Abstract
Deregulation of cap-dependent translation is associated with cancer initiation and progression. The rate-limiting step of protein synthesis is the loading of ribosomes onto mRNA templates stimulated by the heterotrimeric complex, eukaryotic initiation factor (eIF)4F. This step represents an attractive target for anticancer drug discovery because it resides at the nexus of the TOR signaling pathway. We have undertaken an ultra-high-throughput screen to identify inhibitors that prevent assembly of the eIF4F complex. One of the identified compounds blocks interaction between two subunits of eIF4F. As a consequence, cap-dependent translation is inhibited. This compound can reverse tumor chemoresistance in a genetically engineered lymphoma mouse model by sensitizing cells to the proapoptotic action of DNA damage. Molecular modeling experiments provide insight into the mechanism of action of this small molecule inhibitor. Our experiments validate targeting the eIF4F complex as a strategy for cancer therapy to modulate chemosensitivity.
Keywords: chemical genetics, eIF4E:eIF4G inhibitor, lymphoma, translation inhibitor
Eukaryotic translation initiation is tightly regulated at the step of ribosome recruitment. This process involves binding of eukaryotic initiation factor (eIF) 4F complex to mRNA cap structures (m7GpppN; where N is any nucleotide). eIF4F is composed of: eIF4E, the cap-binding protein; eIF4A, a DEAD-box RNA helicase; and eIF4G, a scaffolding protein that bridges the interaction between eIF4E and eIF4A (1). Binding of eIF4F to the cap structure (via eIF4E) delivers eIF4A to the 5′ end of the mRNA template, an event required to disrupt RNA structure or RNP complexes to prepare a ribosome landing pad. eIF4E is the least abundant initiation factor, is rate-limiting for eIF4F complex assembly, and its availability for translation is regulated by the PI3K/Akt/mTOR pathway (1). The binding of one of three eIF4E negative regulators (eIF4E-Binding Proteins [4E-BPs]) to eIF4E is controlled by mTOR—with mTOR-dependent phosphorylation of 4E-BPs leading to their dissociation from the binary 4E-BP:eIF4E complex. Because 4E-BPs and eIF4G (there are two isoforms, called eIF4GI and eIF4GII, that share all structural features and show 46% sequence identity) compete for binding to a common site on eIF4E, 4E-BP binding to eIF4E decreases its availability for incorporation into the eIF4F complex and reduces translation initiation rates—with modest consequences on global translation but more pronounced effects on translation of specific mRNAs (1). This discriminatory effect on translation initiation is dependent on the amount of secondary structure present in the 5′ UTRs of mRNAs—with mRNAs harboring more secondary structure being more dependent on eIF4F for ribosome loading (2, 3). Gene expression profiling has identified mRNA transcripts whose translation is preferentially stimulated by altered eIF4E levels, indicating that eIF4E can affect the expression of a large gene set that impinge on several signaling nodes.
Several lines of evidence indicate that translational regulation is usurped in human tumors. Many cancers harbor lesions in the PI3K/Akt/mTOR pathway, which predictably affect eIF4F activity (4). eIF4E is overexpressed in many human cancers and hyper-phosphorylated 4E-BP is associated with tumor progression and adverse prognosis (5). Ectopic overexpression of eIF4E in vitro (6) and in vivo (7) is oncogenic and associated with chemoresistance. Hence, targeting eIF4F activity has been pursued to determine the consequences on tumor cell growth and chemosensitivity.
