Abstract
Magnetotactic bacteria (MTB) are a phylogenetically diverse group which uses intracellular membrane-enclosed magnetite crystals called magnetosomes for navigation in their aquatic habitats. Although synthesis of these prokaryotic organelles is of broad interdisciplinary interest, its genetic analysis has been restricted to a few closely related members of the Proteobacteria, in which essential functions required for magnetosome formation are encoded within a large genomic magnetosome island. However, because of the lack of cultivated representatives from other phyla, it is unknown whether the evolutionary origin of magnetotaxis is monophyletic, and it has been questioned whether homologous mechanisms and structures are present in unrelated MTB. Here, we present the analysis of the uncultivated “Candidatus Magnetobacterium bavaricum” from the deep branching Nitrospira phylum by combining micromanipulation and whole genome amplification (WGA) with metagenomics. Target-specific sequences obtained by WGA of cells, which were magnetically collected and individually sorted from sediment samples, were used for PCR screening of metagenomic libraries. This led to the identification of a genomic cluster containing several putative magnetosome genes with homology to those in Proteobacteria. A variety of advanced electron microscopic imaging tools revealed a complex cell envelope and an intricate magnetosome architecture. The presence of magnetosome membranes as well as cytoskeletal magnetosome filaments suggests a similar mechanism of magnetosome formation in “Cand. M. bavaricum” as in Proteobacteria. Altogether, our findings suggest a monophyletic origin of magnetotaxis, and relevant genes were likely transferred horizontally between Proteobacteria and representatives of the Nitrospira phylum.
Magnetotactic bacteria (MTB) are widespread aquatic microorganisms that use unique intracellular organelles called magnetosomes to navigate along the earth's magnetic field while searching for growth-favoring microoxic zones within stratified sediments. In strains of Magnetospirillum, it was shown that magnetosomes consist of magnetite (Fe3O4) crystals enclosed by a dedicated phospholipid membrane. The magnetosome membrane (MM) contains a specific set of proteins (1–3), which direct the biomineralization of highly ordered crystals along actin-like cytoskeletal filaments that control the assembly and intracellular positioning of a linear magnetosome chain (4–7). Synthesis of magnetosomes has recently emerged as a model for prokaryotic organelle formation and biomineralization (8–11). The trait of magnetotaxis is widely spread among Proteobacteria including members from the α-, δ- and γ-subdivisions, as well as uncultivated species from the deep branching Nitrospira phylum (8). The presence of MTB within unrelated lines of various phylogenetic groups, as well as their stunning diversity with respect to magnetosome shape, composition, and intracellular organization lead to speculations of whether the evolutionary origin of magnetotaxis is polyphyletic. Thus, independent origins and subsequent convergent evolution were proposed for greigite and magnetite producing MTB (12), and it has been suggested that those MTB forming magnetic crystals of divergent shapes or composition may use different mechanisms of magnetosome formation (13, 14).
Despite recent progress, magnetosome formation is not yet fully understood at the molecular and biochemical levels. Essential molecular factors, cellular structures, and processes leading to organelle formation and biomineralization have been characterized mostly in magnetospirilla. In Magnetospirillum gryphiswaldense most genes implicated in magnetosome synthesis were identified within several operons of a genomic magnetosome island (MAI) (15), which encodes functions in magnetosome membrane biogenesis, magnetosomal iron uptake, and control of magnetite crystallization (8, 10). Because of their conservation in other cultivated α-proteobacterial MTB (16, 17), it has been suggested that the MAI may have been transferred horizontally, which was further corroborated by the recent discovery of homologous gene clusters in metagenomic clones (18) and the δ-proteobacterial Desulfovibrio magneticus RS-1 (19). However, the limited genetic information about magnetosome formation that has been confined to a few cultivated MTB mainly of the α-Proteobacteria, is in striking disparity to the fact that MTB are a noncoherent and phylogenetically heterogeneous group. Because of the lack of cultivated representatives it has remained unknown whether homologous mechanisms and structures are used by divergent MTB from deep branching phyla outside the Proteobacteria.
One of the most intriguing systems for studying magnetosome formation in distantly related, nonproteobacterial MTB is the uncultivated “Candidatus Magnetobacterium bavaricum” (Mbav) from the deep branching Nitrospira phylum. Mbav has been identified originally within suboxic sediment layers of Bavarian lakes (20, 21), but a variety of related MTB were subsequently shown to display a wider global distribution (22–24). A recent cultivation-independent analysis of Mbav revealed first insights into its metabolic and genetic characteristics, suggesting that Mbav might be a chemolithoautotroph, obtaining energy from the oxidation of reduced sulfur compounds (21).
Compared to other MTB, Mbav is unique with respect to its large size (3–10 μm) and distinct cell biology, in particular to its numerous (up to 1,000) magnetosomes, which have a bullet-shaped, kinked morphology and are organized in multiple bundles of chains (20, 21, 25). Because previous studies failed to detect a membrane around magnetosomes of Mbav, it was speculated that non-α MTB producing bullet-shaped magnetite crystals might use different biomineralization mechanisms based on “templates” that might be fundamentally divergent from the MM-dependent mechanism in magnetospirilla and related MTB (13, 14).
