Abstract
A recent body of evidence indicates an active role for stromal (mis)-regulation in the progression of neoplasias. Within this conceptual framework, genes belonging to the growing but still poorly characterized class of tumor antagonizing/malignancy suppressor genes (TAG/MSG) seem to play a crucial role in the regulation of the cross-talk between stromal and epithelial cells by controlling malignant growth in vivo without affecting any cancer-related phenotype in vitro. Here, we have functionally characterized the human RNASET2 gene, which encodes the first human member of the widespread Rh/T2/S family of extracellular RNases and was recently found to be down-regulated at the transcript level in several primary ovarian tumors or cell lines and in melanoma cell lines. Although we could not detect any activity for RNASET2 in several functional in vitro assays, a remarkable control of ovarian tumorigenesis could be detected in vivo. Moreover, the control of ovarian tumorigenesis mediated by this unique tumor suppressor gene occurs through modification of the cellular microenvironment and the induction of immunocompetent cells of the monocyte/macrophage lineage. Taken together, the data presented in this work strongly indicate RNASET2 as a previously unexplored member of the growing family of tumor-antagonizing genes.
Keywords: ovarian cancer, xenograft cancer model
Ovarian carcinoma is the leading cause of death from gynecological cancer in the Western world, and the majority (≈70%) of ovarian cancer patients present with an advanced disease at diagnosis, with a dismal 5-y survival rate of 30% (1). At present, cytoreductive surgery combined with platinum/taxanes chemotherapy is the standard treatment (2). Although such multimodality treatment has resulted in increased response rates and overall survival, the cure rate of the disease has not changed substantially. Indeed, whereas more than 75% of patients respond to first-line chemotherapy, ≈70% of them eventually relapse and die. Among human cancers located in the ovary, those derived from the epithelium are the most frequent (3), although very heterogeneous at both morphological and biological levels. Although such heterogeneity has contributed to difficulties in defining the molecular alterations associated with ovarian cancer development (4), all epithelial ovarian cancer subtypes originate from the single layer of epithelial cells covering the surface of the ovaries (OSE cells) (5) and a solid body of data has provided firm evidence that epithelial ovarian cancers arises from OSE cells (6). These cells undergo repeated cycles of proliferation because of the recurrent growth and rupture of ovarian follicles during the ovulatory cycle, and this phenomenon is supported by a well-characterized interaction between ovarian mesenchyme and epithelium (7). Alterations of such microenviromental interactions might therefore contribute to ovarian cancerogenesis and a growing interest has indeed been placed toward the microenvironment, based on a recent body of evidence suggesting an active role for stromal misregulation in the progression of neoplasias (7, 8). Within this conceptual framework, a special class of genes seems to play a crucial role in the regulation of the cross-talk between stromal and epithelial cells. These genes belong to the growing but still poorly characterized class of tumor antagonizing/malignancy suppressor genes (TAG/MSG) (9–12), whose principal feature is their ability to control malignant growth in vivo but not in vitro. Such “asymmetric tumor suppression” is in keeping with earlier studies showing that somatic hybridization of normal and malignant cells can suppress tumorigenicity in vivo but not cell growth in vitro (13). Moreover, recent theoretical and experimental evidences indicate that a rather large proportion of cancer genes endowed with these peculiar features has escaped detection to date (9–12). TAG/MSG genes are postulated to encode for products required to respond to differentiation-inducing signals in vivo, to mediate cellular interactions with the tumor microenvironment, or to negatively regulate angiogenesis (9–12). Significantly, although biologically different, all three mechanisms point to the occurrence of a functional cross-talk between the cancer cell and the local microenvironment as a critical factor for cancer suppression. This observation confers a particular relevance to the identification of novel TAG/MSGs, as the role played by microenvironmental cues in cancer progression has been increasingly appreciated (7, 8). A few years ago, we reported the isolation of the RNASET2 gene from human chromosomal region 6q27. This gene encodes the only human member of the widespread Rh/T2/S family of extracellular RNAses (14) and was found to be down-regulated at the transcript level in several ovarian primary tumors or cell lines, and in melanoma cell lines (15–17). Moreover, RNASET2 down-regulation was reported in other human malignancies, such as lymphomas and gliomas (18, 19), in keeping with the observation that deletions of chromosome 6q27, where the RNASET2 gene is located, are associated with a wide range of human neoplasias (20–23). Neither inactivating mutations nor abnormal CpG methylation patterns in the promoter region were detected for this gene in the ovarian tumor samples under investigation (15). However, functional studies of the human Hey3Met2 ovarian cancer and SK-MEL 28 melanoma cell lines showed that transfection of the RNASET2 cDNA in these cell lines resulted in a strong inhibition of tumor growth in vivo (15–17). Strikingly, disruption of RNASET2’s catalytic activity did not impair the inhibition of tumorigenicity with respect to the wild-type allele, ruling out ribonuclease activity as an essential requirement for the control of malignant growth (16).
Here, we report a thorough in vivo analysis of RNASET2-expressing ovarian cancer cells and compare them to a lack of evident effects in vitro. We also show that control of ovarian tumorigenesis by RNASET2 occurs through modification of the cellular microenvironment and involvement of immunocompetent cells, thus providing evidence for specific modulations of cellular responses induced by RNASET2 that might underlay ovarian tumorigenesis. Because RNASET2 appears to modulate the tumor microenvironment to repress cancer growth, we propose this gene as a previously unexplored member of the TAG/MSG gene family.