Conceptually, the mRNA-ribosome loading step of eukaryotic translation initiation can be blocked at several points, and include inhibiting mRNA cap-eIF4E interaction with cap analogues, interfering with eIF4F subunit interaction (i.e.,—eIF4E:eIF4G or eIF4G:eIF4A), blocking eIF4A RNA helicase activity, and preventing eIF4F subunit synthesis. Although some of these approaches are beginning to be explored, it is not clear that they will exert similar effects on cell proliferation in vitro or that they will allow for a therapeutic response in vivo. In cell culture, sequestration of eIF4E by ectopic overexpression of 4E-BP1 in eIF4E transformed cells can partially reverse tumorigenicity (8). As well, antisense RNA oligonucleotides directed to eIF4E (9), peptides directed to the eIF4E:eIF4G interaction site (10), or small molecule inhibition of the eIF4E:eIF4G interaction (11) suppress transformation and induce apoptosis in vitro, although the potential of these approaches has not been tested in vivo. Importantly, and more clinically relevant, antisense targeting of eIF4E (12) or small molecule inhibition of eIF4A helicase activity (13, 14) show promising efficacy in blocking tumor growth in vivo in several preclinical cancer models. Herein, we explore the consequences of blocking eIF4E:eIF4G interaction in vivo on chemoresistance in a genetically defined preclinical lymphoma model.
Results
Ultra-High-Throughput Screening Identifies eIF4E:eIF4GI Inhibitors.
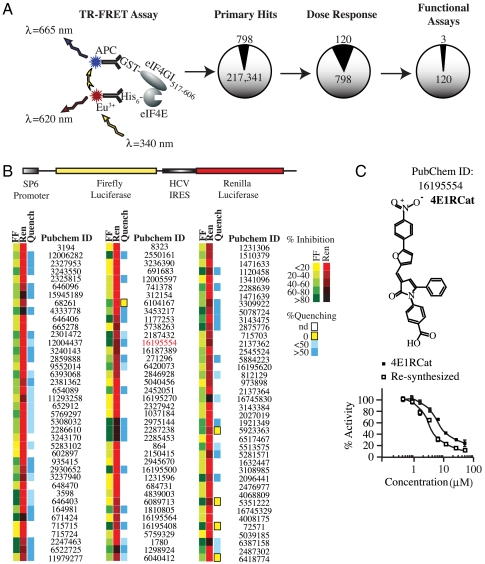
A previous high-throughput screen (HTS) for eIF4E:eIF4G inhibitors probed a small collection of ∼16,000 compounds and identified a molecule that binds to eIF4E, called 4EGI-1 (11). This compound inhibits eIF4E:eIF4G interaction, yet stimulates eIF4E:4E-BP1 interaction (11). To explore the therapeutic potential of uncoupling eIF4E from both eIF4G and 4E-BP, we sought to identify new compounds that could block interaction of eIF4E with eIF4G and 4E-BP1. To this end, a library of 217,341 compounds was screened using a time resolved (TR)-FRET based assay consisting of His-tagged eIF4E and a glutathione S-transferase (GST)-tagged fragment of eIF4GI (GST-eIF4GI517–606) (Fig. 1A). Following dose-response analysis of 798 primary hits, 120 compounds showed an IC50 < 20 μM (Fig. 1A). These compounds were further evaluated in an in vitro translation assay to score for their ability to inhibit cap-dependent but not Hepatitis C Virus internal ribosome entry site (HCV IRES) mediated translation (Fig. 1B). A counterscreen to identify false positives that nonspecifically quenched the luciferase enzymatic reaction rather than inhibiting cap-dependent translation was also performed (Fig. 1B). After testing all other compounds (harboring or lacking quenching activity) in an in vitro translation assay using [35S]-methionine to monitor protein synthesis, we identified three that inhibited cap-dependent translation, one of which we describe in detail herein (called 4E1RCat; PubChem ID 16195554). 4E1RCat displayed an IC50 < 10 μM in the TR-FRET assay (Fig. 1C) and its resynthesis confirmed its activity (Fig. 1C; IC50 ∼ 4 μM). We performed structure-activity relationship (SAR) analysis on 4E1RCat using the TR-FRET assay and tested 16 analogs containing modifications at two positions of the molecule (Fig. S1A). Structure confirmation, resynthesis, and SAR analysis led to the identification of several oxo-pyrrolyl benzoic acid analogs that showed low micromolar potency inhibition for the eIF4E:eIF4G interaction based on the initial hit structure. However, none of the modifications to the “A” or “B” moieties improved the IC50 compared to 4E1RCat in the TR-FRET assay (Fig. S1B).
