserted into a pocket lined by conserved aromatic and hydrophobic residues (Fig. 2D). Intermolecular hydrogen bonds between the phenolic hydroxyls of three tyrosines and the nonbridging phosphate oxygens of the U13-Um14 step anchor the overhang segment within the Hiwi1 PAZ-binding pocket. Specifically, the 2′-OH of U13 hydrogen bonds to Tyr317, while the 3′-OH of Um14 hydrogen bonds to Tyr346 and main-chain oxygen atom of Met381. The sugar moiety of terminal nucleotide Um14 forms van der Waals interactions with the hydrophobic side chains of Leu382 and Ala380, whereas its 2′-oxygen atom forms a hydrogen bond with main-chain amide group of Met381 (Fig. 2D). Among human Piwi proteins, all these interactive residues are conserved (Fig. S1), indicating most likely a similar mode of RNA 3′-end recognition by Hiwi1/2/3 and Hili proteins.
We have also solved the structure of Hiwi1 PAZ domain bound to the same self-complementary RNA containing 2′-OH at its 3′ end. This complex (space group P4122, Table S1) adopts the same structure as its 2′-OCH3 modified counterpart, establishing that the Hiwi1 PAZ-binding pocket is capable of accommodating 3′ ends that are either 2′-OCH3 or 2′-OH.
Piwi PAZ-Binding Pockets in Free and RNA-Bound States.
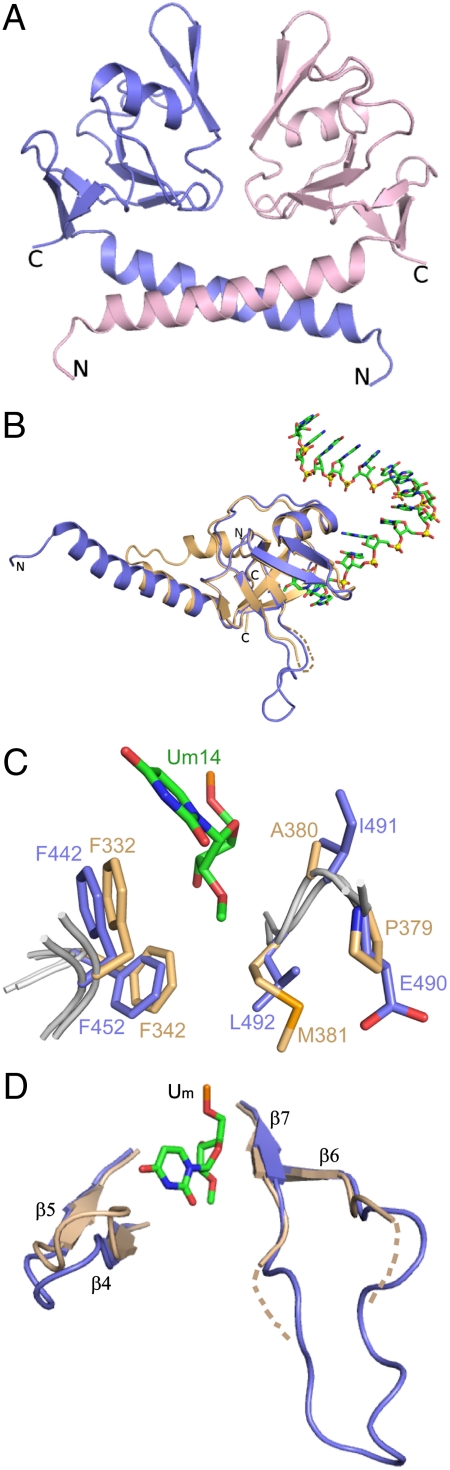
We attempted to grow crystals of Hiwi1/2/3 and Hili-PAZ domains in the free state and were successful for a Hili construct (amino acids 388–525) that diffracted to 2.92 Å. The Hili PAZ (space group C2221, Table S1) forms a symmetric dimer mediated by N-terminal helix–helix interactions as shown in Fig. 3A.
Fig. 3.
