Abstract
The vertebrate hindbrain contains various sensory-motor networks controlling movements of the eyes, jaw, head, and body. Here we show that stripes of neurons with shared neurotransmitter phenotype that extend throughout the hindbrain of young zebrafish reflect a broad underlying structural and functional patterning. The neurotransmitter stripes contain cell types with shared gross morphologies and transcription factor markers. Neurons within a stripe are stacked systematically by extent and location of axonal projections, input resistance, and age, and are recruited along the axis of the stripe during behavior. The implication of this pattern is that the many networks in hindbrain are constructed from a series of neuronal components organized into stripes that are ordered from top to bottom according to a neuron's age, structural and functional properties, and behavioral roles. This simple organization probably forms a foundation for the construction of the networks underlying the many behaviors produced by the hindbrain.
Keywords: interneuron, locomotion, recruitment, topography
The hindbrain contains a diverse set of sensory-motor networks that control movements required for vision, respiration, mastication, and locomotion in all vertebrates (1, 2). Most often these different networks are studied separately from one another, perhaps because the behaviors are distinct, and the regional differentiation of hindbrain suggests that its several networks might have little in common. Thus, we have strong data for the hindbrain control of eye movements, respiration, and locomotion (3–10), but fewer unifying principles of structural and functional organization that apply across the different networks.
Structurally, the hindbrain is divided into segments, called rhombomeres, which differ in the expression of homeotic genes, in the morphological differentiation of neurons, and in their sensory inputs and motor outputs (2, 11). Though there are clear distinctions among rhombomeres, there are indications from previous developmental work using in situ staining for transcription factors and backfilling of hindbrain neurons that there may be structural patterns that cross rhombomere boundaries (12–16). Prior work has also revealed parallels in the development of hindbrain and spinal cord, with the hindbrain sharing features of the now-classic transcription factor code that directs development in spinal cord (17–22). Though these studies did not explore function because they were performed during early development, they raised the possibility of a broader structural-functional patterning that spans rhomobomeres and may underlie the organization of circuits for different behaviors.
Here we show that there is indeed a broad structural and functional patterning of neurons in the hindbrain of young zebrafish. The work was initially prompted by a striking patterning observed in earlier work in which we used in situ staining for markers of neurotransmitter phenotype to reveal putative glycinergic, GABAergic, and glutamatergic neurons in the hindbrain (23). We found that neurons of the same transmitter phenotype were clustered together into stripes when viewed in cross-sections, and that these extended as columns throughout much of the rostrocaudal axis of the hindbrain. The pattern is evident in fish when they are freely swimming and most of the major adult behaviors driven by hindbrain—e.g., swimming, feeding, and eye movements—are functional. This suggests that the diverse networks in hindbrain might share an orderly functional patterning of neurons. We now show that the transmitter stripes reflect an underlying order in the hindbrain in which stripes contain cell types, stacked in order by age as well as structural and functional properties. This pattern is tied to behavior because neurons are recruited along the axis of a stripe as the speed of a motor behavior increases. We conclude that the diverse networks in the hindbrain have at their foundation a common structural and functional plan.
Results
Hindbrain Transmitter Stripes and Transcription Factor Patterning.
We examined the transmitter stripe patterning in more than 20 larval, posthatching fish from each of two BAC transgenic lines expressing green (GFP) and red (DsRed) fluorescent proteins driven by promoter regions of glyt2 and vglut2.1 (vglut2) to mark glycinergic and glutamatergic neurons, respectively (24, 25). Stripes were clearly evident in the hindbrain of dual transgenic lines (vglut:DsRed × glyt2:GFP), as shown in the example from a 4-d postfertilization (dpf) fish in Fig. 1A (n = 16). In cross-sections, these stripes were organized in an interleaved manner from medial to lateral on each side of the brain in all hindbrain segments (rhombomeres), with a glutamatergic stripe located medially followed by alternating glycinergic and glutamatergic stripes that extend predominately dorsoventrally and rostrocaudally. This organization was present in all of the fish. Although the glycinergic and glutamatergic neurons were segregated from one another in a columnar pattern, there were other unlabeled neurons, both scattered within stripes and in large contiguous areas between stripes that are probably neurons with other transmitter phenotypes (cholinergic and GABAergic, e.g.) based upon prior in situ staining (23).
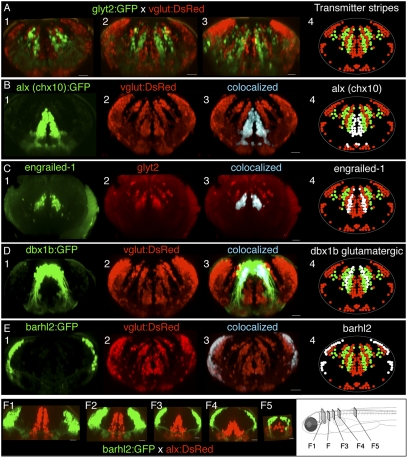
Fig. 1.