Here, we describe an approach for targeted subgenomic and ultrastructural analysis of Candidatus M. bavaricum. By combining whole genome amplification of DNA from few Mbav cells collected by micromanipulation with screening of metagenomic libraries, we demonstrate the presence of a putative genomic magnetosome island with homology to that in Proteobacteria. In addition, the detection of structures such as a magnetosome membrane as well as putative cytoskeletal magnetosome filaments suggests a similar mechanism of magnetosome formation in uncultivated MTB of the deep-branching Nitrospira phylum as in Proteobacteria.
Results
Magnetosomes of “Candidatus M. bavaricum” Are Enclosed by a Membrane and Arranged Along a Cytoskeletal Filamentous Structure.
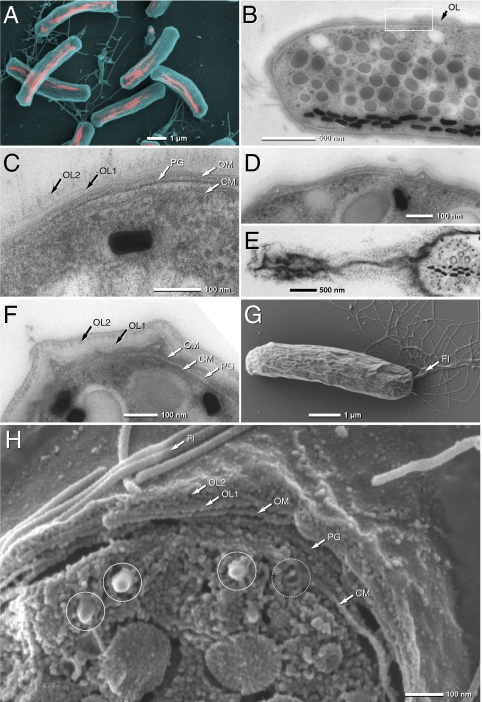
Magnetic mass collections from sediment samples highly enriched in Mbav cells (>40%, Fig. 1A) were subjected to several advanced high-resolution imaging techniques. Transmission electron microscopy (TEM) and SEM of high-pressure frozen and freeze-substituted cells revealed a number of unusual ultrastructural characteristics (Figs. 1 and 2 and Figs. S1 and S2). In addition to a peptidoglycan layer and the outer membrane (OM) and inner membrane (IM), the multilayered cell boundary exhibits an unusually wide pepriplasmic space and a bipartite outer layer resembling a capsular structure, which forms ridges and star-like extensions (Fig. 1; see Fig. S1 for further details). Sulfur globuli and polyhydroxybutyrate (PHB)-like granules are present within the cytoplasm (Fig. 1B and Fig. S2). Cells have a single bundle of ≈40 flagella (15–20 nm in diameter), which originate from different discrete spots of one cell pole (Figs. 1G and 2D). Overall, the cell wall structure resembles that of related representatives of the Nitrospira phylum, such as Nitrospira marina (26), N. moscoviensis (27), and N. defluvii (28), whereas the bipartite outer layer seems to be a distinct feature of the Mbav cell envelope.
Fig. 1.
Scanning (SEM) and transmission electron (TEM) micrographs of “Candidatus M. bavaricum” cells (Mbav). (A) SEM micrograph of Mbav by simultaneous detection of secondary (blue) and backscattered electrons (red). Chains of magnetite crystals are visible (red). (B–D) TEM microcraphs of ultrathin sections of high-pressure frozen and freeze-substituted cells showing the multilayered cell boundary (B, framed area). CM, cytoplasmic membrane; OM, outer membrane; PG, peptidoglycan; OL1, inner part of outer layer; OL2, outer part of outer layer; asterisks, ridges or papillae of periplasmic space. (E) FIB section showing a network of extensions of ridges. (F and G) TEM and SEM of conventionally fixed samples (Fl, flagella). (H) High-resolution SEM of a high-pressure frozen, cryofractured, and frozen hydrated Mbav cell. Solid circle, magnetosome crystals; dotted circle, empty magnetosome membrane vesicles.
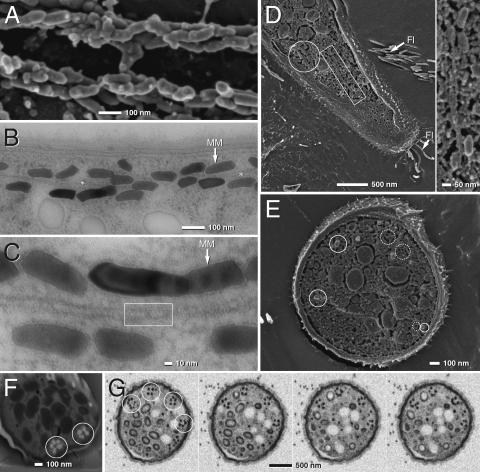
Fig. 2.
TEM and SEM micrographs of Mbav magnetosome chains (see Fig. S3 for an enlarged version at higher resolution). (A) SEM microcraph of a cryofractured cell (after chemical fixation) showing two bundles of magnetosome strands. (B and C) TEM ultrathin sections of high-pressure frozen and freeze-substituted cells showing strands of magnetosomes aligned parallel to a tubular filamentous structure (asterisk, framed area; MM, magnetosome membrane). (D and E) Cryo-SEM (frozen hydrated) of tangential (D) and cross-fractured (E) cells of Mbav (rectangular frame, magnetosomes aligned along MF; solid circles, magnetosomes crystals; dotted circle, empty MM vesicles). (F and G) SEM of focused ion beam (FIB) sections (F), and high-pressure frozen and freeze-substituted (G) Mbav cells. Circles indicate several rosette-like magnetosome bundles. Different micrographs in G represent selected sections from FIB-milling series (every 10th section is shown from left to right). Each section has a thickness of 8 nm.