Structure of Hili PAZ in the free state and comparison of its binding pocket with Hiwi1 PAZ in RNA-bound state. (A) A view of the crystal structure of Hili PAZ in the free state. Two Hili-PAZ molecules (one in blue and the other in pink) form a symmetrical dimer mediated by helix–helix interactions between N-terminal α-helical segments. (B) Structural superposition of the PAZ domains of Hili PAZ in the free form (in blue) and Hiwi1 PAZ (in biscuit) bound to RNA (in green). (C) Structural superposition of binding pockets of Hili PAZ in the free form (in blue) and Hiwi1 PAZ (in biscuit) bound to RNA. The Um14 residue at the 3′ terminus of the RNA in the complex is shown in green. (D) A view of the relative arrangements of β6-β7 strands and β4-β5 strands of Hili PAZ (in blue) in the free state and Hiwi1 PAZ (in biscuit) in the RNA-bound complex. The 2′-OCH3-modified Um14 residue in the Hiwi1 PAZ-RNA complex is shown in green.
Except for the N-terminal helix, the PAZ domains of Hili in the free state (blue color in Fig. 3B) and Hiwi1 in the RNA-bound state (biscuit color, with bound RNA in green, Fig. 3B) superpose quite well (rmsd of Cα atoms = 0.81 Å, Fig. S6), as do the residues lining their binding pockets (Fig. 3C; same color coding as in Fig. 3B). The walls of the PAZ-binding pocket are generated by β4-β5 segment along one face and by β6-β7 segment on an opposing face. The β4-β5 and β6-β7 segments superpose quite well between the Hili-PAZ domain in the free state (blue color) and Hiwi1 PAZ (biscuit color) in the RNA-bound state (Um14 in green) (Fig. 3D). The loop connecting β6 and β7 can be traced in Hili PAZ in the free state, whereas a 10-aa segment at the tip of the loop is disordered in the Hiwi1 PAZ complex in the RNA-bound state (Fig. 3D).
2′-OCH3 Insertion into Piwi and Ago PAZ-binding pockets.
The structures of Hiwi1 PAZ-RNA (PAZ in biscuit and 3′-terminal nucleotide Um in green) and Ago1 PAZ-RNA (PAZ in cyan and 3′-terminal U in yellow) complexes are superposed in stereo in Fig. S7. The PAZ domains in these two complexes exhibit an rmsd of Cα atoms of 1.77 Å.
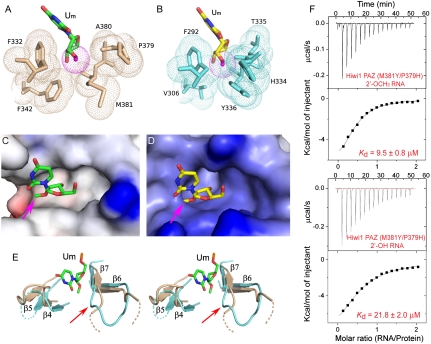
Insertion of the 3′-terminal Um nucleotide containing a 2′-OCH3 group into its Hiwi1 PAZ domain binding pocket is shown in Fig. 4A. The methyl surface of the 2′-OCH3 group (dotted red-colored ball) shows no steric clashes with the surfaces of the amino acid side chains lining the binding pocket (dotted biscuit-colored balls). Insertion of the 3′-terminal U nucleotide containing a 2′-OH group into its Ago1 PAZ domain binding pocket is shown in Fig. 4B, following replacement of the 2′-OH group by a modeled 2′-OCH3 group. We observe steric clashes of the modeled 2′-OCH3 group (dotted red-colored ball) with the surfaces of the amino acid side chains lining the binding pocket (dotted cyan-colored balls). Indeed, electrostatic surface representations show that there is room to accommodate the 2′-OCH3 group within a hydrophobic cleft in the Hiwi1 PAZ-binding pocket (see pink arrow, Fig. 4C), but there is no room to accommodate the modeled 2′-OCH3 group due to steric clashes within the Ago1 PAZ-binding pocket (see pink arrow, Fig. 4D).