Interleaved transmitter stripes of neurons in hindbrain and their overlap with transcription factor expression patterns. (A) Cross-sections of hindbrain in vivo in rhombomeres 4, 6, and 8 (A, panels 1–3, respectively) from a glyt2:GFP × vglut:DsRed four dpf fish, showing interleaved stripes of glycinergic (green) and glutamatergic neurons (red). (B–E) Transcription factor stripes are present in hindbrain and overlap glutamatergic and glycinergic stripes. (B) Cross-sections from rhombomere 8 of a dual transgenic alx (chx10):GFP and vglut: DsRed three dpf fish shows that alx expression colocalizes with the most medial glutamatergic stripe. (C) Engrailed-1 immunostaining in cross-sections from a glyt2 transgenic line from rhombomere 7 in a 5-dpf fish showing that all medial glycinergic stripe neurons express engrailed-1. (D) Cross-section from rhombomere 7 in a four dpf vglut:DsRed × dbx1b:GFP transgenic fish shows that dbx colocalizes with both the middle glutamatergic stripe as well as a more lateral stripe in a region known to contain glycinergic neurons. (E) Cross-sections from rhombomere 8 in a 4-dpf vglut:DsRed × barhl2:GFP fish indicate that barhl2 staining overlaps the lateral portion of the most lateral glutamatergic populations. (F) Cross-sections of the transformation of transcription factor stripes from hindbrain (F1) to spinal cord (F5) in a cross of barhl2GFP and alx:DsRed lines. White regions in panel 4 in B–E indicate the location of the transcription factor expression relative to the neurotransmitter stripes. All cross-sections are maximum-intensity projections of 20- to 50-μm volumes from a confocal image stack of the entire hindbrain. (Scale bars, 20 μm.)
We next used transgenic lines and immunostaining to investigate the relationship between the transmitter stripes and the transcription factors alx (called chx10 in mammals), dbx1b, engrailed-1, and barhl2. The transcription factors were expressed in stripe-like patterns resembling the patterning of transmitter stripes. Neurons expressing the alx (chx10) transcription factor were clustered medially and overlapped the most medial glutamatergic stripe (Fig. 1B). Three-dimensional colocalization revealed that most, if not all, medial glutamatergic stripe neurons express the alx transcription factor (Fig. 1B, panel 3; n = 2 fish). Immunostaining for alx protein in the alx:GFP transgenic line confirmed that this line reliably marked the alx positive neurons in the hindbrain stripes (n = 9). Engrailed-1 immunostaining was located in a stripe just lateral to the alx (medial glutamatergic) stripe and overlapped the most medial glycinergic stripe (Fig. 1C). Colocalization indicated that most, if not all, of the neurons in this glycinergic stripe express engrailed-1 (Fig. 1C, panel 3; n = 4). Dbx1b is a transcription factor expressed in progenitor cells that is also observed in differentiated neurons at early times. In Fig. 1D, panel 1, a cross-section of dbx1b:GFP expression in rhombomere 8 showed that these cells formed a more laterally positioned stripe than both engrailed-1 and alx stripes. The dorsalmost cells were present within the ventricular zone; however, the majority of the expression was located in the parenchyma and overlapped two stripes—a glutamatergic stripe just lateral to the engrailed-1 stripe (cells in blue/white), and more lateral neurons in a region containing a glycinergic stripe (the lateral population of green cells; Fig. 1D, panels 3 and 4; n = 2). The dbx1b area thus encompassed at least a medial glutamatergic domain and, more lateral, most likely, glycinergic one. Neurons expressing the barhl2 transcription factor were located in a band at the lateral edge of the hindbrain (Fig. 1E; n = 10), overlapping a portion of a lateral crescent-like stripe of glutamatergic neurons throughout hindbrain (Fig. 1E, panel 3). In summary, neurons expressing particular transcription factors are clustered into bands that align with neurotransmitter stripes and, depending on the transcription factor, can be coextensive with a transmitter stripe (alx, engrailed-1), overlap multiple transmitter stripes (dbx1b), or overlap only a restricted, spatially segregated portion of a stripe (barhl2).
We examined how the distribution of neurons expressing these transcription factors changes from spinal cord (where they are also expressed) into the hindbrain by examining optical cross-sections at different rostrocaudal locations from confocal image stacks from live fish in which neurons expressing two transcription factors (alx and barhl2) were labeled in different colors. A dorsoventral segregation of the different transcription factors in spinal cord gradually changed at its rostral end into a mediolateral segregation in hindbrain, indicating a topological transformation in expression domains between the two regions (Fig. 1F).
Morphology of Neurons in the Transmitter Stripes.
We used stochastic expression of membrane targeted proteins (mMCherry, mGFP, or Brainbow-1.1) to randomly label neurons in one color in 5- to 6-dpf fish in which the transmitter stripes were labeled in a different color. We then reconstructed the neurons in 3D and sorted them by stripe membership. Reconstructed neurons from four stripes—the medial glutamatergic stripe and the three glycinergic stripes—are shown in dorsal view in Fig. 2.
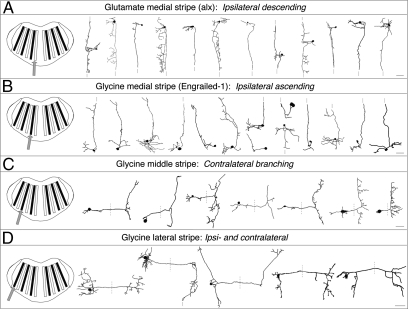
Fig. 2.
Morphology of neurons in the neurotransmitter stripes. All panels show horizontal projections of single-neuron reconstructions from different fish sorted by stripe location. (A) Neurons in the medial glutamatergic stripe. Their descending axons are truncated at the dashed line. (B) Neurons in the most medial glycinergic stripe. (C) Neurons in the middle glycinergic stripe. (D) Neurons in the lateral glycinergic stripe. Reconstructions are shown in a dorsal view, with rostral to the top. In C and D, dotted lines indicate the approximate position of the midline of the fish or, in one instance, the continuation of an axon. In each panel, the left column illustrates the stripe containing the cells. (Scale bars, 20 μm.)