Fig. 1.
UltraHTS TR-FRET identifies inhibitors of eIF4E:eIF4GI interaction. A. Schematic diagram of TR-FRET based assay and attrition rate from secondary and functional assays. B. Schematic representation of the FF/HCV/Ren bicistronic construct used to characterize inhibitory potential of primary hits in in vitro translation extracts. Heat map showing results of secondary in vitro translation assay and of the counterscreen to identify compounds that nonspecifically inhibited luc enzyme activity. Experiments were performed in duplicates and average values obtained were normalized to DMSO controls. nd, not done. C. Structure of 4E1RCat and its dose response in the TR-FRET assay.
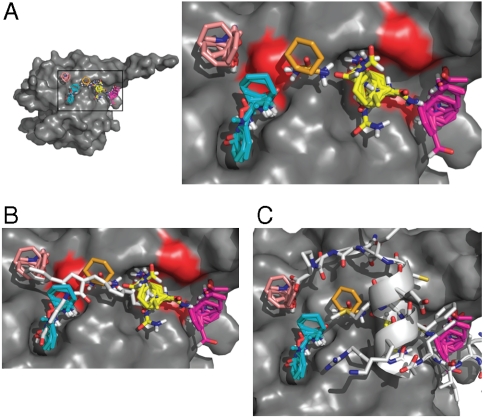
Modeling Binding of 4E1RCat to eIF4E Predicts Interference with eIF4G and 4E-BP Binding.
In silico probing of the three-dimensional surface of eIF4E for potential drug binding sites by computational solvent mapping (15) revealed the presence of five shallow pockets that form an elongated binding site and could support small molecule interactions (Fig. 2A). 4E1RCat shows potential for binding to four of these pockets (Fig. 2B). Both 4E-BP1 and eIF4G bind to a common site on the dorsal side of eIF4E (modeled in Fig. 2C) and it is clear that 4E1RCat is predicted to clash with this interaction (Fig. 2C). This modeling data offers a potential mechanism by which 4E1RCat inhibits eIF4E:eIF4G and eIF4E:4E-BP1 interaction.
Fig. 2.
Modeling of 4E1RCat bound to eIF4E. A. Location of the largest hot spots of eIF4E. Results are shown for the eIF4E structure cocrystallized with segment 47-66 of 4E-BP1 (PDB code 1WKW), but are essentially identical for the other two available X-ray structures (PDB codes 1IPB and 2W97). The largest consensus site, CS1, (shown in yellow), binds 24 probe clusters and defines the main hot spot. The other large consensus sites are CS2 (magenta, 22 probe clusters), CS3 (cyan, 19 probe clusters), and CS4 (salmon, 10 probe clusters). Consensus site CS6 is small (ochre, 5 probe clusters), but indicates a shallow channel connecting the consensus sites CS1 and CS3. The close-up of the hot spots also shows (in red) the location of residues V69, L131, and I138 on the surface of eIF4E. B. The most likely binding pose of 4E1RCat. The predicted hot spots are superimposed for reference. C. Segment 47-66 of 4E-BP1 from the eIF4E:4E-BP1 complex (PDB code 1WKW) superimposed on the hot spots. For 4E-BP1, the side chain of Y54 of the motif extends toward CS2, L59 is deep in the pocket that binds CS1, confirming the importance of the main hot spot, and the side chain of M60 overlaps with CS6.
Inhibition of Cap-Dependent Translation Initiation by 4E1RCat.