As revealed by different imaging techniques, cells contained multiple chains of magnetosomes (Figs. 1A and 2). In contrast to previous studies, we found that the morphology and size of crystals was more variable including bullet-shaped, conical, or blunted polyeders, which had kinked or bent appearances. In SEM micrographs of fractured cells, magnetosomes appeared in densely packed bundles of several individual strands (Fig. 2A). SEM of focused ion beam sections of cryopreparations (Fig. 2F), and high-pressure frozen and freeze-substituted (Fig. 2G) cells revealed that the magnetosome bundles consist of three to six (mostly five) individual magnetosome strands that are arranged around a central core and form a regular rosette-like bundle (Fig. 2F), which is situated 20–50 nm beneath the cytoplasmic membrane. In cross-sections, between two and six of such magnetosome bundles appeared to be distributed preferentially within a roughly semicircular segment along the periphery of cells (Fig. 2G). Individual strands maintain nearly identical positions in 3D reconstructions by serial focused ion beam (FIB) sectioning (Fig. 2G and Movie S1). This argues against a twisted “braid-like” helical structure, which was described in previous studies (e.g., ref. 25). Instead, individual strands within a bundle appear aligned parallel to each other.
Intriguingly, TEM of ultrathin sections of high-pressure frozen and freeze-substituted cells revealed that strands are aligned parallel to a filamentous structure (Fig. 2B, asterisks). At higher magnification this filament is bound by two electron dense layers, which suggests that filaments may form a tubular structure with a diameter of 12–13 nm (Fig. 2C). Filaments closely adjacent to magnetosome particles were also seen in cryo-SEM of tangential-fractured cells (Fig. 2D) and 3D reconstructions of FIB “sections” (Movies S1 and S2). In its appearance, intracellular position, and dimensions these filamentous structures are strongly reminiscent of the cytoskeletal magnetosome filament (MF) that has been previously discovered in cultivated magnetospirilla (4–6).
Most notably, in TEM thin sections, individual crystals were found to be surrounded by a membranous structure that displays a laminate appearance (Fig. 2 B and C). Its thickness of 3–4 nm appeared somewhat lower than that of the cytoplasmic membrane (CM) (6–7 nm), which might be due to the fact that the innermost electron dense layer cannot be discerned against the dark background of the adjacent magnetite crystals. This membranous layer was found in all analyzed thin sections of Mbav and resembles the MM of magnetospirilla detected by the same method (29). Hollow, concave membrane-like structures were also visible by cryo-SEM of cross-fractured cells of Mbav (Figs. 1H and 2E). Because of their size and close vicinity to magnetosome crystals, they are likely to represent empty MM vesicles from which the magnetite core was lost during freeze fracturing.
Altogether, these data suggest that Mbav has a complex and distinct subcellular structure with respect to cell wall architecture and organization of the magnetosome chains. However, structures highly reminiscent of the MM and the MF in α-proteobacterial MTB are clearly present.
Single-Cell Sorting and Whole Genome Amplification (WGA).
To identify putative magnetosome genes, we initially attempted a similar approach as was successfully used for the identification of MAI clusters from metagenomic large-insert libraries (18). However, screening of more than 10,000 clones from six independently constructed fosmid libraries based on magnetically highly enriched Mbav cells failed to detect any clones harboring genes with similarity to known magnetosome proteins. Although in our environmental MTB collections Mbav was the most abundant morphotype (>40%), 16S rRNA gene libraries revealed that Mbav was only poorly represented in fosmid libraries constructed from these collections (<1%). Possibly, this bias was caused by poor DNA recovery due to the unusual cell structure of Mbav (21). All our attempts to increase the relative proportion of Mbav DNA by fluorescence-assisted cell sorting, filtration, and selective lysis were unsuccessful (experimental details available on request). Therefore, we had to develop an alternative strategy, which combined WGA of individually sorted Mbav cells with highly stringent PCR screening of metagenomic fosmid libraries with Mbav-specific primers deduced from WGA sequences (Fig. S4). In addition to Mbav cells, the only other abundant morphotypes present in our magnetic collections were magnetotactic cocci that could be easily distinguished from the conspicuous large (5–10 μm) rod-like Mbav cells (Fig. S5). This facilitated their strictly selective separation using micromanipulation with microscopic control over every sorting step (Figs. S5 and S6 A–D and Movie S3). Mbav cells were collected from a 5-μL droplet, containing the heterogeneous mixture of magnetically collected MTB and subjected to washing steps by two subsequent transfers into water droplets to eliminate potential extracellular DNA contaminations. Between 1 (Movie S3) and 1,000 (Movie S4) Mbav cells were collected into a single capillary, in which cells remained intact and viable upon release into water droplets (Movie S5). For WGA, washed cells were transferred to 0.75-μL droplets of lysis buffer on Ampligrids (Fig. S6C). Five to 10 individually sorted Mbav cells per WGA reaction yielded sufficient amounts of DNA for subsequent sequence analysis. A total of 3.8 μg of DNA was amplified from 158 cells sorted in 10 independent reactions, which were pooled to reduce stochastic amplification bias. Analysis of the corresponding 16S rRNA gene library (25 clones) before pyrosequencing exclusively revealed 100% identical Candidatus M. bavaricum sequences. This indicates that the separation was highly specific for target cells, which were free from contaminating DNA.