The morphology of the neurons differed systematically among stripes. All 34 labeled medial glutamatergic stripe (alx) neurons (12 shown in Fig. 2) imaged and reconstructed from hindbrain rhombomeres 4–8 had ipsilaterally projecting axons that always had a descending portion that often extended into spinal cord (Fig. 2A); there was variation in whether rostral axonal branches were evident. The medial (engrailed-1) glycinergic stripe neurons were predominately cells with ipsilateral and ascending axonal projections (13 of 15 neurons in rhombomeres 3–8; Fig. 2B) that sometimes projected beyond rostral hindbrain. All eight neurons labeled in the middle (putative dbx1b) glycinergic stripe (rhombomeres 6–8) had contralaterally extending axons with both ascending and descending branches within hindbrain, often of similar length (Fig. 2C). All seven neurons reconstructed from the third, lateral glycinergic stripe (Fig. 2D) had both ipsilateral and contralateral axonal projections that distinguished them from those belonging to other stripes; there was, however, variation in the extent of ascending or descending projections on the two sides of the hindbrain.
Although these 63 reconstructed single cells from four of the transmitter stripes represent only a small fraction of the neurons in hindbrain, they reveal a general pattern consistent with the idea that these hindbrain stripes contain neurons with different morphological features. This conclusion is also supported by a series of backfilling experiments designed to more specifically examine projection patterns within these transmitter stripes (26).
Age-Related Patterning of Neurons in the Stripes.
We next examined neurons within a single stripe to determine if there was any organization within stripes. We focused on the medial glutamatergic stripe of neurons expressing the alx transcription factor, for which we had the most morphological data, transgenic lines marking the stripe, and evidence that some neurons in the stripe might be involved in swimming.
We examined whether neurons differed by age in an orderly manner along the axis of a stripe by photoconverting the color change protein Kaede, in an alx-Kaede transgenic line, at different time points during development to generate fish with the earliest differentiating neurons labeled in red, and the latest differentiating cells in green (Fig. 3 A–D).
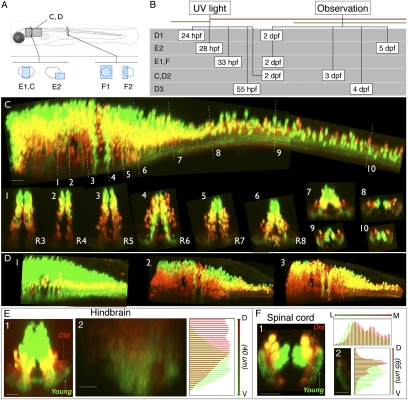
Fig. 3.
Age-related patterning within the medial glutamatergic, alx hindbrain stripe. (A) Location of images. (B) Timing of experiments. (C) Lateral view of alx:Kaede expression in a photoconverted fish shown from hindbrain through to spinal cord. White dashed lines indicate the rostrocaudal locations of the numbered cross-sections from different rhombomeres (R3–R8) and into spinal cord. (D, panels 1–3) Lateral views of hindbrain/spinal cord regions of alx:Kaede transgenic fish photoconverted/imaged at different times (timing shown in B). In this example we assigned colors so that green cells on the figure had no red in them at the imaging time, and neurons colored red on the figure are those with any red staining (they might also have had green but it was removed to make the youngest, pure green cells obvious). The yellow color here thus does not represent colabeling, but instead red and green neurons that overlap in the z direction. (E, panel 1) Reconstructed confocal cross-section shows that neuropil for older neurons (red) in hindbrain tends to be dorsal to that for younger neurons (green). (E, panel 2) Quantification of relative red/green expression at different dorsoventral locations within a cross-section of neuropil in rhombomere 7 from a different fish than E, panel 1; ventral is at the bottom. Red expression is shifted dorsally relative to green. (F) Photoconversion of an alx:Kaede transgenic fish imaged in spinal cord also shows age-related separation in the neuropil. In F, panel 1, a reconstructed confocal cross-section shows that neuropil for older neurons (red) tends to be medial and dorsal to that of younger neurons (green). (F, panel 2) Quantification of the dorsoventral distribution of red and green expression in the neuropil. (Scale bars, panel 2 in E and F, 10 μm; all other images, 20 μm.)
We found a broad and consistent pattern of age-related order in which the oldest neurons were ventrally positioned and the youngest ones populated more-dorsal regions within the alx stripe throughout hindbrain, as in Fig. 3C. The exceptions were two bands in which young alx neurons were ventrally positioned in rhombomere 6, and one group of alx neurons that was located medially and outside of the stripe region in rhombomere 8. The broad age order was present across multiple fish photoconverted and imaged at different times (n = 10, photoconverted between 24 and 55 hpf and imaged between 33 hpf and 8 dpf). In an additional 22 experiments we crossed a fish expressing Kaede under a general neuronal promoter (Huc:Kaede) into transgenic lines with neurotransmitter and transcription factor stripes labeled in green to determine the location of the young and old neurons in the various stripes (glyt2, n = 4; vglut, n = 5; dbx1b, n = 1; barhl2, n = 6; Huc:Kaede alone, n = 6). These experiments supported the conclusion that the pattern in which the oldest neurons are located ventrally, with the younger ones stacked above them, is present broadly across multiple hindbrain stripes.