Fig. 4.
Methyl group of 2′-OCH3 can be accommodated within the Hiwi1 but not Ago1 PAZ-binding pockets. (A and B) Binding pocket residues surrounding the methyl group of 2′-OCH3 at the RNA 3′ end in Hiwi1 PAZ (A) and Ago1 PAZ (B, modeled 2′-OCH3 to replace 2′-OH observed in the crystal structure) RNA complexes. Binding pocket residues in Hiwi1 PAZ (in biscuit, A) and Ago1 PAZ (in cyan, B) complexes are highlighted as dotted balls in a van der Waals radius representation. The 2′-OCH3 residues (dotted ball) are shown in green for the Hiwi1 PAZ-RNA complex (A) and in yellow for the Ago1 PAZ-RNA complex (B). (C and D) Electrostatic surface representation of Hiwi1 PAZ (C) and Ago1 PAZ (D) binding pockets in their RNA complexes containing 2′-OCH3 and 2′OH (replaced by 2′-OCH3) at their 3′ ends respectively. The Hiwi1 PAZ-binding pocket can readily accommodate the methyl group (shown by pink arrow) of 2′-OCH3 of Um at the 3′ end, but the more constricted Ago1 PAZ-binding pocket is unable, due to steric clashes, to accommodate the methyl group. (E) A stereo view of the relative arrangements of β6-β7 strands and β4-β5 strands of Hiwi1 PAZ (in biscuit) and Ago1 PAZ (in blue) in their RNA-bound complexes. The 2′-OCH3-modified Um14 residue in the Hiwi1 PAZ-RNA complex is shown in green. (F) ITC measurements for binding of Hiwi1 (M381Y/P379H) PAZ to self-complementary 14-mer RNA (12-bp + 2-nt overhang) containing 2′-OCH3 (Top) and 2′-OH (Bottom) at the 3′ end.
In addition, the β4-β5 and β6-β7 segments are further apart in Hiwi1 PAZ (biscuit) in the RNA-bound state (Um14 in green) than they are in the Ago1 PAZ (cyan) in the RNA-bound state, as can be observed in the stereo view in Fig. 4E (U9 in the Ago1 PAZ-RNA complex superposes well on Um14 of the Hiwi1 PAZ-RNA complex, and is not shown), indicative of a wider binding pocket to accommodate the 2′-OCH3 group in the former compared to the latter complex.
Impact of Binding Pocket Mutants.
An examination of the positioning of the 2′-OCH3 group in Hiwi1 PAZ and Ago1 PAZ-binding pockets in their RNA complexes indicates that a backbone carbonyl oxygen (of Pro379) is ≈3.5 Å from the methyl carbon of the 2′-OCH3 group in the Hiwi1 PAZ-RNA complex (Fig. S8A), whereas the separation between the same atoms is reduced to ≈2.1 Å, indicative of a steric clash, in the modeled Ago1 PAZ-RNA complex (Fig. S8B).
Based on this observation, we attempted to engineer the methyl group-binding pocket in Hiwi1 PAZ by introducing a double mutant (M381Y/P379H), where Met and Pro residues lining the pocket (Fig. S8A) were replaced by their Tyr and His counterparts, as observed for the Ago1 PAZ-binding pocket (Fig. S8B). We measured the binding affinities using ITC for the M381Y/P379H double mutant of Hiwi1 PAZ for 14-mer RNAs containing 2′-OCH3 and 2′-OH at the 3′ end and found no significant difference in the relative preference between wild-type (Fig. 1A) and double mutant (Fig. 4F) Hiwi1 PAZ (Table 1). This result implies that the backbone conformation rather than the amino acid composition of the binding pocket contributes to preferential binding of Hiwi1 PAZ for RNAs with 2′-OCH3 over 2′-OH at 3′ ends.
Comparison of RNA Alignments in Piwi and Ago PAZ-RNA Complexes.