Identification of a Magnetosome Gene Cluster of Candidatus M. bavaricum by WGA-Enabled Screening of Metagenomic Libraries.
Pyrosequencing generated a total of 118.95 Mb of sequence information. However, only 39% of the obtained sequence data could be assembled (1,061 contigs; average size 556 bp) resulting in about 0.6 Mb. This is likely to represent only a fraction of the Mbav genome, if a genome size between 2 Mb (as for its closest sequenced relative Thermodesulfovibrio yellowstonii NC_011296), and about 5 Mb (like most other MTB) is assumed. Most likely the limited amount of DNA available for the pyrosequencing approach (3.8 μg) resulted in short read lengths and poor assembly, which prevented the analysis of entire genes or operons. However, two contigs of 529 bp (no. 1) and 570 bp (no. 2) were found to display partial similarity to known magnetosome proteins MamE (no. 1; 50% identities in 60 aa) and MamP (no. 2; 38% identities in 76 aa). Using primers targeting these regions, about 10,000 clones from metagenomic fosmid libraries (18, 21) were PCR screened. Five clones, which previously had escaped our initial screening because of the lack of endsequence similarities, were identified from which either identical sequences of mamE (1 clone), or mamP (1 clone), or both (3 clones) could be amplified.
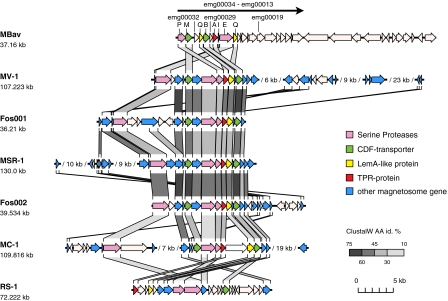
Sequencing, assembly, and gene prediction of mamE and mamP-positive clones yielded a contig of 37 kb with 34 putative genes [Fig. 3 and National Center for Biotechnology Information (NCBI) FQ377626]. The mismatch-free overlap (99.99% sequence identity) of five individual fosmids with an insert size of about 35 kb, 24 kb, 35 kb, 24 kb, and 27 kb, respectively, indicates a clonal origin of the amplified Mbav DNA. This was further verified in two independent control experiments (Fig. S7). The G+C content of 49.7% is within the range of 46.9–50.2% as determined for metagenomic fosmid clones containing phylogenetic marker genes (16S rRNA) from Mbav-like MTB (21, 30), but differs significantly from the related T. yellowstonii (34%), although it is well below the range of percentage of G+C found in MTB thus far (54.8–65.1%) (8). Out of the 34 identified genes, 14 encode hypothetical proteins, and 2 share no similarity to the NCBI database (Table S1). Among the genes related to proteins of known functions, 2 encode transposases, 3 are related to transcriptional regulation, whereas 4 might be related to metabolic functions (lyase, oxidoreductase, polysaccharide biosynthesis, and pyruvate phosphorylation; Table S1). Four genes encode proteins with the highest identity to T. yellowstonii (Table S1).
Fig. 3.
Molecular organization of sections from putative magnetosome islands of Mbav and selected other MTB (Mbav, Candidatus M. bavaricum; MV-1, magnetic vibrio; Fos001+002, metagenomic MTB clones; MSR-1, Magnetospirillum gryphiswaldense; MC-1, magnetic coccus; RS-1, Desulfovibrio magneticus). The black arrow on Top indicates the extension of a putative magnetosome operon in Mbav. Different colored arrows indicate characteristic features of proteins encoded by known magnetosome genes, whereas equivalent genes are connected by stripes of various shadings, which indicate different degrees of identity (id) as calculated from ClustalW alignments of encoded proteins. Alignments were generated with TRAPPIST, a Python-based toolbox for alignment, analysis, and visualization of genomes or genome segments.
Most remarkably, the contig contains a 18-kb cluster of 22 genes of which some display striking homology to known magnetosome genes of proteobacterial MTB (Fig. 3 and Tables S1 and S2). Their short intergenic distance and identical transcriptional direction suggest that they may be part of a common putative operon as in all other MTB analyzed so far. Whereas 8 of the 22 genes encode proteins with greatest similarity to MTB (on the basis of BlastP analysis against the NCBI database), only 5 of them (MamEIBMP) are most similar to known magnetosome genes (Table S1 and Fig. S8). Among them, MamI, which has been implicated in the formation of magnetosome vesicles in α-proteobacterial magnetospirilla (10) is least conserved (23–35% identity; Fig. S9), but shares significant and exclusive similarity to MamI homologs from all other analyzed MTB including Desulfovibrio magneticus, in which it escaped detection in previous studies (10, 19, 31).