The processes of the alx neurons in the neuropil appeared ordered by age as well. In the hindbrain, processes of older ventral neurons (red and yellow) are located dorsally in the neuropil, which lies just below the stripe of somata, whereas younger, dorsal neurons (green) have processes located more ventrally in the neuropil (Fig. 3E, panels 1 and 2). In contrast, in the spinal cord, which contains processes of alx neurons from both hindbrain and spinal cord, the older neuropil lies medial and dorsal to the younger neuropil (Fig. 3F, panels 1 and 2). We quantified the intensities of red and green fluorescence within different regions of the neuropil following photoconversion/imaging at different times and found the age-related order with the neuropil to be present in all four cases quantified in spinal cord and in multiple hindbrain rhombomeres (photoconvert 2 dpf/image 3 dpf; photoconvert 33 hpf/image 2 dpf in spinal cord; photoconvert 28 hpf/image 5 dpf; and photoconvert 2 dpf/image 4 dpf for R4–8 in hindbrain; n = 1 for each; Fig. 3, E2 and F2). This age-related patterning of projections raises the possibility that there might be age-related connectivity.
Structural and Functional Organization of Neurons Within the alx Stripe in Hindbrain.
We suspected that some alx-positive neurons in hindbrain would be active during swimming, based upon evidence for involvement of spinal alx neurons in swimming (27, 28). To examine this, we developed an approach to do targeted patch recording from neurons deep in the brain in vivo in 4- to 6-dpf fish to allow us to explore their activity patterns. When we patched from alx-positive neurons in the caudal hindbrain in rhombomere 7 while recording from ventral roots to monitor the motor pattern, we found that some (but not all) of the neurons were rhythmically active following light or electrical stimulation, in a pattern that matched the swimming motor output recorded from the ventral roots (Fig. 4A, panels 1 and 2).
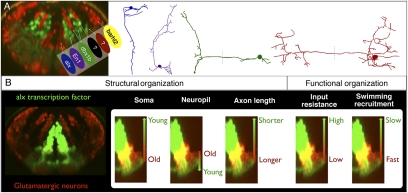
Fig. 4.
Structural and functional patterning within the alx stripe. (A) Patch recording of an alx:GFP+ neuron in hindbrain segment 7 of a 5-dpf fish, while simultaneously recording motor activity in a ventral root. (A, panel 1) Cross-section view of the patched cell (red) within the alx stripe (green). (A, panel 2) Recording from the alx cell on the left is shown below the simultaneous ventral root recording. (B) Variation in axonal projections into spinal cord of hindbrain alx neurons. (B, panel 1) An example of a reconstruction of a single alx neuron (red) within the alx:GFP transgenic line (lateral view). (A, panel 2) Lateral views of 3D reconstructions of neurons from different dorsoventral locations in the medial glutamatergic stripe in rhombomere 7, with the most dorsal one at the top (normalized dorsoventral positions: 0.51, 0.28, and 0.16 from ventral edge of stripe, rostral to the left). (C) A plot of the total 3D axonal length of 10 labeled alx neurons (two of them overlap, shown with white circle) vs. their dorsal ventral location in the stripe. More ventral neurons have systematically longer axonal length (P < 0.0001). Red, green, and blue points correspond with the three neurons in B. (D) Plot of the input resistance of a neuron vs. its dorsoventral location in a stripe. More-dorsal neurons have systematically higher input resistances in both more- (black dots, P < 0.05) and less-exposed brain preparations (gray squares, P < 0.01). See text for further details. (E) Examples of calcium imaging of alx neurons from different locations in the stripe during different frequencies of swimming in 5-dpf fish. (E, panels 1–3) Cross-sections images of alx:DsRed fish with locations of the neurons (dots) shown relative to the stripes. (E, panels 4–6) Calcium responses of the neurons on the left in two example trials at swimming frequencies near those when the neurons are first recruited. (F) Plot of the minimum swimming frequency at which a neuron responds vs. the dorsoventral location of the neuron, including both alx:DsRed+ alx:DsRed− neurons in the region of the alx stripe (P < 0.05 for each correlation). Neurons from Fig. 4B are shown in color. (Scale bars, 200 μm.)
We then looked at structural variation of alx cells along the axis of their stripe. We examined the morphology of 19 alx neurons labeled stochastically with Brainbow-1.1m at different positions along the axis of the stripe in different hindbrain rhombomeres (Fig. 4B). We quantified the total axonal branching for a subset of 10 neurons that were located in the same region (rhombomeres 6–8) of 6-dpf fish by reconstructing them in three dimensions and measuring the total length of their axon. A plot of location vs. axonal length for these neurons showed a significant (P < 0.0001) correlation between their total axonal length and their location (Fig. 4C), with the length increasing systematically, and nearly linearly, from dorsal to ventral along the stripe axis. Tracking the growth of axons from dorsal and ventral neurons in the alx stripe as they descend into spinal cord indicates that the axons of ventral neurons grow along cord faster after their extension than those of more dorsal cells, suggesting that the differences in axonal extent are not just a result of dorsal neurons having less time to grow, and raising the possibility that the differences might persist later in life.