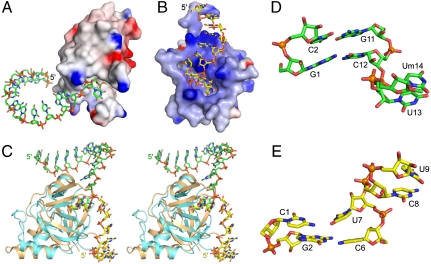
The structure of Hiwi1 PAZ bound to the overhang-containing strand of a self-complementary 14-mer RNA (12-bp and 2-nt 3′-overhang) containing a 2′-OCH3 group at its 3′ end (protein in electrostatic surface view and bound ssRNA in green, Fig. 5A) is compared with the previously reported structure (9) of Ago1 PAZ bound to the overhang-containing strand of a self-complementary 9-mer RNA (7-bp and 2-nt 3′-overhang) containing a 2′-OH group at its 3′ end (protein in electrostatic surface view and bound ssRNA in yellow, Fig. 5B). The face of the PAZ domain that includes the binding pocket is primarily hydrophobic (silver color) in the Hiwi1 PAZ-RNA complex (Fig. 5A), whereas it is strongly basic (blue color) in the Ago1 PAZ-RNA complex (Fig. 5B). Thus, the RNA segment positioned outside the PAZ-binding pocket is directed away from the hydrophobic surface of the Hiwi1 PAZ domain in its complex (Fig. 5A). By contrast, the RNA sugar-phosphate backbone interacts with basic side chains lining the surface of the Ago1 PAZ domain in its complex (Fig. 5B) (9).
Fig. 5.
Orientation of bound RNA with respect to Ago1 and Hiwi1 PAZ domains. (A) Electrostatic surface representation of Hiwi1 PAZ domain with bound RNA (in green). (B) Electrostatic surface representation of Ago1 PAZ domain with bound RNA (in yellow) (9). (C) Stereo view of superposed complexes of Hiwi1 PAZ (in biscuit) with bound 14-mer RNA (in green) and Ago1 PAZ (in cyan) with bound 9-mer RNA (in yellow). (D) Relative orientation of 2-bp duplex and 2-nt overhang (U13-Um14) segments in the Hiwi1 PAZ complex with 14-mer RNA (12-bp and 2-nt overhang) with 2′-OCH3 group at 3′ end. (E) Relative orientation of 2-bp duplex and 2-nt overhang (U8-U9) segments in the Ago1 PAZ complex with 9-mer RNA (7-bp and 2-nt overhang) with 2′-OH group at 3′ end (9).
The different trajectories adopted by the RNA in the two complexes following superposing of their PAZ domains (Hiwi1 PAZ in biscuit and Ago1 PAZ in cyan) are shown in stereo in Fig. 5C. These differences reflect alternate conformational alignments at the junctional phosphate between duplex and overhang segments between the two complexes. The 2-nt overhangs adopt similar stacked conformations in the Hiwi1 PAZ complex with the 14-mer RNA (green color, positions U13 and Um14, Fig. 5D) and the Ago1 PAZ complex with the 9-mer RNA (yellow color, positions C8 and U9, Fig. 5E). The orientations are distinctly different between junctional bases C12 and U13 in the Hiwi1 PAZ-RNA complex (Fig. 5D) and U7 and C8 in the Ago1 PAZ-RNA complex (Fig. 5E) (9).
We have superpositioned the overhang-containing strands in the Ago1 PAZ-RNA (9) and the Hiwi1 PAZ-RNA complexes with the bound 21-mer guide strand in the binary full-length Thermus thermophilus Ago complex (19) in Fig. S9 A and B, respectively. It is apparent that the trajectory of the overhang-containing strand in the Ago1 PAZ-RNA complex is closest to the conformation of guide RNA bound in full-length Ago structure.
Discussion
Piwi PAZ Domain Contains a Preformed RNA-Binding Pocket.