Results
RNASET2 Suppresses Tumorigenesis in Vivo but Not in Vitro Proliferation.
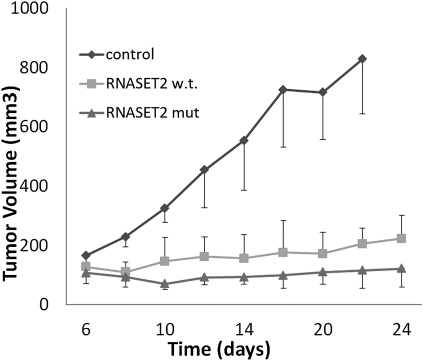
The Hey3Met2 cell line, derived from a highly metastatic subclone of the HEY4 ovarian cancer-cell line (16), shows highly decreased endogenous RNASET2 expression levels when compared with normal human ovarian epithelial cells. Moreover, because this cell line displayed the lowest levels among several other ovarian cancer cell lines (Fig. S1), it appeared to represent an ideal recipient for RNASET2 re-expression experiments. Thus, Hey3Met2 cells were stably transfected with plasmids encoding either wild-type RNASET2 or a catalytically dead form (whose cDNA has been previously mutagenized in the two CAS catalytic sites) (16) or with the empty vector as a control. Several clones transfected with RNASET2 expression vectors showed significantly increased levels of the RNASET2 protein and were thus selected for subsequent analyses. The effects of RNASET2 expression on several cancer-related parameters were subsequently investigated in vitro. In all these assays, the effect of both wild-type and catalytically dead RNASET2 expression in Hey3Met2 cells was unremarkable (Fig. S2 and Table S1). To evaluate the role of RNASET2 in vivo, we then turned to a xenograft assay. Nude mice were injected with the same Hey3Met2 clones previously investigated in vitro and followed for up to 24 d. As expected, large tumors developed in mice inoculated with vector-only transfected cells, reaching a volume 800 mm3 after 24 d. Interestingly, clones expressing either wild-type or the catalytically dead form of RNASET2 were clearly suppressed in their tumorigenic potential (Fig. 1). These notable in vivo findings clearly contrast with what was observed in the in vitro context, where RNASET2 was shown to have no effect whatsoever. To verify that expression of RNASET2 was maintained in vivo, immunohistochemical (IHC) assays with anti-RNASET2 antibodies were carried out on sections from excised tumor samples. As shown in Fig. 2 A–C, RNASET2 was clearly detected in sections from tumors inoculated with cells previously transfected with the RNASET2 expression vectors, whereas expression of the protein in the tumors bearing empty vector-transfected control cells was barely detectable. The results from both in vitro and in vivo functional assays thus showed that RNASET2 behaves similarly to those tumor-suppressor genes whose biological function is carried out in vivo but not in vitro (9–12). Under the assumption that these tumor suppressors exert their action within the tumor microenvironment, we next investigated several components that define the architectural structure of the tumor tissue.
Fig. 1.
Expression of RNASET2 suppresses tumor growth in vivo. Following subcutaneous inoculation of nude mice with Hey3Met2 clones stably transfected with pcDNA3 control plasmid vector or RNASET2-expressing vectors, tumors were collected at days 22 to 24 for histological and IHC analyses. At least five mice were inoculated for each tested clone. Bars represent SD values.
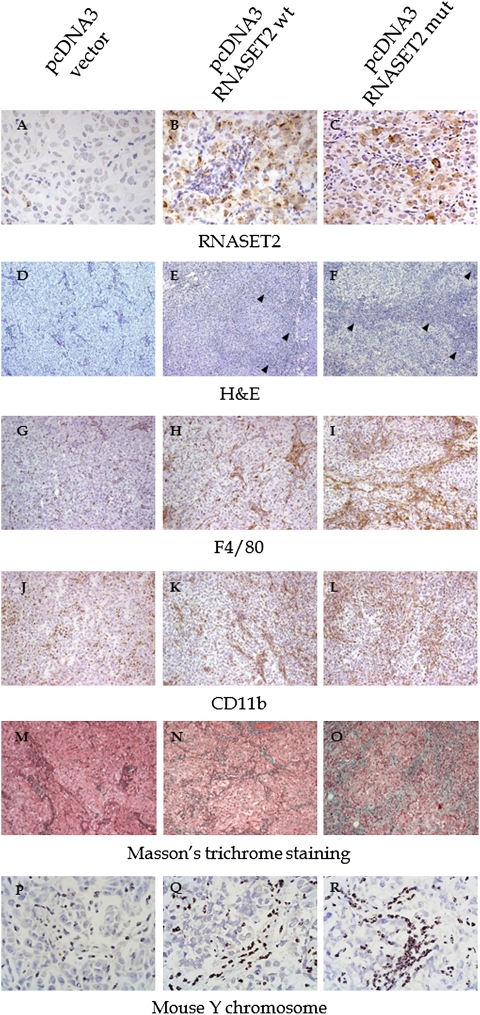
Fig. 2.