We also explored whether physiological properties might vary systematically along the axis of a stripe. Using hyperpolarizing current steps while patch recording from single alx-positive neurons in five dpf alx:GFP transgenic fish, followed by labeling and 3D reconstruction, we examined whether the input resistance of alx neurons varied systematically with location in a stripe. We used both an exposed-brain preparation as well as a more intact preparation and sampled neurons from the hindbrain in R7–8 across the dorsoventral extent of the alx stripe. Neurons positioned very dorsally had small amplitude, relatively long duration action potentials, a depolarized resting membrane potential, and very high input resistances in both preparations, suggesting that they were very young, in accord with the Kaede photoconversion data (see above). Interneurons above ∼70% of the way up the stripe showed persistent activity that was not correlated with behaviors such as swimming. Consequently, we focused our input resistance and functional analysis on the ventral 70% of the stripe, which included older neurons that were already clearly incorporated into networks based upon their rhythmic activation during swimming (see below).
Neurons with the lowest input resistance values were consistently located in the most ventral stripe regions at 5 dpf, both in the preparations with massive exposure of the brain (black dots on Fig. 4D) and those with reduced exposure (gray squares on Fig. 4D). There was a significant correlation between the position of a neuron along the stripe axis and its input resistance within both preparations with either massive or reduced exposure of the brain (Pearson correlation: P < 0.01 for reduced exposure, P < 0.05 for massive exposure, P < 0.05 for both together). The input resistance was lowest at the bottom of the stripe and increased in more dorsal locations in both cases, although the measurements from the more exposed preparation were more scattered, probably because of damage during exposure.
The presence of an orderly pattern of structural and physiological properties along the stripe axis by age raised the question of whether the neurons were recruited during behavior in an orderly way along a stripe. To explore the involvement of neurons at different dorsoventral locations in the alx stripe at different frequencies/speeds of swimming in a minimally invasive way, we used calcium imaging with Oregon Green BAPTA-1 dextran (Invitrogen) electroporated into the neurons in the alx:DsRed transgenic line. We recorded from ventral roots to monitor the frequency of swimming produced by light or electrical stimulation and determined, based upon the calcium response, the lowest swimming frequency at which a neuron was activated (25). We subsequently collected confocal image stacks through the hindbrain in the region of labeled neurons so that we could measure the position of the imaged cells relative to the axis of the stripe.
We collected many trials (averaging about 100) over a range of frequencies of swimming for each neuron to determine its minimum recruitment frequency. Some examples of calcium responses of neurons at different dorsoventral locations are shown in Fig. 4E. In these examples, the top neuron exhibited a fluorescence increase in a bout with a peak frequency of 24 Hz, but not in one at 22 Hz; the middle neuron at 28Hz, but not 26Hz; and the bottom cell at 31 but not 29 Hz. We obtained recruitment patterns and positions for 29 neurons from 20 fish. Eleven of the neurons expressed the alx transcription factor and 18 did not, although the 18 non-alx neurons were in the general region of the alx stripe, so their position along the stripe could be measured. A plot of recruitment frequency vs. position for both alx and non-alx neurons, in Fig. 4F, showed a consistent pattern in which more-dorsal, younger cells were recruited at the lowest swimming frequencies and increasingly more-ventral neurons were recruited at increasingly higher swimming frequencies (P < 0.05 for both). Thus, the neurons are recruited from dorsal to ventral along the axis of the stripe as the frequency of swimming increases, indicating a systematic relationship between position, age, and recruitment that maps onto the axis of the alx stripe and extends to other non-alx neurons in hindbrain as well.
Discussion
Our experiments reveal that the striking organization into transmitter stripes in hindbrain reflects a broad patterning of neurons by cell type, morphology, age, projections, cellular properties, and activity patterns (summarized in Fig. 5). The fish we studied are freely swimming animals with functional motor networks for swimming, escape, jaw, and eye movements, all located in a hindbrain with a strikingly regular organization. The transmitter stripes extend throughout the hindbrain and across the well-studied rhombomeres (2, 11, 13, 15, 29), through regions containing many different networks. The implication is that the many neural circuits in hindbrain are built from a set of neuronal types, which might be viewed as circuit components, with particular morphologies, neurotransmitters, and transcription factor phenotypes. The stripes represent these different neuronal types. Within a given stripe, the neurons are arranged in an orderly way by structural and functional properties (and, importantly, age) so that those with more extensive projections and higher thresholds for activation, which are involved in the fastest movements, are at the bottom of a stripe, with the neurons involved in increasingly refined movements stacked in order above them. We infer that hindbrain networks are constructed in an orderly manner by drawing cells from organized stripes, much like selecting components from a set of parts where the parts are arranged in order according to their structural or functional properties.
Fig. 5.
Summary of the structural and functional rules governing hindbrain patterning. (A) Interleaved neurotransmitter stripes overlap with transcription factor expression patterns. Different neuronal morphologies (Right) are associated (by matching colors here) with the different stripes. (B) Within an individual stripe (alx stripe, Left), the somata of neurons are ordered along the axis of the stripe by age, the location of their processes in the neuropil, and their total axonal length. This corresponds to systematic changes in input resistance and recruitment along the axis of the stripe.