The core fold of the Hiwi1 PAZ domain in its RNA complex consists of a twisted β-barrel formed by six β-strands (β1-β3, β6-β8) connected to an αβ-module (β4-β5-α3) (Fig. 2C). The overall structure of Hili PAZ in the free state superposes well with that of Hiwi1 PAZ in the RNA-bound state (Fig. 3 B–D), consistent with formation of an essentially preformed Piwi PAZ-binding pocket. The main difference is away from the binding pocket, where the N terminus contains two short α-helices (α1 and α2), with α1 folding back toward the β-barrel, for Hiwi1 PAZ in its RNA-bound state (Figs. 2C and 3B), while a single long α-helix is observed at the N terminus for Hili PAZ in the free state (Fig. 3 A and B).
Common Hydrophobic RNA-Binding Pocket Amongst Piwi PAZ Domains.
The PAZ domain binding pockets are lined by hydrophobic residues in the superposed structures of Hili PAZ in the free state and Hiwi1 PAZ in complex with 2′-OCH3 RNA (Fig. 3C). Thus, Phe332 and Phe342 in Hiwi1 PAZ are replaced by Phe442 and Phe452 in Hili PAZ (Fig. 3C), whereas consecutive residues within the loop between β6 and β7, Ala380 and Met381 in Hiwi1 PAZ are replaced by their hydrophobic counterparts Ile491 and Leu492 in Hili PAZ (Fig. S1). In addition, the corresponding residues are similar and hydrophobic in nature in Hiwi2 and Hiwi3 PAZ domains (Fig. S1), indicative of a common hydrophobic binding pocket amongst Piwi PAZ domain family members.
Recognition of 2′-OCH3 Modification by Piwi PAZ Domains.
The electron density for the methyl group at the 2′-OCH3 position is very well defined in the structure of the Hiwi1 PAZ complex with 2′-O-methylated RNA (Fig. 2B). Notably, this methyl group projects toward a hydrophobic groove formed by β6-β7 strands on one side and β4-β5 strands on the other side (Fig. 4C). In the Hiwi1 PAZ complex with 2′-OCH3 RNA, this methyl group is surrounded by Phe332, Phe342, Pro379, Ala380, and Met381 (Fig. 4A), with the conformation of these residues remaining the same in the Hiwi1 PAZ complex with the 2′-OH RNA.
These observations are supported by the ITC binding data that establish that human Piwi PAZ domains bind RNAs that are 2′-OCH3 at the 3′ end with three- to sixfold higher binding affinity compared to RNAs that are 2′-OH at their 3′ ends (Fig. 1 A and B and Fig. S2). This modest preference for 2′-O-methylated RNA most likely reflects the contributions of additional hydrophobic interactions formed between the methyl group and hydrophobic residues lining the Piwi PAZ-binding pocket.
Factors Dictating Dimensions of Ago PAZ Domain Binding Pockets.
An examination of the binding pocket in the Ago1 PAZ-RNA complex clearly highlights several clashes between the superposed methyl group (2′-OCH3 replacing 2′-OH) and surrounding residues (Phe292, Val306, His334, Thr335, and Tyr336) lining the Ago1 PAZ-binding pocket (Fig. 4 B and D).
Unlike Piwi PAZ domains, the human Ago1 PAZ domain has no Piwi PAZ-specific insertion element (Fig. S1), resulting in a sharp turn between β6 and β7, which in turn leads to formation of a main-chain bulge protruding toward the 2′-O site (see red arrow, Fig. 4E). The binding pocket is also narrower in the Ago1 PAZ-RNA complex relative to the Hiwi1 PAZ-RNA complex due to the movement of the β4-β5 strands toward the 2′-O site (Fig. 4E). Both these factors make any modification at the 2′-O position unfavorable for binding within the Ago1 PAZ pocket.
Factors Dictating Dimensions of Piwi PAZ Domain Binding Pockets.