IHC, histological, and CISH analysis of the cell infiltrate in tumor samples. (A–C) Hey3Met2-derived tumors grown in nude mice were paraffin-embedded and tissue sections were processed for IHC assay with anti-RNASET2 antibody. Photomicrographs show the distribution of RNASET2 in tumor xenograft derived from pcDNA3/RNASET2-transfected cells in comparison with control cells. (D–F) Tissue sections were also stained with H&E. The arrowheads mark the massive cellular infiltrate within RNASET2-expressing tumors. (G–L) Sections were also processed for IHC with anti-F4/80 and anti-CD11b (G–I and J–L, respectively). As shown in the photomicrographs, both wild type and mutant RNASET2-expressing tumors showed increased infiltration of F4/80 and CD11b+ cells compared with control tumors. (M–O) Tumor sections processed for Masson's trichrome staining showed an abundant collagen deposition within RNASET2-expressing tumors. (P–R) A CISH assay was carried out to confirm that cells infiltrating the tumor mass were host-derived. A DNA probe from mouse chromosome Y was used to discriminate male-host murine cells from the injected human female tumor cells. All photomicrographs shown are representative images of multiple fields examined in at least four sections derived from tumor excised in two to three independent animals for each experimental group. (Magnifications: A–C and P–R, 100×; D–O, 20×.)
Morphological Characterization of RNASET2-Expressing Tumors Grown in Vivo.
To compare the morphological features of RNASET2-expressing and control tumors, they were excised 23 d after implantation, sectioned, and stained with H&E. As shown in Fig. 2 D–F, an irregular architectural pattern was observed in both wild-type and mutant RNASET2-expressing tumors compared with control tumors. The latter were mostly filled by vital and rapidly dividing cells, whereas in RNASET2-expressing tumors scattered cells showing cytological features of apoptosis with condensation of nuclear chromatin and cytoplasm were also observed (see section below). Moreover, in wild-type and particularly in mutant RNASET2-expressing tumors, neoplastic cells were entrapped by bridges of accumulated connective tissue containing an increased population of host-derived cells (Fig. 2 D–F). Morphological analyses indicated that the latter were mostly granulocytes/mononuclear cells segregating within connective strands in the neoplastic mass. In contrast, a few granulocytes in a diffuse pattern could be seen in control tumors. IHC assays for F4/80 and CD11b markers showed that the cellular population infiltrating the neoplastic mass within RNASET2-expressing tumors was mostly represented by cells from the monocyte/macrophage lineage (Fig. 2 G–L). Accordingly, a Masson's trichrome staining indicated the prevalence of connective structures in the RNASET2-expressing tumors, likely derived from nonmalignant infiltrating mononuclear cells (Fig. 2 M–O).
To confirm the murine origin of the cell population infiltrating the tumors, a chromogenic in situ hybridization (CISH) assay was performed by taking advantage of the XX chromosomal complement of the inoculated Hey2Met3 ovarian cells in the context of a XY complement of the recipient male nude mice. This assay clearly showed that the infiltrating cellular component in RNASET2-expressing tumors was indeed of murine origin (Fig. 2 P–R).
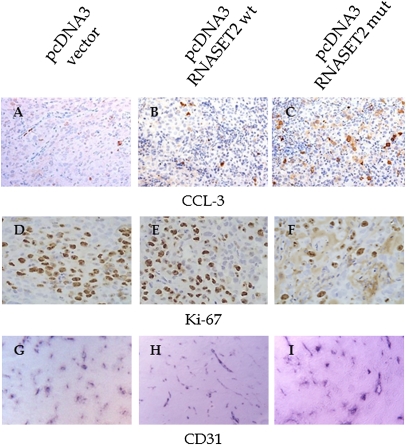
To further characterize the tumors in vivo, we then evaluated their proliferation and apoptosis rates by IHC with Ki67 and cleaved caspase-3 (CCL-3) antibodies, respectively. As shown in Fig. 3, in RNASET2-expressing tumors, and particularly in those expressing the mutant form, Ki67 staining showed a decreased proliferation rate compared with control tumors, whereas CCL-3 immunoreactive neoplastic cells showed an opposite pattern, being increased when compared with control tumor (Fig. 3 A–F and Fig. S3). In clear contrast, no apparent changes in blood-vessel density and morphology could be observed between RNASET2-transfected tumors and controls (Fig. 3 G–I).
Fig. 3.
IHC detection of tumor apoptotic/proliferating and endothelial cells in tumor samples. Sections from tumor grown in nude mice were analyzed by IHC with antiactivated caspase 3 (A–C), anti-Ki-67 (D–F), and anti-CD31 (G–I) antibodies, to detect apoptotic and proliferating cells and intratumor vessels, respectively. Photomicrographs shown are representative images of multiple fields examined in at least four sections derived from tumor excised in two to three independent animals for each experimental group. (Magnifications: A–C, 40×; D–F, 100×; G–I, 10×.)
Comprehensive Characterization of the Macrophage Infiltrating Population.