Two genes of this putative operon (emg00025 and emg00031) were annotated as mamQ-I and mamQ-II, respectively, because they share similarity to all MamQ homologs, although the database comparison shows the highest similarity to a LemA-like protein from non-MTB. For the same reasons, the TPR domain protein (emg00028) was annotated as MamA, which was reported to be involved in magnetosome “activation” in M. magneticum (32). MamE and MamP are PDZ-containing putative serine proteases (33). Whereas MamP was implicated in the control of magnetite crystal size and number, MamE is thought to be involved in magnetosome formation by directing the proper localization of a subset of magnetosome proteins (10). MamM and MamB share homology with cation diffusion facilitator (CDF) transporters (34, 35) and are assumed to mediate iron transport into the magnetosome compartment (8). Multiple sequence comparisons were performed with all identified Mam homologs of Mbav to analyze their phylogenetic relation to magnetosome proteins from other MTB (Figs. S8 and S11). As an example, regardless of the algorithm used, Mbav MamB, and MamM CDF transporters cluster together with MamB and MamM proteins of MTB, which form a phylogenetically distinct branch separately from other CDF proteins, such as iron transporting FieF-like proteins (35) (Fig. S11). All other analyzed magnetosome genes display comparable branching patterns (Fig. S8). This demonstrates homology of magnetosome proteins from the Nitrospira phylum with those of proteobacterial origin, suggesting a horizontal gene transfer (HGT) between both phyla.
In addition to the 8 genes with clear homology to known α-proteobacterial magnetosome proteins, the Mbav magnetosome cluster comprises 14 further genes, which either intersperse the mam homologs (emg00032–emg00029) or are mostly located downstream of them (emg00024–emg00013). The majority of them (10 genes) encode hypothetical proteins, or share only weak similarity (22–27%) to proteins of known function, such as proteins involved in polysaccharide biosynthesis and chromosome condensation. Interestingly, 3 of these 14 genes encode proteins that share the highest similarity to Desulfovibrio magneticus (28–29%), whereas one of them (emg00029) encodes a hypothetical membrane protein and is localized between mamA and mamB. One further putative membrane protein encoded by orf emg00019 shares the highest identity with a bacterium not known to produce magnetosomes.
Discussion
On the basis of their unusual characteristics such as magnetosome arrangement, distinct cell biology and distant phylogenetic position, and lacking any genetic information, it has been questioned whether nonproteobacterial MTB from deep-branching phyla such as Nitrospira share homologous structures and genetic mechanisms of biomineralization with magnetotactic Proteobacteria (13).
Whereas our magnetic mass collection enabled the application of advanced imaging techniques, the poor representation and unknown identity of Mbav DNA in multispecies large-insert libraries from those enrichments hampered the metagenomic analysis of magnetosome formation. Therefore, for the confident identification and targeted analysis of magnetosome genes from Mbav, we had to resort to an alternative strategy based on WGA of individually sorted cells. Such targeted WGA approaches have recently been demonstrated to be functional for other cultivated and uncultivated MTB (36).
Genomes from several uncultured bacteria have been partly sequenced from single or few cells (for instance ref. 37, 38). However, despite recent improvements in genome recovery and normalization procedures (39), single-cell genome sequencing has not yet become a routine technique, and only recently the first gap-free reconstruction of an entire genome from a single polyploidic bacterium was demonstrated (40). Unlike the large contigs of >0.5 Mb reported in some of these studies, our WGA approach yielded only relatively short average contig lengths, most likely due to poor DNA recovery from lysis-recalcitrant Mbav cells (21) and consequent amplification bias due to limiting amounts of template DNA. However, the obtained sequences allowed the identification of true Mbav clones in existing multispecies metagenomic libraries. Whereas neither of the two complementary approaches (WGA and metagenomics) alone was successful, their combination led to first insights into the genetic control of magnetosome formation in a nonproteobacterial MTB from the deep-branching Nitrospira phylum. Similar strategies could be applied as an efficient and economical approach for targeted genomic analysis of further MTB or other low-abundant environmental bacteria with conspicuous morphologies.
Whereas in one study organic material and several polypeptides with homology to known magnetosome proteins copurified with isolated magnetite crystals of the δ-protebacterium D. magneticum RS-1 (3), other in situ TEM studies failed to detect a MM surrounding the individual magnetite crystals in this organism and Mbav, which are only distantly related to the prototypical magnetospirilla. This led to speculations that bullet-shaped magnetosomes are generally not formed within a MM, and that these MTB may have evolved a divergent molecular mechanism of magnetite biomineralization (13, 14).
However, our discovery of similar structures and genes in this study is strongly indicative of homologous mechanisms of biomineralization also in distantly related MTB outside the Proteobacteria. By using several advanced electron-microscopic techniques including focus ion beam sectioning of cryofixed cells, which to our knowledge has been successfully applied for the first time for the 3D reconstruction of a bacterial cell, an unprecedented insight into the subcellular organization of Mbav was obtained. The complex structure of its cell envelope and its intricate magnetosome architecture represent one of the highest structural levels found in a bacterial cell. In particular, we were able to demonstrate that, contrary to previous observations (13), magnetite crystals of Mbav are surrounded by a MM, which strongly resembles the well-analyzed structure in α-proteobacterial MTB, such as M. gryphiswaldense (4, 33). However, the relatively large distance (20–50 nm) of the magnetosome bundles from the CM in Mbav makes it unlikely that the MM remains permanently contiguous with the CM, but may become detached from it during magnetosome assembly.