The electron density corresponding to the 10-aa segment (amino acids 368–377) between β6 and β7 strands is missing in the structures of Hiwi1 PAZ complexes with bound RNA containing either 2-OCH3 or 2′-OH at the 3′ end, whereas the same segment can be precisely traced in the structure of Hili-PAZ domain in the free state (Fig. 3D). Our structural studies suggest that this 10-aa segment, which partially overlaps with the Piwi PAZ-specific insertion element (amino acids 370–378) (Fig. S1), could facilitate the accommodation of the methyl group of 2′-OCH3 at the 3′ end following insertion into its Piwi PAZ-binding pocket (Fig. 4E). Although residues present in the Piwi PAZ-specific insertion element are not conserved among human Piwi proteins, the flexible nature of this linker loop between β6 and β7 in the Hiwi1 PAZ-RNA complex most likely prevents protrusion of structural elements toward the 2′-O site, thereby providing enough space for accommodation of the inserted 2′-OCH3 group.
Trajectories of RNA Bound to Piwi and Ago PAZ Domains.
The role of Ago1 PAZ is to target double-stranded RNA containing 2-nt overhangs ending in 2′-OH at the 3′ end, whereas Hiwi PAZ targets single-stranded RNA containing 2′-OCH3 at the 3′ end. The 2-nt 3′ ends of the bound RNAs are anchored in a similar manner within the PAZ-binding pockets in the Hiwi1 PAZ and Ago1 PAZ complexes, but the remainder of the RNA takes very different trajectories in these two complexes (stereo view in Fig. 5C). There is minimal contact of the RNA (in green) with the hydrophobic surface (colored silver, Fig. 5A) of the Hiwi1 PAZ domain. By contrast, there is extensive contact of the RNA (in yellow) along its entire length, with the positively charged basic surface (colored blue, Fig. 5B) of the Ago1 PAZ domain, mediated through lysine-phosphate interactions (9). Thus, there are more intermolecular contacts observed in the Ago1 PAZ-RNA complex than in the Hiwi1 PAZ-RNA complex, which may account for the difference in trajectories. We also cannot rule out the contributions of crystal packing interactions, because the PAZ domains are bound to both ends of the siRNA duplexes in both complexes. In this regard, the PAZ domains are further apart in the Ago1 PAZ-RNA complex (9) compared to the Hiwi1 PAZ-RNA complex.
Functional Implications of 2′-O-Methylation at piRNA 3′ Ends.
The 3′ end of RNA is known to be susceptible to degradation by RNase and modification by RNA-modifying enzymes. Methylation of piRNA by HEN1 may play a protective role by preventing undesirable decay from this end, which is known to occur for miRNAs in plants (30, 31). The present study provides insights into how human Piwi proteins have evolved to recognize 2′-O-methylated piRNA. Recent structural studies on thermophilic eubacterial Argonautes bound to guide and target strands have established that the 2-nt overhang at the 3′ end of the guide strand gets released from the PAZ pocket once the propagation step of guide-target pairing exceeds a certain duplex length (3). Unlike most miRNAs, piRNAs require perfect complementarity with the target strand for target cleavage (13, 32, 33). It is possible that in the case of Piwi proteins, this requirement for perfect complementarity could lead to release of the 2-nt overhang at the 3′ end of piRNA from the PAZ pocket as a prerequisite for slicer activity. Protection of piRNA by methylation at the 3′ end would result in a much longer-lived RNA, making it less susceptible to degradation.
Experimental Procedures
Protein Preparation.
The human Piwi PAZ domains of Hiwi1 (residues 277–399), Hiwi2 (residues 271–407), Hiwi3 (residues 341–477), and Hili (residues 388–525) were generated using a standard PCR-based cloning strategy. They were cloned into a modified pET-28b vector (EMD Biosciences) with a His6-SUMO (small ubiquitin-related modifier) tag at the N terminus. All clones were overexpressed in Escherichia coli strain BL21 (DE3) cells. The proteins were purified from the soluble fraction by a nickel-chelating affinity column, followed by overnight treatment with SUMO protease Ulp1 at 4 °C to cleave the His6-SUMO tag, with the cleaved tag removed using a second nickel-chelating column. Proteins were further purified by gel filtration chromatography using HiLoad 16/60 Superdex-75 prep grade column (Amersham). Peak fractions obtained from gel filtration experiments were pooled and concentrated using a Centricon (Vivaspin) centrifugal concentrator with a 10 kDa cutoff. Protein purity was assessed by SDS-PAGE, with the purified proteins immediately frozen in liquid nitrogen and stored at -80 °C. The V361M and M381Y/P379H mutant-containing Hiwi1-PAZ domains were generated by a two-step PCR-based overlap extension method and confirmed by DNA sequencing. The l-SeMet derivatized protein was produced using the feedback-inhibition of methionine synthesis pathway. SeMet-containing protein was purified using the same protocol as for the native protein. A 1 mM DTT concentration was maintained during protein purification to prevent oxidation of SeMet.