Because cells from the monocyte/macrophage lineage were shown to represent the main host cellular population infiltrating RNASET2-expressing tumors, we aimed at evaluating whether these cells could be functionally responsible for RNASET2-mediated suppression of tumorigenicity, as already described in other cancer models (24). To this aim, we exploited a different xenograft model based on Rag2−/−γc−/− mice, which lack both lymphocytes and NK cells. As shown in Fig. S4, wild-type and catalytically dead mutant RNASET2-expressing cells were again clearly suppressed in tumorigenicity. Indeed, whereas large tumors developed in mice inoculated with control cells, reaching a 1,400 mm3 volume in 19 d, clones expressing either wild-type or mutant RNASET2 produced just a slight increase in the tumor mass over time. Thus, we used this experimental model to evaluate the role played by macrophages in the control of cancer growth in vivo. To this end, Rag2−/−γc−/− mice were pretreated with the macrophage-depleting agent clodronate before inoculation of the RNASET2-expressing Hey3Met2 clones.
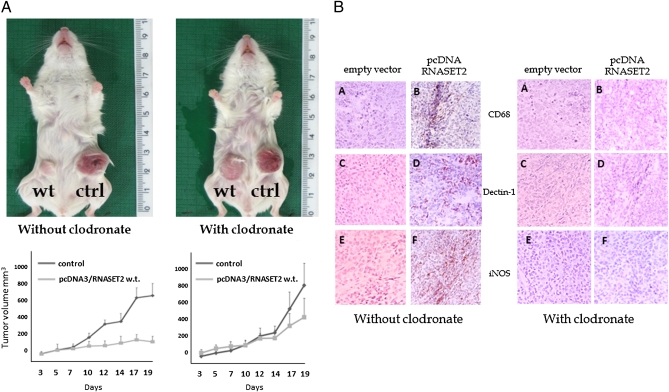
Noteworthy, the tumor suppressing activity of wild-type RNASET2 turned out to be largely impaired in clodronate-treated mice, as shown in Fig. 4A. Indeed, in contrast to the remarkable difference in tumor size between RNASET2-overexpressing vs. control clones that was observed in untreated mice, clodronate-treated animals developed RNASET2-expressing tumors whose size was very similar to that observed in control clones (Fig. 4A). To confirm the efficient depletion of host macrophages in clodronate-treated animals, IHC assay with anti-CD68 antibodies was carried out on xenograft tumor sections. As shown in Fig. 4B, the treatment was highly effective because no CD68+ cells could be detected in RNASET2-expressing tumors from clodronate-injected animals. Unfortunately, this assay was not informative with mutant RNASET2-expressing tumors, because macrophages were still detected in clodronate-treated mice following injection of mutant RNASET2-expressing cells (Fig. S5), probably as a result of the much prominent macrophages infiltration triggered by mutant versus wild-type RNASET2 from neighboring immunocompetent regions (compare Fig. 2 H and I). Taken together, these data strongly suggest that recruitment of murine host cells from the monocyte/macrophage lineage is a critical step in RNASET2-mediated tumor suppression.
Fig. 4.
RNASET2-mediated tumor suppression requires host-derived macrophages. (A) To investigate the functional role of host macrophages, Rag/γ-chain −/− mice were either mock-treated or injected with the macrophage-depleting agent clodronate before being inoculated subcutaneously with control or RNASET2-expressing Hey3Met2 clones. (Upper) A representative image of mock- or clodronate-treated single animals is shown for wild-type RNASET2-expressing tumors. The tumor growth kinetics (Lower) shows a clear increase in tumorigenicity for wild-type RNASET2-expressing tumors upon clodronate treatment. (B) To evaluate the effectiveness of clodronate, tumor sections were analyses by IHC with the macrophage lineage-specific anti-CD68 antibody (subpanels A and B). The relative proportion of M2 and M1 macrophage subpopulations in the tumors was also investigated by IHC with anti-dectin1 (subpanels C and D) and anti-iNOS (subpanels E and F) antibodies, respectively. (Left) Tumors from mock-treated animals. (Right) Tumors from clodronate liposomes-treated animals.
Tissue macrophages are known to be endowed with a wide functional plasticity, which allows them to carry out both protumoral and antitumoral activities depending on microenvironmental cues (24). Therefore, we investigated the pattern of macrophage polarization within RNASET2-expressing tumor xenografts by carrying out IHC assays with markers known to discriminate the differentiation pattern of macrophages into either M1 and M2 subtypes. Interestingly, as shown in Fig. 4B (subpanels C–F), most of the infiltrating macrophages were of the M1, inducible nitric oxide synthase-positive type, in keeping with the suggested role of this macrophage subpopulation in tumor suppression (24). These data further support the crucial role for macrophage recruitment in RNASET2-mediated tumor suppression. As a last point, a preliminary study aimed at defining the mechanism by which RNASET2 affects the behavior of the monocyte/macrophage cell lineage was carried out. To this end, in vitro chemotaxis assays were performed with peripheral blood lymphocyte-derived human monocytes, which were cultured in the top insert of Boyden chambers in the presence of conditioned media from RNASET2-expressing or control Hey3Met2 clones added to the bottom wells. Significantly, as shown in Fig. S6, conditioned media of wild-type RNASET2-expressing Hey3Met2 clones turned out to provide a strong chemotactic signal to human monocytes when compared with media conditioned by empty vector-transfected Hey3Met2 clones. These results add further evidence for a mechanistic crosstalk between extracellular RNASET2 and immunocompetent inflammatory cells.