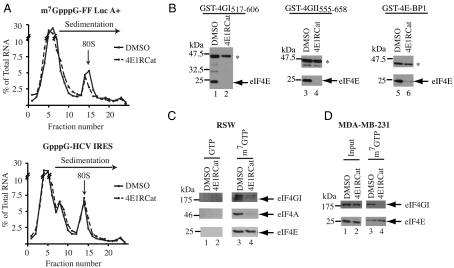
Much of our current understanding of factor assembly on mRNA templates during translation initiation is based on reconstitution and biochemical approaches that have dissected various steps of this process. One very useful assay for monitoring ribosome binding to mRNA is the visualization of 80S complexes on radiolabeled mRNA templates following sedimentation velocity centrifugation. In this assay, 4E1RCat reduced 80S ribosome complex formation (Fig. 3A), albeit not to the same extent as observed for m7GTP (Fig. S2). The inhibition of ribosome recruitment to mRNA by 4E1RCat was cap-specific, because this compound showed no inhibitory effect on 80S complex formation on the GpppG-HCV IRES (Fig. 3A).
Fig. 3.
4E1RCat inhibits cap-dependent translation initiation. A. Inhibition of cap-dependent 80S complex formation by 4E1RCat. 32P-labeled m7GpppG-FF Luc A+ or GpppG-HCV mRNA was incubated with cycloheximide (CHX) and either vehicle (1% DMSO) or 50 μM 4E1RCat in RRL. Total counts recovered from each gradient and the percent mRNA bound in 80S complexes were—m7GpppG-FF/mRNA + 1% DMSO [58895 cpm, 14.2% binding], m7GpppG-FF/mRNA + 4E1RCat [60503 cpm, 10.6% binding], GpppG-HCV/mRNA + 1% DMSO [64426 cpm, 14.2% binding], and GpppG-HCV/mRNA + 4E1RCat [67592 cpm, 17% binding]. B. Effect of 4E1RCat on the interaction between eIF4E and GST-eIF4GI517–606 (left), GST-eIF4GII555–658 (center), and GST-4E-BP1 (right). GST-pull downs were performed in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of 4E1RCat. Glutathionine eluents were probed for the presence of GST-tagged proteins (denoted by an asterisk) and eIF4E by Western blotting. C. 4E1RCat inhibits eIF4F complex assembly in vitro. Pull-down experiments from RSW were performed as described in the Materials and Methods. GTP and m7GTP eluents were fractionated by SDS-PAGE and probed for the presence of eIF4E, eIF4A, and eIF4GI by Western blotting. D. 4E1RCat inhibits eIF4F complex assembly in vivo. Pull-down experiments from cell extracts were performed as described in the Materials and Methods. Input and m7GTP eluents were analyzed by SDS-PAGE and probed for the presence of eIF4E and eIF4GI by Western blotting.
4E1RCat blocked eIF4E:eIF4G and eIF4E:4E-BP1 interaction (Fig. 3B). This compound also inhibited the ability of full-length eIF4GI to bind to GST-eIF4E (Fig. S3). To determine if 4E1RCat could disrupt preformed eIF4F complexes, we used m7GTP-agarose to purify the eIF4F complex from Ribosome Salt Wash (RSW) (Fig. 3C) and from extracts prepared from 4E1RCat treated MDA-MB-231 cells (Fig. 3D). Western blot analysis probed for the presence of eIF4E and the copurifying subunits, eIF4A and eIF4GI, in the m7GTP eluents. The amount of eIF4GI present in the eIF4F complex was reduced in vitro (Fig. 3C) and in vivo (Fig. 3D).