RNA Preparation.
The 5′-GCGAAGCUUCGCUU-3′ 14-mer sequence containing a 2′-OH 3′ end was chemically synthesized on a 3400 DNA synthesizer (Applied Biosystems), and subsequently deprotected and purified by denaturing polyacrylamide gel electrophoresis. The 5′-GCGAAGCUUCGCUUm-3′ 14-mer sequence containing a 2′-OCH3 at the 3′ end was purchased from Dharmacon. The self-complementary RNAs were dissolved in diethyl-pyrocarbonate-treated water and annealed by heating to 95 °C for 1 min followed by temperature reduction in a 37 °C water bath over a period of 1 h.
Crystallization and Data Collection.
Crystals of Hili PAZ in the free state were grown using the hanging-drop vapor-diffusion method by mixing the protein (20 mg/mL in 100 mM NaCl, 20 mM Tris•HCl, pH 8.0) with an equal volume of reservoir solution containing 4–8% PEG4000 and 50 mM MgSO4 in 50 mM MES buffer, pH 5.6 to 6.1. Small crystals appeared overnight at 20 °C and grew to full size within 1 wk.
Hiwi1 PAZ was mixed with RNA containing 2′-OH or 2′-OCH3 at the 3′ end in a molar ratio of 1∶1.2 and incubated at 4 °C for at least 1 h. The complex crystals were grown using the hanging-drop vapor-diffusion method by mixing the protein-RNA complex (≈5mg/mL in 100 mM NaCl, 20 mM Tris•HCl, pH 8.0) with an equal volume of reservoir solution containing 1–5% PEG3350, 50 mM NaCl in 50 mM Na-acetate buffer, pH 4.6 at 4 °C. The crystals grew to full size in ≈10 d.
For data collection, crystals were flash frozen (100 K) in the above reservoir solutions supplemented with 30% (vol/vol) ethylene glycol. The single-wavelength anomalous dispersion (SAD) data were collected for the SeMet-labeled crystals at Se-peak wavelength. Diffraction datasets were collected on 24-ID-C and 24-ID-E beamlines at the Advanced Photon Source and X29 beamline at Brookhaven National Laboratory. The collected datasets were integrated and scaled using the HKL2000 suite (34).
Structure Determination and Refinement.
Crystals of Hili-PAZ in the free state belonged to the space group C2221 (Table S1) and contained four molecules of the protein in the asymmetric unit. The structure of the Hili-PAZ was determined by the SAD method using Se anomalous scattering data, with the selenium sites located and refined for phasing using SHARP (35). Phase improvement with SOLOMON, as implemented in SHARP (36), produced a clearly interpretable electron density map, from which an initial model was built manually using COOT (37). The structure was completed through several rounds of manual model fitting in COOT and refinement coupled with noncrystallographic symmetry averaging using PHENIX.REFINE (38).
The crystal of Hiwi1 PAZ in complex with 2′-O-methylated RNA belonged to space group P4122 (Table S1), with the asymmetric unit containing one molecule each of protein and 14-mer single-stranded RNA. Molecular replacement programs using the structure of Hili PAZ in the free state as a search model failed to give any solution. Our attempt to solve the structure of the complex using multiwavelength anomalous dispersion datasets collected on SeMet-labeled protein-containing crystals of the complex was also unsuccessful due to lack of sufficient phasing power. Based on sequence alignment among Piwi proteins, we mutated Val361 present in Hiwi1 PAZ into Met, so as to improve the phasing power. Unexpectedly, the V361M mutation not only increased the phasing power resulting in an interpretable electron density map, but also improved the resolution from 2.9 to 2.1 Å. The structure of Hiwi1(V361M) PAZ in complex with 2′-O-methylated 14-mer RNA was solved by SAD method using the data collected at Se-peak wavelength by a procedure similar to that described above for Hili PAZ in the free state.