Discussion
In this work, we report an extensive analysis of the biological properties of RNASET2 using ovarian cancer as an experimental model, and show that the remarkable tumor-suppressive activity of this gene is carried out in vivo, but not in vitro. RNASET2-mediated in vivo tumor suppression was observed in two independent xenograft models and was not dependent on the catalytic activity of RNASET2, in keeping with other reports focusing on the biological activities of T2 RNase family members (25). Noteworthy, the control of tumor growth apparently relies on the establishment of a cross-talk between RNASET2-expressing cancer cells and the tumor microenvironment. More specifically, cells of the monocyte/macrophage lineage turned out to represent the target of RNASET2 in vivo.
The failure of RNASET2 to suppress tumor growth in vitro is not unprecedented. Indeed, the lack of any RNASET2 in vitro activity was also reported by Liu et al. in a panel of several ovarian cancer cell lines, although the authors did not consider any in vivo role for the RNASET2 gene (26). On the other hand, in vivo control of tumorigenicity by RNASET2 has already been reported in human colon cancer (27) and malignant melanoma cell lines (17). However, our group is unique in reporting a comparative study of the in vivo and in vitro properties of RNASET2, and such comprehensive analyses allowed us to define this gene as a previously unrecorded member of the TAG/MSG gene family. The conceptual bases for the identification of the TAG family stem from the pioneeristic work of H. Harris and G. Klein (9, 12, 13) with somatic hybrids generated by fusion of normal and malignant cells. These hybrids lost their tumorigenicity as long as they maintained a nearly full set of chromosomes from both parental cells, but the subsequent loss of chromosomes from the normal parent cell was associated with reappearance of tumorigenicity. Indeed, later studies showed that single chromosomes from noncancer cells could suppress tumorigenicity by themselves (12, 13). Strikingly, the normal parent-derived chromosomes that were lost in the subclones that regained a tumorigenic phenotype were not those harboring P53, RB, or any other tumor-suppressor gene known at that time (9, 11, 13). Indeed, because these studies entailed the selection of in vitro permanently growing somatic hybrid lines that were then tested in vivo for tumorigenicity, the experimental design itself could not select tumor suppressor genes that would induce growth arrest or apoptosis in vitro (9–12). The genes belonging to the TAG/MSG class are postulated to encode for products required to respond to differentiation-inducing signals in vivo, to mediate normal cellular responses to microenvironment controls, or to negatively regulate the process of angiogenesis (11). Initial examples of this emerging category of genes came from the work on human chromosomal region 3p12-26 (9, 11). Among the TAGs most recently discovered are the hyaluronidase HYAL1 and HYAL2 genes (28) and the lactoferrin and LIMD1 genes, also mapping to chromosome 3p (9, 29). The hypoxia-regulated transcription factor DEC1 mapping in 9q32 and more recently the WWOX gene mapping in 16q23 have also been recently added to the TAG/MSG gene list (30, 31).
In this article, the biological properties of RNASET2 allowed us to ascribe it to the TAG/MSG gene class. Indeed, besides the observed asymmetric tumor-suppressive activity (which functionally define the TAG/MSG family), the fact that RNASET2 overexpression triggers a marked response of cells from the monocyte/macrophage lineage suggests a biological mechanism based on a microenvironmental control of tumorigenesis, which represents one of the hallmarks of the TAG family members. The crucial role of macrophage infiltration in the control of tumorigenicity in our model was demonstrated by treating Rag/γ-chain double knock-out mice with the macrophage-depleting agent clodronate. These experiments confirmed that macrophage recruitment was crucially important for the control of tumorigenicity mediated by RNASET2, because their effective ablation was associated with the reappearance of tumors indistinguishable from those of the control clones. We have also investigated the polarization pattern of the macrophage cell population infiltrating RNASET2-expressing tumors, as the M1 or M2 subtypes are associated with rather different biological responses in the context of tumor progression. As expected, most of the infiltrating macrophages were shown to express M1 markers, in keeping with a solid body of evidence pointing to this macrophage subtype as an important player in the control of cancer growth (24).
Noteworthy, a role for T2 RNases in modulating the host immune response is not unprecedented, as epitomized by the RNASET2 ortholog gene from Schistosoma mansoni, whose gene product is responsible for priming dendritic cells to trigger a T2 polarization pattern in CD4+ cells during human infection by this parasite (32). Moreover, although the mechanistic details are unknown, an involvement of the innate immune response with potential downstream consequences on neurodevelopment has also been postulated in an autosomal-recessive form of human cystic leukoencephalopathy characterized by mutations in the RNASET2 gene (33).