The presence of a MM is consistent with the identification of genes that encode homologs of magnetosome proteins MamI, -P, -A, -M, -B, -Q, and -E, which are all known to be associated with the MM and are implicated in its biogenesis as well as magnetosomal transport of iron in Magnetosopirillum species (8, 10, 33). mamI, -P, -A, -M, -B, -Q, and -E belong to the group-specific “signature genes” of magnetotaxis, which has recently been identified by comparative genomics (41), i.e., they are conserved and located within the MAI of all MTB, but exhibit no (MTB-specific, mamI), or only remote (MTB-related, mamPAMBQE) similarity to any genes from nonmagnetotactic organisms. Despite general conservation, homologous genes within the Mbav mam cluster are less conserved with respect to sequences and synteny than among the proteobacterial MTB. For example, a peculiarity not found in other magnetobacterial genomes is the presence of two divergent mamQ copies (Fig. 3), which might be related to a somewhat distinct mechanism of MM formation in this organism. However, a number of the previously identified magnetosome genes, such as mamHRSTNOLKJ, which are conserved in most or all proteobacterial MTB, is missing. One possible reason might be a slightly distinct genetic control of biomineralization in Mbav. On the other hand, however, the cluster is bound by a mamP-like gene at one end of the assembled contig sequence, but on the basis of the usual central position of mamP within the MAI of other MTB, it can be expected that a significant portion of the operon containing additional magnetosome genes extends further upstream beyond the boundary of the contig and might be part of an even larger MAI.
One example for such a “missing” gene is mamK, which is conserved in all MTB and encodes an actin-like protein that in magnetospirilla forms the cytoskeletal MF and is involved in intracellular assembling, aligning, and positioning of the linear magnetosome chains (4–7). We have demonstrated by cryo-SEM and -TEM the presence of a similar filamentous structure in Mbav, which is aligned along the magnetosome chains in close vicinity. Unlike the MF in magnetospirilla, which form a network of filament bundles, filaments found in Mbav appear to form an ordered tubular structure with a hollow interior, around which the individual magnetosome strands are arranged. However, its general resemblance suggests that this structure might be homologous to the MF of magnetospirilla and is possibly also formed by a MamK-like protein encoded elsewhere in the genome.
On the other hand, the intricate 3D tubular organization of magnetosome chains in multiple discrete bundles is significantly more complicated than the single or double linear chains found in magnetospirilla. Therefore, it is very likely to be governed by a much more elaborate mechanism of assembly, which, in addition to constituents of the magnetosome pathway that are generally conserved among all MTB, involves further specific genetic determinants. These might be encoded elsewhere in the genome, or for example, by the genes that follow the identified mam genes immediately downstream. Although the function of these gene is not known, their colocalization with known mam genes within a single putative operon and their partial conservation in other MTB suggests that they might be involved in magnetosome formation and biomineralization as well.
Our discovery of structures and genes in MTB from the deep-branching Nitrospira phylum similar to those in distantly related proteo-MTB also raises another important question, which is the phylogenetic origin and evolution of magnetotaxis. In contrast to previous postulations (13, 14), our results provide further evidence that the mechanism of magnetite biomineralization (apart from some species-specific variations) might be universal. Recently, we have demonstrated that horizontal gene transfer rather than independent evolution is likely to account for the emergence of magnetotaxis among diverse lines of proteobacterial MTB (16), which has been confirmed by the detection of homologous genes in RS-1 (19). Similar studies on greigite-forming uncultivated MTB can be expected to reveal the genetic basis and evolution of greigite-based magnetotaxis in the future.
As revealed by phylogenetic trees, putative magnetosome proteins of Mbav, such as the CDF transporters MamB and MamM, branch together with their proteo-MTB relatives, whereas homologs from non-MTB form a distinct phylogenetic group (Fig. S11), suggesting that proteins within MTB-specific branches have distinct functions and are universally conserved in all MTB. Because Mbav and the proteobacterial MTB are not closely related and phylogenetically separated by numerous bacterial species lacking the magnetotactic trait (8), there are two possible scenarios to explain the presence of homologous magnetosome genes. One would be the assumption of a common magnetotactic ancestor of the Nitrospira and Proteobacteria. However, this scenario would require multiple events of loss of magnetosome genes in all other nonmagnetotactic proteobacterial descendants. Although spontaneous loss of MAI genes at rather high frequency has been demonstrated in cultivated MTB (15), and therefore cannot be entirely dismissed, secondary loss in almost all other proteobacteria seems rather unlikely. In contrast, a second scenario, in which horizontal gene transfer of magnetosome genes between proteobacterial MTB and the Nitrospira phylum provides a more likely explanation for the occurrence of highly conserved magnetotactic signature genes in Mbav. Similar examples of horizontal gene transfer between the Nitrospira phylum and other bacterial phyla have been already demonstrated for genes encoding other metabolic pathways, such for instance as sulfite reductase genes (42), or genes involved in nitrite oxidation (43). The observed compositional differences, such as the percentage of G+C content of the MAI (Mbav 49.7%, M. gryphiswaldense 61.1%) argue against recent HGT, but indicate a rather ancient event of transfer. However, whether or not magnetotaxis has originated from the deep branching Nitrospira phylum and thus, formation of magnetosomes is an evolutionary ancient trait, requires more sequencing efforts that have to include further distantly related MTB from other groups.
Materials and Methods
Electron Microscopy.
TEM and SEM of samples prepared by chemical fixation and high-pressure freezing was performed with an EM 912 transmission microscope (Zeiss) and a Zeiss Auriga SEM equipped with a focused ion beam consisting of Ga+ ions for “sectioning.” See SI Materials and Methods for details.