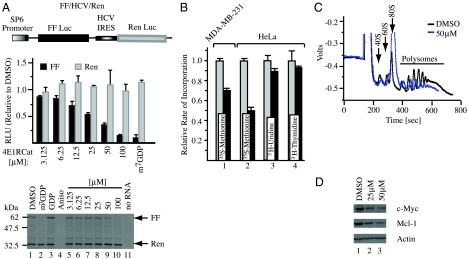
In vitro translations were performed to assess the effects of 4E1RCat on bicistronic mRNAs harboring IRESes that do not require eIF4F for ribosome recruitment (HCV and CrPV) (16) (Fig. 4A and Fig. S4) and on a bicistronic mRNA harboring an IRES that requires eIF4G but not eIF4E for ribosome recruitment (17) (EMCV, Fig. S4). As expected, m7GDP inhibited cap-dependent firefly (FF) expression, whereas the general translation inhibitor, anisomycin, inhibited production of both FF and renilla (Ren) from FF/HCV/Ren mRNA (Fig. 4A, lanes 2 and 4, respectively). 4E1RCat inhibited cap-dependent translation from FF/HCV/Ren in a dose dependent manner and did not affect Ren expression (Fig. 4A, compare lanes 5–10). 4E1RCat also inhibited cap-dependent production of Ren Luc from Ren/CrPV/FF although the extent of cap-dependent inhibition was not as strong as seen with FF/HCV/Ren (Fig. S4A). At higher concentrations of compound, CrPV IRES mediated translation was stimulated—an effect that could be due to increased ribosome availability and has been previously reported with other inhibitors of initiation (18). 4E1RCat inhibited cap-dependent translation from FF/EMCV/Ren but did not affect translation initiation mediated by the EMCV IRES (Fig. S4B). 4E1RCat inhibited protein synthesis in vivo in MDA-MB-231 and HeLa cells, but did not significantly affect RNA or DNA synthesis (Fig. 4B). Inhibition of protein synthesis by 4E1RCat in vivo was readily reversible (Fig. S5). 4E1RCat decreased polysomes, increased the fraction of 80S ribosomal subunits (Fig. 4C), and decreased levels of Mcl-1 and c-Myc proteins, two eIF4E-dependent mRNAs (19, 20) (Fig. 4D).
Fig. 4.
Effects of 4E1RCat on translation. A. Effect of 4E1RCat on in vitro translations performed in Krebs extracts programmed with FF/HCV/Ren. A schematic representation of FF/HCV/Ren mRNA is provided (top). In vitro translations were performed in the presence of 35S-Met and a representative autoradiograph of the products after fractionation by 10% SDS-PAGE is provided (bottom). Reactions contained vehicle (1% DMSO) (lane 1), 500 μM m7GDP (lane 2), 500 μM GDP (lane 3), and 50 μM anisomycin (lane 4), the indicated concentrations of 4E1RCat (lanes 5–10), or lacked input mRNA (lane 11). Center: FF and Ren RLU values (relative to DMSO controls) from two independent experiments with the SEM indicated. B. 4E1RCat inhibits protein synthesis in vivo. The rate of incorporation of each radioisotope tracer into TCA-insoluble material was monitored and is expressed relative to vehicle (DMSO) treated cells, which is set at 1. Results are the average of three experiments with the error of the mean shown. C. Polysome profiling analysis of Jurkat cells treated with 50 μM 4E1RCat. D. 4E1RCat inhibits c-Myc and Mcl-1 production. Jurkat cells were treated with 4E1RCat for 1 h, cell extracts prepared, and analyzed by Western blotting for c-Myc (Santa Cruz SC-40), Mcl-1 (Rockland), and actin (Sigma A5441) expression.
4E1RCat Reverses Chemoresistance.
The Eμ-Myc lymphoma model is a powerful, genetically defined system for studying drug action in vivo. Activation of mTOR signaling in this preclinical model through constitutive activation of the serine/threonine-specific kinase AKT or loss of Pten accelerates tumorigenesis and promotes chemoresistance—effects that have been attributed to increased eIF4F activity (7, 21). In this model Pten+/-Eμ-Myc and Tsc2+/-Eμ-Myc tumors are resistant to doxorubicin and we have previously shown that inhibition of mTOR signaling (7, 22) or eIF4A activity (13) can dramatically impact on this and significantly extend tumor-free survival. We therefore used this model to assess the consequences of 4E1RCat on chemoresistance (Fig. 5A).
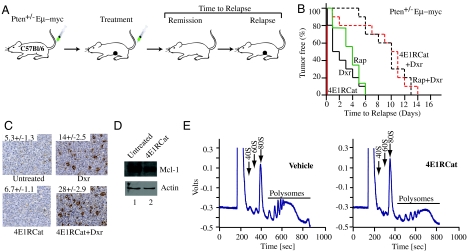
Fig. 5.