Finally, although our in vitro studies have not shown any particular effect mediated by RNASET2, we cannot rule out that this gene could also play a cell-autonomous role in specific in vivo conditions. In this context, it is worth mentioning a recent work on the yeast orthologue of RNASET2, Rny1p, which is involved in a conserved intracellular circuit controlling cytosolic RNA cleavage and, separately, cell fate (34–36). As a response to oxidative stress, Rny1p is induced to translocate from the vacuole into the cytosol, where it cleaves tRNA (and also rRNA or potentially some mRNAs) but also activates a downstream pathway that promotes cell death irrespective of the protein's catalytic activity (34–36). Significantly, the latter finding was also observed in our in vivo model, although it is uncertain whether the apoptotic phenotype could only be ascribed to macrophage recruitment and activation or might also be attributed to RNASET2 cell-autonomous effects within tumor cells, as observed in yeast cells (34). This topic, together with further studies to better define the mechanisms by which RNASET2 triggers tumor suppression in vivo, will be the focus of our future investigations. In conclusion, the data presented in this work altogether indicate RNASET2 as a unique member of the growing family of tumor-antagonizing genes.
Materials and Methods
Chemicals.
Cl2MDP (clodronate) was a gift of Roche Diagnostics, Mannheim, Germany.
In Vitro Assays.
The Hey3Met2 ovarian cancer cell line (16) was maintained in DMEM/F12 medium with 10% FBS and 1% GlutaMax (Gibco). Expression vectors with human RNASET2 cDNA cloned in pcDNA3 (16) were transfected into Hey3Met2 cells with Lipofectamine 2000 (Invitrogen). Single stably transfected clones were picked 14 d after selection in 300 μg/mL G418. A Western blot with anti-RNASET2 antibody on 30 μg protein extract per lane was carried out to evaluate the level of RNASET2 expression in each clone. To evaluate the proliferation rate, stably transfected Hey3Met2 clones were plated in a 24-well culture plate and grown for 8 d. Cell samples were counted daily using a hemocytometer. A growth curve was obtained by plotting the log of cell number against time and the doubling time was calculated by standard methods. To carry out colony-formation assays, 2 × 102 cells per well were plated in six-well culture plates. Following a 10- to 14-d incubation, clones were stained with 1% methylene blue/50% ethanol and colonies were manually counted. The adhesion of control and RNASET2-expressing cells to ECM components was assessed with the ECM cell adhesion kit (Chemicon International). To evaluate the apoptotic rate, the DeadEnd colorimetric TUNEL System (Promega) assay was carried out on the Hey3Met2 cell clones. To investigate anchorage-independent growth, a two-layer soft-agar test was performed. Briefly, for each clone, 100 cells were resuspended in 0.3% Bacto-Agar in DMEM/F12 supplemented with 20% FBS and then plated over a layer of 0.6% Bacto-Agar in DMEM/F12 supplemented with 20% FBS in 12-well culture plates. Colonies were counted after 14 d. For in vitro chemotaxis assay, human monocytes from healthy donors were isolated from buffy coats by standard protocols (37). Freshly prepared human monocytes (1.5 × 105) were placed in the top chamber of Boyden chambers equipped with 3-μm polycarbonate filters, whereas the bottom wells were filled with 200 μL of conditioned medium from RNASET2-expressing or control Hey3Met2 clones. Following 2 h of incubation at 37 °C, cells attached to the upper side of the filter were removed by scraping. Thereafter, cells that had actively migrated through the filter were fixed in ethanol and stained with 7.5 μg/mL DAPI before being visualized under a fluorescence microscope.
Histological and IHC Assays.
Harvested tumors were either fixed in 4% formalin or embedded in OCT and frozen in liquid nitrogen. Paraffin sections (3 μm) were either stained with H&E or Masson's trichrome for morphological analysis or immunostained with rat anti-F4/80 or rabbit anti–CCL-3, anti–Ki-67, and anti-RNASET2 (16) antibodies after antigen retrieval. Cryosections were fixed with 4% PFA and incubated with the rat anti-CD11b and rat anti-mouse CD68 (Pharmingen). The immunoreactions were revealed either by biotinylated-conjugated anti-rat antibody (Vector), horseradish peroxidase-conjugated streptavidin either by rabbit or rat on rodent HRP-polymer (Biocare Medical) and using DAB as chromogen (Biogenex). Slides were counterstained with Hematoxylin. Photomicrographs were taken using the AxioCam HRc (Zeiss) with the AxioVision System 6.4.
To evaluate blood-vessel density in tumor samples, cryosections were treated with 50 mM Levamisole to block endogenous alkaline phosphatase. A mouse anti-human PECAM-1 (CD31) antibody (1: 20; DAKO) was then incubated for 1 h at 37 °C, followed by a 45-min treatment with a secondary goat anti-mouse AP-conjugated antibody (1:200). The signal was detected with BCIP/NBT Substrate Solution (Sigma). Control reactions were carried out by omitting the primary antibody. For CISH analysis, the ZytoDot probe kit (Histo-Line) was used to hybridize a biotinylated murine chromosome Y-specific probe to serial sections of Hey3Met2-derived tumors.
In Vivo Xenograft Studies.