Though the larval fish has nearly all of the same motor behaviors as an adult, the brain of the adult is considerably larger and the neurons are clustered into nuclei located among the fiber tracts—an arrangement much different from the simple patterning in the young fish. This raises the question of the relationship between the networks in the young fish and those in the adult. A wholesale reorganization of the connectivity of the networks with growth seems unlikely, because the patterns of motor output in the young fish parallel those in adults (e.g., the alternation and rostrocaudal delays characteristic of swimming are present throughout life) (30–32). A reconstruction of the networks would mean dispensing with a motor circuitry that already worked. The more likely possibility is that the essentials of the larval pattern of network organization and recruitment are retained even as the neurons disperse to reorganize into nuclei. This would imply that circuits are set up early when brain patterning is simple, but that the somata then migrate with growth to form nuclei, obscuring a simple underlying network patterning. If so, we predict that if we could track neurons as the brain differentiated further, the recruitment order and the basic patterns of connectivity established early would be retained later in life, after migration, and be related to the age of the neuron, which corresponded to its location in the younger fish. This migration may occur to minimize the extent of axonal and dendritic arbors in a much larger brain, given the evidence for placement of neurons to minimize wiring in other systems (33).
This early organization in zebrafish probably extends to the hindbrain of other vertebrates. Longitudinal bands of some of the transcription factors we studied, and others, extend through the hindbrain of frogs, chicks, and mice, and their locations relative to the midline parallel the patterns in zebrafish (12–14, 16, 34, 35). Morphological data from backfilling studies in embryonic chicks indicates that neurons with different morphological features occupy systematic locations relative to the midline (15), consistent with an early orderly disposition of cell types. Though these studies provide structural indications of a patterning like that in fish, there is no information about the functional organization at early stages to allow comparison with the zebrafish patterning. We would predict, however, that networks are set up when the patterning is simple, with the later function of the neurons mapping systematically onto their age as in zebrafish.
The organization of hindbrain has clear similarities to the patterning in spinal cord, suggesting that the two regions giving rise to all of the motor output from the nervous system have elements of the same structural and functional network plan (17–22, 25, 27, 28). Though there are undoubtedly changes with growth, nervous system plasticity, and divergence in the networks for specific tasks, the basic features of structural and functional organization that we describe may lie at the foundation of the construction of the many sensory-motor circuits throughout the hindbrain, which produces a much broader range of sensory-motor outputs than spinal cord. The challenge now is to understand how many different networks in hindbrain are constructed from this basic ground plan and how the early pattern is reorganized as the brain continues to grow into its adult form. We explore one of these specialized hindbrain networks, and its relationship to the patterning, in the study by Koyama et al. (26).
Experimental Procedures
Fish Care.
All experiments were performed on zebrafish (Danio rerio) between 1 and 8 dpf obtained from a laboratory stock of wild-type and transgenic adults. All procedures conform to the National Institutes of Health guidelines regarding animal experimentation and were approved by Cornell University's Institutional Animal Care and Use Committee.
Immunostaining.
Standard whole-mount antibody staining procedures were used as described previously (36, 37).
Stochastic Labeling.
Stochastic labeling was performed as described previously (23).
Transgenic Lines.
The transgenic lines we used included ones described in prior studies (24, 25, 38), as well as two new ones with the promoters dbx1b and barhl2. These new lines, Tg(dbx1b:GFP) and Tg(barhl2:GFP), were constructed with the BACs zK17G17 and zC15L16 using a previously described method (38). The detailed approaches for generating these lines are described elsewhere.
Confocal Imaging.
Confocal imaging was performed as described previously (28, 39).
Colocalization.
Colocalization was assessed using the colocalization add-on in the Imaris software package (Bitplane).
Neuronal Tracing and Location Measurements.
Neurons were traced using the filament reconstruction feature provided in Imaris.
Photoconversion and Analysis.
Huc:Kaede and alx:Kaede transgenic embryos (38, 40) were illuminated with UV light from a mercury bulb source for 10–40 s within their chorion. Immediately afterward, photoconversion was confirmed by observation of the presence of red expression and an absence of green expression. Photoconverted fish were then kept at 28.5 °C in a light-tight container until the day of imaging.
Targeted Whole-Cell Patching of alx Neurons in an Exposed Brain Preparation for Input Resistance Measurements.
Larvae were anesthetized in 3-aminobenzoic acid ethyl ester (0.02% in HBSS) and then immobilized using α-bungarotoxin (Sigma-Aldrich; 0.1% in HBSS). To measure input resistance of alx neurons in hindbrain, we initially used a dissection procedure similar to one described previously (41). We later switched to a less-extensive exposure of the brain, leaving more of the head, including the eyes, intact to minimize potential damage to the neurons. After the electrophysiological measurements, the preparation was fixed with 4% formaldehyde with the pipette in place to avoid the movement of the cell body during the retraction of pipette. Then a z stack was acquired with a confocal microscope (LSM 510 META; Zeiss) for the measurement of the soma position relative to the alx stripe.
In Vivo Whole-Cell Recordings in the Hindbrain.
Whole-cell recordings were done in current clamp mode in 5-dpf alx:GFP nacre transgenic larvae using methods similar to those described previously (25, 30, 41).
Calcium Imaging.
Transgenic 4-dpf alx:DsRed Casper fish (42) were first anesthetized using 3-aminobenzoic acid ethyl ester (MS-222, 0.02% in HBSS) and embedded in low-melting-point agarose (1.6% in HBSS; Sigma). A patch electrode (5–10 MΩ resistance) was filled with 20% Oregon Green BAPTA-1 (10,000 Mr; Invitrogen/Molecular Probes), and the indicator was electroporated along the dorsoventral axis of the alx stripe in caudal hindbrain by using a single-cell Axoporator (−4 V, 20-ms duration square pulse; Molecular Devices). Larvae were then removed from the agarose, placed in a Petri dish containing HBSS, and stored in an incubator (28.5 °C) overnight.