4E1RCat alters chemosensitivity of Pten+/-Eμ-Myc tumors in vivo. A. Representation of Eμ-Myc model and treatment response. B. Kaplan-Meier plot showing tumor-free survival of mice bearing Pten+/-Eμ-Myc tumors following treatment with doxorubicin (Dxr, solid black line; n = 10), rapamycin (Rap, solid green line; n = 9), Rap and Dxr (dashed black line; n = 10), 4E1RCat (4E1RCat, solid red line; n = 10), or 4E1RCat and Dxr (dashed red line; n = 10). C. Combination treatment of 4E1RCat and doxorubicin increases the percentage of apoptotic cells. Representative TUNEL staining on sections of Pten+/-Eμ-Myc tumors following treatments (original magnification × 20-fold). The percentage of cells that stained positive represents the average of four different fields, where 500 cells were counted per field. D. 4E1RCat inhibits Mcl-1 production in vivo. E. 4E1RCat inhibits protein synthesis in vivo.
Treatment of mice bearing Pten+/-Eμ-Myc or Tsc2+/-Eμ-Myc lymphomas with 4E1RCat alone was not effective in inducing any noticeable remission, whereas doxorubicin (Dxr) or rapamycin (Rap) induced a short-lived remission (Fig. 5B and Fig. S6A). Dxr and 4E1RCat synergized in mice and extended tumor-free remissions for up to 14 d (Fig. 5B, p < 0.001; Fig. S6B, p < 0.001), similar to what was observed with Dxr and Rap. This effect was unlikely due to 4E1RCat nonspecifically increasing Dxr efficacy because we did not observe synergy between 4E1RCat and Dxr in mice bearing Eμ-Myc lymphomas (Fig. S6B). Nor did we observe synergy in vitro on Eμ-Myc lymphomas, in contrast to the effects of 4E1RCat and Dxr on Tsc2+/-Eμ-Myc lymphomas (Fig. S6C). Analysis of Pten+/-Eμ-Myc tumor samples 6 h after treatment revealed an increase in the number of apoptotic cells for 4E1RCat + Dxr treated samples, compared to Dxr or 4E1RCat only samples (Fig. 5C). As expected, levels of Mcl-1 were decreased in tumors following treatment of mice with 4E1RCat (Fig. 5D). These results indicate, that 4E1RCat sensitizes Pten+/-Eμ-Myc and Tsc2+/-Eμ-Myc lymphomas to the cytotoxic effects of Dxr by inhibition of a prosurvival pathway. 4E1RCat was targeting translation in vivo as determined by polysome profiles analysis (Fig. 5E).
Discussion
We have identified and characterized a small molecule inhibitor that blocks interaction of eIF4E with two of its binding partners, eIF4G and 4E-BP1. Molecular modeling of 4E1RCat indicates that it binds to eIF4E to the region that is also utilized by eIF4G and 4E-BP1 for binding (Fig. 2). The binding site is also located at residues that show preferential broadening of heteronuclear single quantum coherence peaks by the small molecule inhibitor 4EGI-1 (11). However, the inhibitory properties of 4E1RCat are different from 4EGI-1, because the latter blocks eIF4E:eIF4G but paradoxically increases eIF4E:4E-BP1 interaction (11). These results indicate that 4E1RCat and 4EGI-1 share overlapping but nonidentical sites.
We note that 4E1RCat appeared to be a weaker inhibitor than m7GTP in preventing 80S complex formation (Fig. 3A and Fig. S2). The lower efficiency in inhibition may be due to differences in binding affinities, the inability of 4E1RCat to efficiently disrupt all preformed eIF4F complex, or the ability of the newly released eIF4G/eIF4A dimers to partially compensate for loss of eIF4F activity. Along these lines, we note that eIF4G can function in stimulating mRNA translation independent of eIF4E. Truncated mutants of eIF4G that lack the eIF4E-binding site have been shown to stimulate translation of uncapped mRNAs in rabbit reticulocyte lysate (RRL) (23), restored translation of capped mRNAs in eIF4F-depleted RRL (24), and in vivo can stimulate initiation of translation (25). In reconstituted systems, eIF4G (lacking the eIF4E binding site) and eIF4A can efficiently load 48S complexes on capped and uncapped ß-globin mRNA (26). These observations are consistent with reports demonstrating that translation initiation is reduced, but not abolished, by removal of the cap structure (27). Hence, we do not expect complete inhibition of translation by 4E1RCat upon exposure to cells, which is what was observed (Fig. 4B).