The structures of native Hiwi1 PAZ in complex with 2′-OCH3 and 2′-OH containing 14-mer RNAs was solved by difference Fourier using the structure of the SeMet-labeled Hiwi1(V361M) PAZ-RNA complex. These structures were subject to several rounds of refinement in REFMAC5 (39) and manual model building using COOT (37). Water molecules were included near the end of refinement. The majority of the model has a clear and well-interpretable electron density map with the exception of a few solvent-exposed side chains, which were omitted in the final model. The geometry of the final model was checked using PROCHECK (40). The final data collection and refinement statistics are given in Table S1.
Figures were prepared using the programs PyMol (41) and ESPript (42).
ITC Binding Assays and Data Analysis.
All human Piwi PAZ domains and RNAs were dialyzed extensively against running buffer (100 mM NaCl, 10 mM Tris•HCl, pH 8.0). Protein concentration was measured using absorbance at 280 nm, whereas RNA concentration was measured using absorbance at 260 nm. Prior to titration, both protein and RNA were centrifuged at 18,000 × g at 25 °C for more than 10 min to remove any debris and air bubbles. The calorimetric titrations were carried out at 25 °C on MicroCal ITC200 instrument with 16 successive injections of 2.4 μL (400–600 μM) 2′-OCH3 or 2′-OH containing RNA, spaced 180 s apart, into the sample cell containing a solution of ∼200 μL (20–40 μM) different human Piwi PAZ proteins. A control experiment was performed by titrating RNA into a sample cell that contained buffer alone, with no significant heat release observed in a control experiment. The data were analyzed using the ORIGIN software. The association constant (Ka), enthalpy change (ΔH), and the stoichiometry (N) were calculated by fitting the thermograms to one set of binding sites. The dissociation constant (Kd), free energy change (ΔG), and the entropy change (ΔS) were calculated using the equation
Supplementary Material
Acknowledgments.
We thank Zhanxin Wang of the Patel laboratory and the staff at the Advanced Photon Source beamline ID-24 and Brookhaven beamline X-29 for assistance with data collection. The research was supported by National Institutes of Health Grant AI068776 (to D.J.P.).
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected in 2009.
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PBD ID codes Hili PAZ in the free state (3O7X), Hiwi1(V361M) PAZ with 2′-OCH3 RNA (3O7V), Hiwi1 PAZ with 2′-OCH3 RNA (3O6E), and Hiwi1 PAZ with 2′-OH RNA (3O3I)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017762108/-/DCSupplemental.
References
- 1.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 2.Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siomi MC, Kuramochi-Miyagawa S. RNA silencing in germlines—exquisite collaboration of Argonaute proteins with small RNAs for germline survival. Curr Opin Cell Biol. 2009;21:426–434. doi: 10.1016/j.ceb.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 7.Song JJ, et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 8.Yan KS, et al. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 9.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 11.Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210.1–210.8. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 15.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 16.Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: Short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 17.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 18.Filipowicz W. RNAi: The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker JS. How to slice: Snapshots of Argonaute in action. Silence. 2010;1:3.1–3.10. doi: 10.1186/1758-907X-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 22.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 23.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau NC, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 27.Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 28.Horwich MD, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 29.Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 33.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: Recent developments in and around SHARP 2.0. Acta Crystallogr, Sect D: Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 36.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr, Sect D: Biol Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Afonine PV, Grosse-Kunstleve RW, Adams PD. The Phenix refinement framework. CCP4 Newsletter on Protein Crystallography 42. 2005. contribution 8.
- 39.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr, Sect D: Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck—a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 41.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 42.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.