Environmental Sampling, Magnetic Enrichments, DNA Extraction, Fosmid-Library Construction, and Screening.
Sediment sample collection, magnetic Mbav enrichment, DNA extraction and fosmid library construction was performed as previously described (18, 21). Six fosmid libraries were screened via endsequencing and PCR as previously described (18, 21). See SI Materials and Methods for details.
Single Cell Sorting and WGA.
Single cell sorting was achieved via micromanipulation and subsequent phi29-mediated whole genome amplification as explained in detail in SI Materials and Methods.
Sequence Analysis.
WGA DNA was analyzed by the Genome Sequencer FLX system (GS FLX Titanium chemistry) and reads were assembled via MIRA as previously described (44). Sequence determination of entire fosmids as well as phylogenetic analysis were performed as previously described (21).
Supplementary Material
Acknowledgments
We thank Roberto Kolter, René Uebe, Wei Lin, Ramon Egli, Kerstin Reimer, Frank Oliver Glöckner, Hanno Teeling, Jost Waldmann, and Hera Vlamakis for valuable scientific discussions. The help of Geraldine A. Van der Auwera for the preparation of Fig. 3 is highly appreciated. We thank Tobias Salge for X-ray element mapping, Andreas Schertel and Udo Graf for support with cryo-SEM preparations, and Silvia Dobler for skillful technical assistance. We thank Hans von Besser, Andreas Tögel, and Theis Stüven for help with developing the single-cell sorting and amplification workflow. This project was funded by the Deutsche Forschungsgemeinschaft (DFG Schu1080/11-1) and the Max Planck Society (project Envi-Tools).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. FQ377626).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012694108/-/DCSupplemental.
References
- 1.Jogler C, Schüler D. Genetic and biochemical analysis of magnetosomes in Magnetospirillum gryphiswaldense. In: Baeuerlein E, editor. Handbook of Biomineralization Vol Biological aspects and structure formation. Germany: Wiley-VCH, Weinheim; 2007. pp. 145–162. [Google Scholar]
- 2.Schüler D. Biochemical and genetic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. In: Baeuerlein E, editor. Biomineralization. 2nd Ed. Weinheim: Wiley-VCH; 2004. pp. 61–74. [Google Scholar]
- 3.Matsunaga T, Nemoto M, Arakaki A, Tanaka M. Proteomic analysis of irregular, bullet-shaped magnetosomes in the sulphate-reducing magnetotactic bacterium Desulfovibrio magneticus RS-1. Proteomics. 2009;9:3341–3352. doi: 10.1002/pmic.200800881. [DOI] [PubMed] [Google Scholar]
- 4.Scheffel A, et al. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- 5.Katzmann E, Scheffel A, Gruska M, Plitzko JM, Schüler D. Loss of the actin-like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense. Mol Microbiol. 2010;77:208–224. doi: 10.1111/j.1365-2958.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- 6.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 7.Pradel N, Santini CL, Bernadac A, Fukumori Y, Wu LF. Biogenesis of actin-like bacterial cytoskeletal filaments destined for positioning prokaryotic magnetic organelles. Proc Natl Acad Sci USA. 2006;103:17485–17489. doi: 10.1073/pnas.0603760103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jogler C, Schüler D. Genomics, genetics, and cell biology of magnetosome formation. Annu Rev Microbiol. 2009;63:501–521. doi: 10.1146/annurev.micro.62.081307.162908. [DOI] [PubMed] [Google Scholar]
- 9.Faivre D, Schüler D. Magnetotactic bacteria and magnetosomes. Chem Rev. 2008;108:4875–4898. doi: 10.1021/cr078258w. [DOI] [PubMed] [Google Scholar]
- 10.Murat D, Quinlan A, Vali H, Komeili A. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc Natl Acad Sci USA. 2010;107:5593–5598. doi: 10.1073/pnas.0914439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komeili A. Molecular mechanisms of magnetosome formation. Annu Rev Biochem. 2007;76:351–366. doi: 10.1146/annurev.biochem.74.082803.133444. [DOI] [PubMed] [Google Scholar]
- 12.Delong EF, Frankel RB, Bazylinski DA. Multiple evolutionary origins of magnetotaxis in bacteria. Science. 1993;259:803–806. doi: 10.1126/science.259.5096.803. [DOI] [PubMed] [Google Scholar]
- 13.Hanzlik M, Winklhofer M, Petersen N. Pulsed-field-remanence measurements on indivdual magnetotactic bacteria. J Magn Magn Mater. 2002;248:258–267. [Google Scholar]
- 14.Byrne ME, et al. Desulfovibrio magneticus RS-1 contains an iron- and phosphorus-rich organelle distinct from its bullet-shaped magnetosomes. Proc Natl Acad Sci USA. 2010;107:12263–12268. doi: 10.1073/pnas.1001290107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J Bacteriol. 2005;187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jogler C, et al. Comparative analysis of magnetosome gene clusters in magnetotactic bacteria provides further evidence for horizontal gene transfer. Environ Microbiol. 2009;11:1267–1277. doi: 10.1111/j.1462-2920.2009.01854.x. [DOI] [PubMed] [Google Scholar]