We find that 4E1RCat can reverse chemoresistance in a Myc-driven lymphoma model, consistent with the idea that deregulated translation plays a role in this phenomenon (7). Because 4E1RCat prevents eIF4E from interacting with two known protein partners, our data does not allow us to discriminate between which interaction is responsible for the biological effects observed. However, we favor the interpretation that 4E1RCat acts through disruption of eIF4E:eIF4G interaction because this would be consistent with results demonstrating that blocking the eIF4A subunit of eIF4F from loading onto mRNA templates (13, 14) shows similar chemosensitizing properties. By targeting eIF4E:eIF4G interaction, 4E1RCat is predicted to uncouple eIF4A binding to the mRNA from the eIF4E-cap recognition step, because cap-dependent eIF4A binding a priori requires eIF4E-cap recognition. One potential target that has been implicated in the chemosensitizing response is Mcl-1 (19, 22), whose levels we find decreased in the presence of 4E1RCat (Figs. 4 and 5).
4E1RCat represents a starting pharmacophore upon which to improve biological activity—both with respect to potency (Fig. 1C) and selectivity (http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=16195554&loc=ec_rcs). Indeed, the modeling results presented herein (Fig. 2) also suggests a framework for achieving this improvement—by extension of 4E1RCat into an adjacent binding groove (Fig. 2B). We are currently exploring this avenue. 4E1RCat also offers a pharmacological approach by which to interrupt eIF4E-dependent signaling nodes deregulated in neoplasia and provides a chemical genetic tool with which to explore translational control.
Materials and Methods
In Vitro Translations.
In vitro transcriptions and translations of bicistronic mRNA reporters were performed as previously described (28). Firefly (FF) and renilla (Ren) luciferase (luc) activity (RLU) was measured using a Berthold Lumat LB 9507 luminometer. To eliminate luc quenchers, in vitro translation reactions were performed with FF/HCV/Ren in the absence of compounds. After 1 h at 30 °C, compounds were added to the translations and FF and Ren luc activity measured. To visualize in vitro translated products, reactions were performed in micrococcal nuclease treated Krebs extracts in the presence of [35S]-methionine. Translations were analyzed on a 10% SDS-polyacrylamide gel, treated with  (PerkinElmer), dried, and exposed to X-Omat film (Kodak).
(PerkinElmer), dried, and exposed to X-Omat film (Kodak).
eIF4F Pull-Down Experiments.
Pulldown experiments of eIF4F were performed as previously described (14). In the case of RSW, this was treated for 1 h with either 1% DMSO or 50 μM 4E1RCat, whereas for cell extracts, MDA-MB-231 extracts prepared from cells treated with either 1% DMSO or 50 μM 4E1RCat for 4 h. Primary antibodies used were anti-eIF4E (Santa Cruz), anti-eIF4GI (Bethyl), and anti-eIF4A (29). Secondary antibodies were from Jackson Immuno Research.
Treatment Studies.
One million secondary Pten+/-Eμ-Myc, Tsc2+/-Eμ-Myc, or Eμ-Myc lymphoma cells were injected into the tail vein of 6–8 w old female C57BL/6 mice. When tumors were palpable, mice were treated with rapamycin (4 mg/kg daily for 5 d), 4E1RCat (15 mg/kg daily for 5 d), or doxorubicin (once at 10 mg/kg). Compounds were administered via intraperitoneal (i.p.) injection in 5.2% PEG 400/ 5.2% Tween 80. For combination studies, rapamycin or 4E1RCat were injected i.p. daily for five consecutive days, with doxorubicin being administered once on day two. Animals were palpated daily to monitor for the onset of tumors. Tumor-free survival was defined as the time between disappearance and reappearance of tumors. Data was analyzed using the log-rank (Mantel-Cox) test for statistical significance (SigmaStat software) presented in Kaplan-Meier format.