Male nude (five animals per group) or female Rag2−/−γc−/− (three animals per group) mice were challenged subcutaneously in the left flank with 5 × 106 Hey3Met2 cells (parental or Hey3Met2 cells expressing wild-type RNASET2 or RNASET2 catalytically dead cells) in 0.15 mL of medium and 0.15 mL matrigel (Sigma). Animals were monitored twice a week for weight and tumor growth (measuring three perpendicular diameters), and killed when the mean tumor volume of control mice reached a dimension of ≥1,000 mm3. Tumors were formalin-fixed, paraffin-embedded, cut at 5-μm thick sections and stained with H&E. Histological sections were evaluated in a double-blinded fashion. Macrophages depletion in Rag2−/−γc−/− mice was carried out using clodronate liposomes (38). Four animals per group were injected subcutaneously with control Hey3Met2 cells in the left flank and with Hey3Met2 cells expressing either wild-type or catalitically impaired mutant RNASET2 in the right flank. Each group of transplanted animals was either macrophage-depleted or not by subcutaneous injection in both flanks of clodronate liposomes (50 μL per flank) every 6 d, starting at day −2 of the tumor challenge. Animals were monitored twice a week for weight and tumor growth, and killed when the mean tumor volume reached a dimension of ≥800 mm3. Animals were maintained in a pathogen-free facility and treated in accordance with the European Union guidelines under the approval of the Ethical Committee of the Istituto San Raffaele (IACUC #418).
Macrophages populations in clodronate-treated and control animals were detected on cryosections obtained as described above, and treated with 0.3% H2O2 in PBS to remove endogenous peroxidase. Washed sections were incubated for 1 h at 37 °C with the following biotinylated primary antibodies: rat anti-mouse CD68 (1:100; HyCult Biotechnology b.v.), rat anti-mouse Dectin-1 (1:50; Cell Sciences), and rabbit anti-mouse Nos-2 (1: 10; Santa Cruz Biotechnology). The washed specimens were incubated for 30 min at room temperature with streptavidin-peroxidase (Dako), washed, and signal was detected with 0.05% DAB and 0.03% H2O in PBS. Sections were counterstained with Ematoxylin. Coverslips were mounted and slides were examined with a microscope Olympus. Images were combined with Adobe Photoshop.
Statistical Analysis.
The results from in vitro assays were evaluated by one-way ANOVA, whereas the Student t test was used to analyze in vivo data from the xenograft assays for statistical significance.
Supplementary Material
Acknowledgments
This work was partly supported by Federico Ghidoni's memorial funds. F.A. was supported by a grant from the Fondazione Comunitaria del Varesotto Organizzazione Non Lucrativa di Utilita' Sociale (ONLUS).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013746108/-/DCSupplemental.
References
- 1.Runnebaum IB, Stickeler E. Epidemiological and molecular aspects of ovarian cancer risk. J Cancer Res Clin Oncol. 2001;127(2):73–79. doi: 10.1007/s004320000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGuire WP., 3rd Current status of taxane and platinum-based chemotherapy in ovarian cancer. J Clin Oncol. 2003;21(10, Suppl):133s–135s. doi: 10.1200/JCO.2003.01.066. [DOI] [PubMed] [Google Scholar]
- 3.Feeley KM, Wells M. Precursor lesions of ovarian epithelial malignancy. Histopathology. 2001;38(2):87–95. doi: 10.1046/j.1365-2559.2001.01042.x. [DOI] [PubMed] [Google Scholar]
- 4.Shih IeM, Kurman RJ. Ovarian tumorigenesis: A proposed model based on morphological and molecular genetic analysis. Am J Pathol. 2004;164:1511–1518. doi: 10.1016/s0002-9440(10)63708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auersperg N, Maines-Bandiera SL, Dyck HG, Kruk PA. Characterization of cultured human ovarian surface epithelial cells: Phenotypic plasticity and premalignant changes. Lab Invest. 1994;71:510–518. [PubMed] [Google Scholar]
- 6.Murdoch WJ, McDonnel AC. Roles of the ovarian surface epithelium in ovulation and carcinogenesis. Reproduction. 2002;123:743–750. doi: 10.1530/rep.0.1230743. [DOI] [PubMed] [Google Scholar]
- 7.Mueller MM, Fusenig NE. Friends or foes—Bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 8.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 9.Klein G, Imreh S, Zabarovsky ER. Why do we not all die of cancer at an early age? Adv Cancer Res. 2007;98:1–16. doi: 10.1016/S0065-230X(06)98001-4. [DOI] [PubMed] [Google Scholar]
- 10.Klein G. Are there large uncharted regions of tumor suppressor genes? IUBMB Life. 2001;51(2):83–85. doi: 10.1080/15216540117178. [DOI] [PubMed] [Google Scholar]
- 11.Imreh S, Klein G, Zabarovsky ER. Search for unknown tumor-antagonizing genes. Genes Chromosomes Cancer. 2003;38:307–321. doi: 10.1002/gcc.10271. [DOI] [PubMed] [Google Scholar]