- 17.Schübbe S, et al. Complete genome sequence of the chemolithoautotrophic marine magnetotactic coccus strain MC-1. Appl Environ Microbiol. 2009;75:4835–4852. doi: 10.1128/AEM.02874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jogler C, et al. Toward cloning of the magnetotactic metagenome: Identification of magnetosome island gene clusters in uncultivated magnetotactic bacteria from different aquatic sediments. Appl Environ Microbiol. 2009;75:3972–3979. doi: 10.1128/AEM.02701-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakazawa H, et al. Whole genome sequence of Desulfovibrio magneticus strain RS-1 revealed common gene clusters in magnetotactic bacteria. Genome Res. 2009;19:1801–1808. doi: 10.1101/gr.088906.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spring S, et al. Dominating role of an unusual magnetotactic bacterium in the microaerobic zone of a freshwater sediment. Appl Environ Microbiol. 1993;59:2397–2403. doi: 10.1128/aem.59.8.2397-2403.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jogler C, et al. Cultivation-independent characterization of ‘Candidatus Magnetobacterium bavaricum’ via ultrastructural, geochemical, ecological and metagenomic methods. Environ Microbiol. 2010;12:2466–2478. doi: 10.1111/j.1462-2920.2010.02220.x. [DOI] [PubMed] [Google Scholar]
- 22.Flies CB, Peplies J, Schüler D. Combined approach for characterization of uncultivated magnetotactic bacteria from various aquatic environments. Appl Environ Microbiol. 2005;71:2723–2731. doi: 10.1128/AEM.71.5.2723-2731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin W, Li J, Schüler D, Jogler C, Pan Y. Diversity analysis of magnetotactic bacteria in Lake Miyun, northern China, by restriction fragment length polymorphism. Syst Appl Microbiol. 2009;32:342–350. doi: 10.1016/j.syapm.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Lefèvre CT, et al. Moderately thermophilic magnetotactic bacteria from hot springs in Nevada. Appl Environ Microbiol. 2010;76:3740–3743. doi: 10.1128/AEM.03018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanzlik M, Winklhofer M, Petersen N. Spatial arrangement of chains of magnetosomes in magnetotactic bacteria. Earth Planet Sci Lett. 1996;145:125–134. [Google Scholar]
- 26.Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U. Nitrospira marina gen. nov., sp. nov.: A chemolithotrophic nitrite-oxidizing bacterium. Arch Microbiol. 1986;144:1–7. [Google Scholar]
- 27.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol. 1995;164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 28.Off S, Alawi M, Spieck E. Enrichment and physiological characterization of a novel Nitrospira-like bacterium obtained from a marine sponge. Appl Environ Microbiol. 2010;76:4640–4646. doi: 10.1128/AEM.00320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheffel A, Gärdes A, Grünberg K, Wanner G, Schüler D. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J Bacteriol. 2008;190:377–386. doi: 10.1128/JB.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin W, Jogler C, Schüler D, Pan Y. Metagenomic analysis reveals unexpected subgenomic diversity of magnetotactic bacteria within the Nitrospira phylum. Appl Environ Microbiol. 2010 doi: 10.1128/AEM.01476-10. 10.1128/AEM.01476-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jogler C, Schüler D. Genetic analysis of magnetosome biomineralization. Magnetoreception and Magnetosomes in Bacteria, Microbiology Monographs. In: Schüler D, editor. Vol. 3. Heidelberg: Springer; 2006. pp. 133–162. [Google Scholar]
- 32.Komeili A, Vali H, Beveridge TJ, Newman DK. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc Natl Acad Sci USA. 2004;101:3839–3844. doi: 10.1073/pnas.0400391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grünberg K, et al. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl Environ Microbiol. 2004;70:1040–1050. doi: 10.1128/AEM.70.2.1040-1050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montanini B, Blaudez D, Jeandroz S, Sanders D, Chalot M. Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: Improved signature and prediction of substrate specificity. BMC Genomics. 2007;8:107. doi: 10.1186/1471-2164-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grass G, et al. FieF (YiiP) from Escherichia coli mediates decreased cellular accumulation of iron and relieves iron stress. Arch Microbiol. 2005;183:9–18. doi: 10.1007/s00203-004-0739-4. [DOI] [PubMed] [Google Scholar]
- 36.Arakaki A, Shibusawa M, Hosokawa M, Matsunaga T. Preparation of genomic DNA from a single species of uncultured magnetotactic bacterium by multiple-displacement amplification. Appl Environ Microbiol. 2010;76:1480–1485. doi: 10.1128/AEM.02124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woyke T, et al. Assembling the marine metagenome, one cell at a time. PLoS ONE. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Podar M, et al. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol. 2007;73:3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrigue S, et al. Whole genome amplification and de novo assembly of single bacterial cells. PLoS ONE. 2009;4:e6864. doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woyke T, et al. One bacterial cell, one complete genome. PLoS ONE. 2010;5:e10314. doi: 10.1371/journal.pone.0010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richter M, et al. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J Bacteriol. 2007;189:4899–4910. doi: 10.1128/JB.00119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein M, et al. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J Bacteriol. 2001;183:6028–6035. doi: 10.1128/JB.183.20.6028-6035.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lücker S, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA. 2010;107:13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevreux B, Wetter T, Suhai S. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics. Vol. 99. Hannover, Germany: German Conference on Bioinformatics; 1999. Genome sequence assembly using trace signals and additional sequence information; pp. 45–56. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.