Supplementary Material
Acknowledgments.
We thank Marilyn Carrier and Isabelle Harvey for excellent technical assistance. Fellowship support was from a Canadian Institutes of Health Research Cancer Consortium Training Grant Award and a Cole Foundation Award to R.C. This work was supported by a grant from the National Institutes of Health (1 R01 CA114475) and the Canadian Cancer Society Research Institute (#17099) to J.P.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011477108/-/DCSupplemental.
References
- 1.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 2.Svitkin YV, et al. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68:631–634. doi: 10.1158/0008-5472.CAN-07-5635. [DOI] [PubMed] [Google Scholar]
- 4.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2:pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 5.Armengol G, et al. 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 2007;67:7551–7555. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- 6.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 7.Wendel HG, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 8.Rousseau D, Gingras AC, Pause A, Sonenberg N. The eIF4E-binding proteins 1 and 2 are negative regulators of cell growth. Oncogene. 1996;13:2415–2420. [PubMed] [Google Scholar]
- 9.DeFatta RJ, Nathan CA, De Benedetti A. Antisense RNA to eIF4E suppresses oncogenic properties of a head and neck squamous cell carcinoma cell line. Laryngoscope. 2000;110:928–933. doi: 10.1097/00005537-200006000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Herbert TP, Fahraeus R, Prescott A, Lane DP, Proud CG. Rapid induction of apoptosis mediated by peptides that bind initiation factor eIF4E. Curr Biol. 2000;10:793–796. doi: 10.1016/s0960-9822(00)00567-4. [DOI] [PubMed] [Google Scholar]
- 11.Moerke NJ, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 12.Graff JR, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117:2638–2648. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bordeleau ME, et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest. 2008;118:2651–2660. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cencic R, et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS ONE. 2009;4:e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenke R, et al. Fragment-based identification of druggable “hot spots” of proteins using Fourier domain correlation techniques. Bioinformatics. 2009;25:621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pestova TV, et al. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robert F, et al. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNAiMet ternary complex availability. Mol Biol Cell. 2006;17:4632–4644. doi: 10.1091/mbc.E06-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wendel HG, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin CJ, Cencic R, Mills JR, Robert F, Pelletier J. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res. 2008;68:5326–5334. doi: 10.1158/0008-5472.CAN-07-5876. [DOI] [PubMed] [Google Scholar]
- 21.Wendel HG, et al. Determinants of sensitivity and resistance to rapamycin-chemotherapy drug combinations in vivo. Cancer Res. 2006;66:7639–7646. doi: 10.1158/0008-5472.CAN-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mills JR, et al. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci USA. 2008;105:10853–10858. doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Gregorio E, Preiss T, Hentze MW. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA. 1998;4:828–836. doi: 10.1017/s1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali IK, McKendrick L, Morley SJ, Jackson RJ. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 2001;20:4233–4242. doi: 10.1093/emboj/20.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Gregorio E, Preiss T, Hentze MW. Translation driven by an eIF4G core domain in vivo. EMBO J. 1999;18:4865–4874. doi: 10.1093/emboj/18.17.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunnery S, Maivali U, Mathews MB. Translation of an uncapped mRNA involves scanning. J Biol Chem. 1997;272:21642–21646. doi: 10.1074/jbc.272.34.21642. [DOI] [PubMed] [Google Scholar]
- 28.Novac O, Guenier AS, Pelletier J. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 2004;32:902–915. doi: 10.1093/nar/gkh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edery I, et al. Involvement of eukaryotic initiation factor 4A in the cap recognition process. J Biol Chem. 1983;258:11398–11403. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.