The following day, we imaged the calcium indicator in hindbrain on a Zeiss LSM 510 inverted confocal microscope while recording from a ventral root using methods similar to those applied previously (25, 27).
Analysis of calcium imaging and ventral root recordings was performed using custom-written MATLAB software with an approach similar to McLean et al. (25).
Image stacks were then reconstructed in 3D using Imaris software (Bitplane). Neurons that were active during the experiment were identified. The length of the left and right stripes (at the position of the cell body) and the location of the neuron were each measured three times and averaged to obtain the location of the neurons in the stripe.
More details of methods are provided in SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank David McLean for comments on the manuscript and Lindsay Heller for maintaining the many lines of fish. This work was supported by National Science Foundation Integrative Graduate Education and Research Traineeship Grant 0333366 (to A.K.), National Institutes of Health Training Grant T32 GM007469 (to A.K. and M.R), the Ministry of Education, Science, Technology, Sports and Culture of Japan (S.-i.H.), the Japan Society for the Promotion of Science (M.K.), and National Institutes of Health Grant NS26539 (to J.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012185108/-/DCSupplemental.
References
- 1.Garcia-Campmany L, Stam FJ, Goulding M. From circuits to behaviour: Motor networks in vertebrates. Curr Opin Neurobiol. 2010;20:116–125. doi: 10.1016/j.conb.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tümpel S, Wiedemann LM, Krumlauf R. Hox genes and segmentation of the vertebrate hindbrain. Curr Top Dev Biol. 2009;88:103–137. doi: 10.1016/S0070-2153(09)88004-6. [DOI] [PubMed] [Google Scholar]
- 3.Sparks DL. The brainstem control of saccadic eye movements. Nat Rev Neurosci. 2002;3:952–964. doi: 10.1038/nrn986. [DOI] [PubMed] [Google Scholar]
- 4.Smith JC, Abdala AP, Rybak IA, Paton JF. Structural and functional architecture of respiratory networks in the mammalian brainstem. Philos Trans R Soc Lond B Biol Sci. 2009;364:2577–2587. doi: 10.1098/rstb.2009.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grillner S, et al. Neural networks that co-ordinate locomotion and body orientation in lamprey. Trends Neurosci. 1995;18:270–279. [PubMed] [Google Scholar]
- 6.Smith JC, Ellenberger HH, Ballanyi K, Richter DW, Feldman JL. Pre-Bötzinger complex: A brainstem region that may generate respiratory rhythm in mammals. Science. 1991;254:726–729. doi: 10.1126/science.1683005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts A, Li WC, Soffe SR, Wolf E. Origin of excitatory drive to a spinal locomotor network. Brain Res Brain Res Rev. 2008;57:22–28. doi: 10.1016/j.brainresrev.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 8.Aksay E, et al. Functional dissection of circuitry in a neural integrator. Nat Neurosci. 2007;10:494–504. doi: 10.1038/nn1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubuc R, et al. Initiation of locomotion in lampreys. Brain Res Brain Res Rev. 2008;57:172–182. doi: 10.1016/j.brainresrev.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Faber DS, Fetcho JR, Korn H. Neuronal networks underlying the escape response in goldfish. General implications for motor control. Ann N Y Acad Sci. 1989;563:11–33. doi: 10.1111/j.1749-6632.1989.tb42187.x. [DOI] [PubMed] [Google Scholar]
- 11.Moens CB, Prince VE. Constructing the hindbrain: Insights from the zebrafish. Dev Dyn. 2002;224:1–17. doi: 10.1002/dvdy.10086. [DOI] [PubMed] [Google Scholar]
- 12.Moreno N, Bachy I, Rétaux S, González A. LIM-homeodomain genes as territory markers in the brainstem of adult and developing Xenopus laevis. J Comp Neurol. 2005;485:240–254. doi: 10.1002/cne.20498. [DOI] [PubMed] [Google Scholar]
- 13.Schubert FR, Dietrich S, Mootoosamy RC, Chapman SC, Lumsden A. Lbx1 marks a subset of interneurons in chick hindbrain and spinal cord. Mech Dev. 2001;101:181–185. doi: 10.1016/s0925-4773(00)00537-2. [DOI] [PubMed] [Google Scholar]
- 14.Cepeda-Nieto AC, Pfaff SL, Varela-Echavarría A. Homeodomain transcription factors in the development of subsets of hindbrain reticulospinal neurons. Mol Cell Neurosci. 2005;28:30–41. doi: 10.1016/j.mcn.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 15.Clarke JD, Lumsden A. Segmental repetition of neuronal phenotype sets in the chick embryo hindbrain. Development. 1993;118:151–162. doi: 10.1242/dev.118.1.151. [DOI] [PubMed] [Google Scholar]
- 16.Gray PA. Transcription factors and the genetic organization of brain stem respiratory neurons. J Appl Physiol. 2008;104:1513–1521. doi: 10.1152/japplphysiol.01383.2007. [DOI] [PubMed] [Google Scholar]
- 17.Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 18.Goulding M. Circuits controlling vertebrate locomotion: Moving in a new direction. Nat Rev Neurosci. 2009;10:507–518. doi: 10.1038/nrn2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ericson J, et al. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- 20.Osumi N, et al. Pax-6 is involved in the specification of hindbrain motor neuron subtype. Development. 1997;124:2961–2972. doi: 10.1242/dev.124.15.2961. [DOI] [PubMed] [Google Scholar]