- 12.Klein G. Toward a genetics of cancer resistance. Proc Natl Acad Sci USA. 2009;106:859–863. doi: 10.1073/pnas.0811616106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris H, Miller OJ, Klein G, Worst P, Tachibana T. Suppression of malignancy by cell fusion. Nature. 1969;223:363–368. doi: 10.1038/223363a0. [DOI] [PubMed] [Google Scholar]
- 14.Trubia M, Sessa L, Taramelli R. Mammalian Rh/T2/S-glycoprotein ribonuclease family genes: Cloning of a human member located in a region of chromosome 6 (6q27) frequently deleted in human malignancies. Genomics. 1997;42:342–344. doi: 10.1006/geno.1997.4679. [DOI] [PubMed] [Google Scholar]
- 15.Acquati F, et al. Cloning and characterization of a senescence inducing and class II tumor suppressor gene in ovarian carcinoma at chromosome region 6q27. Oncogene. 2001;20:980–988. doi: 10.1038/sj.onc.1204178. [DOI] [PubMed] [Google Scholar]
- 16.Acquati F, et al. Tumor and metastasis suppression by the human RNASET2 gene. Int J Oncol. 2005;26:1159–1168. [PubMed] [Google Scholar]
- 17.Monti L, et al. RNASET2 as a tumor antagonizing gene in a melanoma cancer model. Oncol Res. 2008;17(2):69–74. doi: 10.3727/096504008784523658. [DOI] [PubMed] [Google Scholar]
- 18.Steinemann D, et al. Identification of candidate tumor-suppressor genes in 6q27 by combined deletion mapping and electronic expression profiling in lymphoid neoplasms. Genes Chromosomes Cancer. 2003;37:421–426. doi: 10.1002/gcc.10231. [DOI] [PubMed] [Google Scholar]
- 19.Kim TY, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–7501. doi: 10.1158/0008-5472.CAN-05-4552. [DOI] [PubMed] [Google Scholar]
- 20.Morita R, et al. Common regions of deletion on chromosomes 5q, 6q, and 10q in renal cell carcinoma. Cancer Res. 1991;51:5817–5820. [PubMed] [Google Scholar]
- 21.Tibiletti MG, et al. Early involvement of 6q in surface epithelial ovarian tumors. Cancer Res. 1996;56:4493–4498. [PubMed] [Google Scholar]
- 22.Theile M, et al. A defined chromosome 6q fragment (at D6S310) harbors a putative tumor suppressor gene for breast cancer. Oncogene. 1996;13:677–685. [PubMed] [Google Scholar]
- 23.Hauptschein RS, et al. Cloning and mapping of human chromosome 6q26-q27 deleted in B-cell non-Hodgkin lymphoma and multiple tumor types. Genomics. 1998;50(2):170–186. doi: 10.1006/geno.1998.5321. [DOI] [PubMed] [Google Scholar]
- 24.Sica A, et al. Macrophage polarization in tumour progression. Semin Cancer Biol. 2008;18:349–355. doi: 10.1016/j.semcancer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Luhtala N, Parker R. T2 Family ribonucleases: Ancient enzymes with diverse roles. Trends Biochem Sci. 2010;35:253–259. doi: 10.1016/j.tibs.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, et al. Physical and transcript map of the region between D6S264 and D6S149 on chromosome 6q27, the minimal region of allele loss in sporadic epithelial ovarian cancer. Oncogene. 2002;21:387–399. doi: 10.1038/sj.onc.1205067. [DOI] [PubMed] [Google Scholar]
- 27.Smirnoff P, Roiz L, Angelkovitch B, Schwartz B, Shoseyov O. A recombinant human RNASET2 glycoprotein with antitumorigenic and angiogenic characteristics. Cancer. 2006;107:2760–2769. doi: 10.1002/cncr.22327. [DOI] [PubMed] [Google Scholar]
- 28.Wang F, et al. HYAL1 and HYAL2 inhibit tumour growth in vivo but not in vitro. PLoS ONE. 2008;3:e3031. doi: 10.1371/journal.pone.0003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharp TV, et al. LIM domains-containing protein 1 (LIMD1), a tumor suppressor encoded at chromosome 3p21.3, binds pRB and represses E2F-driven transcription. Proc Natl Acad Sci USA. 2004;101:16531–16536. doi: 10.1073/pnas.0407123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, et al. Tumor suppressive role of a 2.4 Mb 9q33-q34 critical region and DEC1 in esophageal squamous cell carcinoma. Oncogene. 2005;24:697–705. doi: 10.1038/sj.onc.1208179. [DOI] [PubMed] [Google Scholar]
- 31.Gourley C, et al. WWOX gene expression abolishes ovarian cancer tumorigenicity in vivo and decreases attachment to fibronectin via integrin alpha3. Cancer Res. 2009;69:4835–4842. doi: 10.1158/0008-5472.CAN-08-2974. [DOI] [PubMed] [Google Scholar]
- 32.Steinfelder S, et al. The major component in schistosome eggs responsible for conditioning dendritic cells for Th2 polarization is a T2 ribonuclease (omega-1) J Exp Med. 2009;206:1681–1690. doi: 10.1084/jem.20082462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henneke M, et al. RNASET2-deficient cystic leukoencephalopathy resembles congenital cytomegalovirus brain infection. Nat Genet. 2009;41:773–775. doi: 10.1038/ng.398. [DOI] [PubMed] [Google Scholar]
- 34.Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009;185(1):43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 37.McNally AK, Chisolm GM, Morel DW, Cathcart MK. Activated human monocytes oxidize LDL by a lipoxygenase-dependent pathway. J Immunol. 1990;145:254–259. [PubMed] [Google Scholar]
- 38.Sunderkötter C, et al. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J Immunol. 2004;172:4410–4417. doi: 10.4049/jimmunol.172.7.4410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.