- 21.Placzek M, Yamada T, Tessier-Lavigne M, Jessell T, Dodd J. Control of dorsoventral pattern in vertebrate neural development: Induction and polarizing properties of the floor plate. Development. 1991;2(Suppl 2):105–122. [PubMed] [Google Scholar]
- 22.Takahashi M, Osumi N. Pax6 regulates specification of ventral neurone subtypes in the hindbrain by establishing progenitor domains. Development. 2002;129:1327–1338. doi: 10.1242/dev.129.6.1327. [DOI] [PubMed] [Google Scholar]
- 23.Higashijima S, Schaefer M, Fetcho JR. Neurotransmitter properties of spinal interneurons in embryonic and larval zebrafish. J Comp Neurol. 2004;480:1–18. doi: 10.1002/cne.20279. [DOI] [PubMed] [Google Scholar]
- 24.Bae YK, et al. Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Dev Biol. 2009;330:406–426. doi: 10.1016/j.ydbio.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 25.McLean DL, Fan J, Higashijima S, Hale ME, Fetcho JR. A topographic map of recruitment in spinal cord. Nature. 2007;446:71–75. doi: 10.1038/nature05588. [DOI] [PubMed] [Google Scholar]
- 26.Koyama M, et al. Mapping a sensory-motor network onto a structural and functional ground plan in the hindbrain. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1012189108. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLean DL, Fetcho JR. Spinal interneurons differentiate sequentially from those driving the fastest swimming movements in larval zebrafish to those driving the slowest ones. J Neurosci. 2009;29:13566–13577. doi: 10.1523/JNEUROSCI.3277-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLean DL, Masino MA, Koh IY, Lindquist WB, Fetcho JR. Continuous shifts in the active set of spinal interneurons during changes in locomotor speed. Nat Neurosci. 2008;11:1419–1429. doi: 10.1038/nn.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heyman I, Kent A, Lumsden A. Cellular morphology and extracellular space at rhombomere boundaries in the chick embryo hindbrain. Dev Dyn. 1993;198:241–253. doi: 10.1002/aja.1001980402. [DOI] [PubMed] [Google Scholar]
- 30.Masino MA, Fetcho JR. Fictive swimming motor patterns in wild type and mutant larval zebrafish. J Neurophysiol. 2005;93:3177–3188. doi: 10.1152/jn.01248.2004. [DOI] [PubMed] [Google Scholar]
- 31.Liu DW, Westerfield M. Function of identified motoneurones and co-ordination of primary and secondary motor systems during zebra fish swimming. J Physiol. 1988;403:73–89. doi: 10.1113/jphysiol.1988.sp017239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fetcho JR, Svoboda KR. Fictive swimming elicited by electrical stimulation of the midbrain in goldfish. J Neurophysiol. 1993;70:765–780. doi: 10.1152/jn.1993.70.2.765. [DOI] [PubMed] [Google Scholar]
- 33.Chen BL, Hall DH, Chklovskii DB. Wiring optimization can relate neuronal structure and function. Proc Natl Acad Sci USA. 2006;103:4723–4728. doi: 10.1073/pnas.0506806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Passini MA, Raymond PA, Schechter N. Vsx-2, a gene encoding a paired-type homeodomain, is expressed in the retina, hindbrain, and spinal cord during goldfish embryogenesis. Brain Res Dev Brain Res. 1998;109:129–135. doi: 10.1016/s0165-3806(98)00069-8. [DOI] [PubMed] [Google Scholar]
- 35.Storm R, et al. The bHLH transcription factor Olig3 marks the dorsal neuroepithelium of the hindbrain and is essential for the development of brainstem nuclei. Development. 2009;136:295–305. doi: 10.1242/dev.027193. [DOI] [PubMed] [Google Scholar]
- 36.Higashijima S, Masino MA, Mandel G, Fetcho JR. Engrailed-1 expression marks a primitive class of inhibitory spinal interneuron. J Neurosci. 2004;24:5827–5839. doi: 10.1523/JNEUROSCI.5342-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLean DL, Fetcho JR. Ontogeny and innervation patterns of dopaminergic, noradrenergic, and serotonergic neurons in larval zebrafish. J Comp Neurol. 2004;480:38–56. doi: 10.1002/cne.20280. [DOI] [PubMed] [Google Scholar]
- 38.Kimura Y, Okamura Y, Higashijima S. alx, a zebrafish homolog of Chx10, marks ipsilateral descending excitatory interneurons that participate in the regulation of spinal locomotor circuits. J Neurosci. 2006;26:5684–5697. doi: 10.1523/JNEUROSCI.4993-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hale ME, Ritter DA, Fetcho JR. A confocal study of spinal interneurons in living larval zebrafish. J Comp Neurol. 2001;437:1–16. doi: 10.1002/cne.1266. [DOI] [PubMed] [Google Scholar]
- 40.Sato T, Takahoko M, Okamoto H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis. 2006;44:136–142. doi: 10.1002/gene.20196. [DOI] [PubMed] [Google Scholar]
- 41.Drapeau P, Ali DW, Buss RR, Saint-Amant L. In vivo recording from identifiable neurons of the locomotor network in the developing zebrafish. J Neurosci Methods. 1999;88:1–13. doi: 10.1016/s0165-0270(99)00008-4. [DOI] [PubMed] [Google Scholar]
- 42.White RM, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.